Abstract
Bcr-Abl tyrosine kinase activity initiates a number of intracellular signaling cascades that result in leukemogenesis. Imatinib mesylate, a specific Bcr-Abl tyrosine kinase inhibitor, has been highly successful in the treatment of chronic myelogenous leukemia (CML). However, the emergence of imatinib resistance and the incomplete molecular response of a significant number of patients receiving this therapy have led to a search for combinations of drugs that will enhance the efficacy of imatinib. We have demonstrated that mycophenolic acid (MPA), a specific inosine monophosphate dehydrogenase (IMPDH) inhibitor that results in depletion of intracellular guanine nucleotides, is synergistic with imatinib in inducing apoptosis in Bcr-Abl-expressing cell lines. Studies of signaling pathways downstream of Bcr-Abl demonstrated that the addition of MPA to imatinib reduced the phosphorylation of both Stat5 and Lyn, a Src kinase family member. The phosphorylation of S6 ribosomal protein was also greatly reduced. These results demonstrate that inhibitors of guanine nucleotide biosynthesis may synergize with imatinib in reducing the levels of minimal residual disease in CML and lay the foundation for clinical trials in which IMPDH inhibitors are added to imatinib in patients who have suboptimal molecular responses to single agent therapy or who have progressive disease. (Blood. 2005; 105:3270-3277)
Introduction
Bcr-Abl, a chimeric oncogene, is formed by a reciprocal translocation between chromosomes 9 and 22 (Philadelphia chromosome, Ph1) that places the c-ABL gene under the transcriptional control of the BCR gene,1,2 resulting in the constitutive activation of c-Abl tyrosine kinase.3,4 Depending on the breakpoint site in the Bcr gene, 2 variants of the Bcr-Abl fusion proteins are generated.5 The p210 Bcr-Abl is present in virtually all cases of chronic myelogenous leukemia (CML), while the p185 product is present in approximately 20% of acute lymphoblastic leukemias.6 The Bcr-Abl tyrosine kinase activates a number of downstream cell signaling pathways, resulting in growth factor-independent cell proliferation, altered cell adhesion, and resistance to apoptosis induced by other chemotherapeutic agents.7 The recently developed inhibitor of Bcr-Abl kinase activity, imatinib mesylate, induces a high percentage of complete hematologic responses and partial or complete cytogenetic remissions in chronic phase CML.8,9 Imatinib has high affinity for Bcr-Abl tyrosine kinase10 and inhibits its activity by binding to its adenosine triphosphate (ATP)-binding pocket.11 Despite a high degree of selectivity, approximately 60% of CML patients retain the Ph1 chromosome after 18 months on imatinib treatment12 and more than 90% remain positive for Bcr-Abl transcripts, as measured by the quantitative real-time polymerase chain reaction assay.13 Residual Bcr-Abl positivity is clearly linked to risk for disease progression and the development of blast crisis. Resistance to imatinib eventually develops in the majority of patients treated in the blast crisis stage of the disease, primarily due to the emergence of point mutations in or amplification of the Bcr-Abl gene.14 The emergence of imatinib resistance creates a strong incentive to find nontoxic approaches to enhance its efficacy, including combining imatinib with a drug that enhances its cytotoxicity.
Inosine monophosphate dehydrogenase (IMPDH; EC1.1.1.205) is an essential, rate-limiting enzyme for the generation of guanine nucleotides through the de novo synthesis pathway. It catalyzes the NAD-dependent conversion of inosine monophosphate to xanthine monophosphate, which is subsequently converted to guanosine monophosphate (GMP) by GMP synthase. The only alternative pathway for guanine nucleotide biosynthesis is through the salvage of guanine to GMP by hypoxanthine-guanine phosphoribosyltransferase. IMPDH enzyme activity is composed of the activities of 2 separate but very closely related IMPDH isoenzymes, termed type I and type II, that share 84% amino acid identity.15 Expression of IMPDH, particularly the type II enzyme, is significantly up-regulated in many tumor cells, including leukemia cells,16-19 presumably to meet a high demand for guanine nucleotides during cell proliferation. Reduction of guanine nucleotides by inhibiting IMPDH activity effectively induces cell-cycle arrest in late G1 phase in lymphocytes20,21 and results in differentiation22-24 or apoptosis25-27 in cultured cell lines depending on the cell type. The fact that these effects on cell growth and differentiation are completely reversed by the addition of guanine or guanosine to the medium demonstrates that the effects of IMPDH inhibitors are specifically due to guanine nucleotide depletion. Inhibitors of IMPDH have been used clinically as immunosuppressive agents28-30 and also have been used to induce hematologic responses in patients with CML in blast crisis31,32 and in Bcr-Abl-positive AML patients.33 However, the reduction in blast count is temporary, and other studies have suggested that IMPDH inhibitors may be better used in combination with other chemotherapeutic drugs.34,35
In the present study, we have demonstrated that a specific IMPDH inhibitor, mycophenolic acid (MPA), can significantly enhance imatinib cytotoxicity in Bcr-Abl-positive cells. We also have investigated the possible Bcr-Abl signaling pathways affected by combining a specific inhibitor of Bcr-Abl tyrosine kinase with guanine nucleotide depletion.
Materials and methods
Recombinant retroviral infection
The pSRMSVtkneo plasmid containing the p185 form of Bcr-Abl was kindly provided by Dr C. Der (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill). Retrovirus was produced in 293T cells using pVpack system (Stratagene, La Jolla, CA). Both 32D and FL5.12 cells were infected with control retrovirus alone or retrovirus encoding the p185 Bcr-Abl fusion gene. Cells were selected and maintained in the presence of 500 μg/mL G418. The p185 Bcr-Abl-expressing cells also were selected for growth independent of interleukin-3 (IL-3).
Cell lines and reagents
The murine myeloid cell line 32D was maintained in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone, Logan, UT) and 10% WEHI-conditioned medium as a source of IL-3. The murine pre-B-cell line FL5.12 (a gift from C. B. Thompson, University of Pennsylvania, Philadelphia) was maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 55 μM 2-mercaptoethonal, and 10% WEHI-conditioned medium. Human leukemia cell lines K562, HL60, and U937 (originally obtained from American Type Culture Collection, Rockville, MD) were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS. An imatinib-resistant K562 derivative cell line K562R and LAMA84 cells were generously provided by Dr S. Grant (Commonwealth University, Richmond, VA). LAMA84 cells were maintained in RPMI-1640 medium (without phenol-red) supplemented with 0.1 mM minimum essential media (MEM) nonessential amino acid (Gibco, Caraday, CA), 2 mM l-glutamine (Gibco), and 1 mM MEM sodium pyruvate (Gibco). All cell cultures were maintained in the presence of 100 μg/mL penicillin and streptomycin.
Mycophenolic acid purchased from Sigma (St. Louis, MO) was dissolved in ethanol to a final concentration of 10 mM. Imatinib obtained from Novartis Pharm (Basel, Switzerland) was dissolved in dimethyl sulfoxide to a final concentration of 10 mM. Both drugs were stored in aliquots in -20°C. In all experiments, control samples received the equivalent amount of solvent used for drugs. Antibodies against total and phospho-Stat5 (Tyr 694), phospho-Src (Tyr 416), and total and phospho-S6 (Ser 235/236) all were purchased from Cell Signaling Technology (Beverly, MA). The anti-Lyn antibody and protein A-beads were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antiphosphotyrosine (4G10) antibody was obtained from Upstate USA (Charlottesville, VA). MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide) was purchased from Sigma and dissolved in phosphate-buffered saline (PBS) to a final concentration of 5 mg/mL and stored aliquots in -20°C.
MTT assay
Cells were seeded at approximately 1 × 104 in a final volume of 100 μLin 96-well flat-bottom microtiter plates (Corning Costar, Corning, NY) with or without various concentrations of drugs. To exclude effects of drugs on the OD reading, the highest concentration of drug was added to the medium without cells to serve as the background control. Plates were incubated at 37°C in a 5% CO2 incubator for the indicated times. At the end of incubation, 15 μL of MTT was added to each well, and plates were incubated at 37°C for another 4 hours, followed by the addition of 100 μL 10% sodium dodecyl sulfate (SDS) solution in 0.01 M hydrochloric acid. Plates were incubated overnight in 37°C, and absorbance was measured at 595 nm against a reference wavelength at 650 nm using a MAXline Microplate Reader (Molecular Devices, Sunnyvale, CA). The mean of quadruplicate for each dose was used to calculate the IC50 and for all the combination index (CI) values.
CI calculation
CI values were calculated using Biosoft CalcuSyn program (Ferguson, MO). The dose-effect relationships analyzed using the median-effect equation was first obtained for each single drug by its serial dilution. A CI value for each combination treatment was then calculated based on the formula D1/Dx1 + D2/Dx2, where Dx1 and Dx2 are the doses of drug 1 and drug 2 alone required to produce x% effect, and D1 and D2 are the doses of drug 1 and drug 2 in combination required to produce the same effect.36,37 CI was used to express synergism (CI < 1), additive effect (CI = 1), or antagonism (CI > 1).
Annexin V staining and cell-cycle analysis
Apoptosis was measured using the Annexin V-FITC Apoptosis Detection Kit I (BD PharMingen, San Diego, CA), following the staining procedure provided by the manufacturer. Briefly, 2 to 3 × 105 cells after single or combination drug treatment were washed twice with cold PBS and resuspended in 100 μL of 1 × binding buffer for annexin V and propidium iodide (PI) staining. For cell-cycle analysis, approximately 5 × 105 cells were harvested after drug treatment, washed once in cold PBS, and fixed in 80% EtOH and stored at 4°C overnight. For PI staining, cells were centrifuged at 1300g for 5 minutes, washed once with PBS containing 0.2% bovine serum albumin (BSA) (BSA/PBS), and pellets were resuspended in 500 μL BSA/PBS containing 100 μg RNaseA and 50 μg PI and incubated at 37°C for 30 minutes. Both annexin V-stained and cell-cycle PI-stained cells were analyzed by flow cytometry. Approximately 30 000 cells were collected using FACScan Flow (Becton Dickinson Immunocytometry, Mountain View, CA).
DNA fragmentation ELISA assay
Mononucleosomes and oligonucleosomes in the cytoplasm of the apoptotic cells were measured using a Cell Death Detection ELISA kit (Roche Diagnostic GmbH, Indianapolis, IN) according to the manufacturer's protocol. Lysate from approximately 1 × 103 cells was used for each well. Absorbance was measured at 405 nm against a reference of 490 nm using a MAXline Microplate Reader (Molecular Devices). All the experiments were performed in duplicate in enzyme-linked immunosorbent assay (ELISA) assays.
Western immunoblotting
To prepare cell lysates for Western immunoblot analysis, cell pellets were lysed in buffer containing 20 mM Tris(tris(hydroxymethyl)aminomethane)- HCl (pH 8.0), 137 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM Na3VO4, 25 mM β-glycerophosphate, 1 × protease inhibitor cocktail, and 1 × phosphatase inhibitor cocktail 1 (Sigma). After centrifugation at 14 000g for 15 minutes, protein concentrations were quantitated in duplicate by the Bradford method (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein were separated on an SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to either polyvinylidene difluoride Immobilon-P membranes (Millipore, Bedford, MA) or Protran nitrocellulose membranes (Schleicher & Schuell, Keene, NH). The membranes were blocked with 5% nonfat milk made in Tris-buffered saline containing 0.1% Tween 20 (TBST0.1) at room temperature for 1 hour, and then incubated with primary antibodies diluted either in 5% BSA for phospho-protein detection or in blocking solution for other antibodies at 4°C overnight. Dilutions of primary antibodies were made following the manufacturer's instructions. Following several washes in TBST0.1, membranes were incubated with appropriate secondary antibodies (1:2000 to 1:5000 dilution, Amersham, Arlington Heights, IL) at room temperature for 1 hour. Detection was performed using the enhanced chemiluminescence system (Amersham).
Immunoprecipitation
Immunoprecipitation (IP) of Lyn was performed following the protocol described by Danhauser-Riedl et al.38 Briefly, 1 mg total lysate was incubated with 5 μg anti-Lyn antibody. After rotating 3 hours at 4°C, 100 μL protein A-Beads (50% beads volume) was added for an additional 2 hours. Precipitated proteins were separated on 10% SDS-PAGE and blotted with antiphosphotyrosine antibody (4G10).
Results
Effects of imatinib and MPA on growth of Bcr-Abl-transformed cell lines
Murine 32D myeloid and FL5.12 pre-B cells were infected with retrovirus carrying the p185 Bcr-Abl fusion protein or an empty vector (as described in “Materials and methods”). Both cell lines appropriately expressed the Bcr-Abl protein, as demonstrated by Western blots (data not shown). Inhibition of cell growth by MPA or imatinib was then analyzed using MTT assays.37 As indicated in Table 1, the IC50 values for MPA are similar across cell lines regardless of Bcr-Abl expression. Consistent with previous results,39 32D and FL5.12 cell lines expressing Bcr-Abl are highly sensitive to imatinib, with IC50 values of approximately 0.6 μM, whereas vector control cells are completely resistant to imatinib-induced growth inhibition up to the highest concentration (8 μM) tested. Growth inhibition data for these 2 drugs also were obtained for 2 human leukemia cell lines expressing p210 Bcr-Abl, K562 and LAMA84, and 2 Bcr-Abl-negative leukemic cell lines, HL-60 and U937. The growth of all 4 cell lines is inhibited by MPA at similar concentrations, albeit at longer incubation times than for the murine cells. In contrast, the 2 Bcr-Abl-negative cell lines are completely resistant to imatinib concentrations up to 8 μM, whereas the K562 and LAMA84 cells are very sensitive to imatinib with IC50 values of 0.5 and 0.1 μM, respectively.
Effect of MPA or imatinib on cell growth in Bcr-Abl-positive and −negative cell lines
Cell lines . | Description . | Incubation time (hrs) . | IC50 MPA (μM) . | IC50 imatinib (μM) . |
|---|---|---|---|---|
| 32D-vector | Murine myeloid | 24 | 0.46 (±0.16) | >8 |
| 32D-p185 | Murine myeloid (p 185 Bcr-Abl) | 24 | 0.56 (± 0.14) | 0.56 (± 0.08) |
| FL5.12-vector | Murine pre-B | 24 | 0.4 (± 0.16) | >8 |
| FL5.12-p185 | Murine pre-B (p185 Bcr-Abl) | 24 | 0.8 (± 0.14) | 0.68 (± 0.2) |
| K562 | Human CML (p210 Bcr-Abl) | 72 | 0.41 (± 0.17) | 0.53 (± 0.07) |
| LAMA84 | Human CML (p210 Bcr-Abl) | 48 | 0.47 (± 0.03) | 0.089 (± 0.01) |
| HL-60 | Human promyelocytic leukemia | 48 | 1.0 (± 0.2) | >8 |
| U937 | Human monocytic leukemia | 48 | 0.65 (± 0.37) | >8 |
Cell lines . | Description . | Incubation time (hrs) . | IC50 MPA (μM) . | IC50 imatinib (μM) . |
|---|---|---|---|---|
| 32D-vector | Murine myeloid | 24 | 0.46 (±0.16) | >8 |
| 32D-p185 | Murine myeloid (p 185 Bcr-Abl) | 24 | 0.56 (± 0.14) | 0.56 (± 0.08) |
| FL5.12-vector | Murine pre-B | 24 | 0.4 (± 0.16) | >8 |
| FL5.12-p185 | Murine pre-B (p185 Bcr-Abl) | 24 | 0.8 (± 0.14) | 0.68 (± 0.2) |
| K562 | Human CML (p210 Bcr-Abl) | 72 | 0.41 (± 0.17) | 0.53 (± 0.07) |
| LAMA84 | Human CML (p210 Bcr-Abl) | 48 | 0.47 (± 0.03) | 0.089 (± 0.01) |
| HL-60 | Human promyelocytic leukemia | 48 | 1.0 (± 0.2) | >8 |
| U937 | Human monocytic leukemia | 48 | 0.65 (± 0.37) | >8 |
IC50 values were obtained using the MTT assays. Values represent the mean ± SD for at least 3 experiments.
To study the possible synergy between imatinib and MPA, approximately equipotent concentrations of each drug were used. Combination index (CI) values were calculated using CalcuSyn software (see “Materials and methods”), where CI = 1 indicates an additive affect, CI less than 1 indicates synergy, and CI more than 1 indicates antagonism calculated using a formula initially proposed by Chou and Talalay.36 The 2 drugs were used in constant ratios in most MTT assays to generate classical isobolograms. In some instances, nonconstant ratios also were tested to determine the effectiveness of a broader range of combinations (data not shown). The CI values at ED50, ED75, and ED90 generated from classical isobolograms for all 4 Bcr-Abl-positive cell lines are summarized in Table 2. Combined treatment with MPA and imatinib demonstrates an additive (at about the ED50) to synergistic (above the ED50) effect on growth inhibition in Bcr-Abl-expressing -32D and -FL5.12 cell lines. Combination treatment is clearly synergistic at and above the ED50 in both human Bcr-Abl-positive cell lines. Since the cell lines not expressing Bcr-Abl are insensitive to imatinib and data obtained for the combination are similar to those obtained with MPA alone, the combination index calculation is not applicable.
Combination index values for growth inhibition
. | . | CI values . | . | . | ||
|---|---|---|---|---|---|---|
| Cell lines . | MPA/Imatinib . | ED50 . | ED75 . | ED90 . | ||
| 32D-p185 | 2:1 | 0.94 (± 0.2) | 0.64 (± 0.03) | 0.50 (± 0.07) | ||
| FL5.12-p185 | 2:1 | 0.88 (± 0.07) | 0.77 (± 0.06) | 0.76 (± 0.03) | ||
| K562 | 4:1 | 0.63 (± 0.1) | 0.27 (± 0.02) | 0.58 (± 0.1) | ||
| LAMA84 | 8:1 | 0.79 (± 0.1) | 0.53 (± 0.09) | 0.58 (± 0.05) | ||
. | . | CI values . | . | . | ||
|---|---|---|---|---|---|---|
| Cell lines . | MPA/Imatinib . | ED50 . | ED75 . | ED90 . | ||
| 32D-p185 | 2:1 | 0.94 (± 0.2) | 0.64 (± 0.03) | 0.50 (± 0.07) | ||
| FL5.12-p185 | 2:1 | 0.88 (± 0.07) | 0.77 (± 0.06) | 0.76 (± 0.03) | ||
| K562 | 4:1 | 0.63 (± 0.1) | 0.27 (± 0.02) | 0.58 (± 0.1) | ||
| LAMA84 | 8:1 | 0.79 (± 0.1) | 0.53 (± 0.09) | 0.58 (± 0.05) | ||
Combination index values were calculated from the MTT assays. Constant ratios of 2 drugs were used to obtain the isobologram ED values. Data represent the mean ± SD of 3 independent experiments.
Effect of MPA and imatinib combinations on the induction of apoptosis
To determine whether combined treatment with MPA and imatinib also enhances apoptosis, 32D-p185 and K562 cells were treated with various concentrations of MPA and/or imatinib followed by annexin V staining and analyzed by flow cytometry. Each drug alone at the doses used induced very little or no apoptosis; however, the combination of the 2 greatly increased the percentage of annexin V-positive cells in both cell lines (Figure 1). To determine the CI values for apoptosis, at least 4 doses of each drug were used in every experiment to generate single drug curves. Highly synergistic effects were observed for multiple combination treatments in both 32D-p185 and K562 cell lines (Table 3). Interestingly, the CI values for apoptosis demonstrate far more synergy than do the cell growth inhibition experiments. This is most likely because each drug alone can significantly inhibit growth but only induce very low levels of apoptosis at the concentrations used. The reduction in live cells, as defined by both annexin V and PI negativity, for each combination treatment as compared to each drug alone also was statistically significant (P < .05) using a paired 2-tailed Student t test analysis on 3 independent experiments (data not shown).
Effect of combined treatment with imatinib and MPA on the induction of apoptosis. (A) 32D-p185 and (B) K562 cells were treated with MPA and/or imatinib at the doses indicated for 24 and 72 hours, respectively. Apoptosis was measured by annexin-V and PI staining. The percentage of annexin V-negative and -positive cells is indicated on each histogram. The annexin V-positive peak shown in the histogram includes both the annexin V-positive/PI-negative, and the annexin V-positive/PI-positive populations. At least 3 experiments with multiple drug combinations were performed on each cell line. One representative experiment at a selected combination of doses is shown.
Effect of combined treatment with imatinib and MPA on the induction of apoptosis. (A) 32D-p185 and (B) K562 cells were treated with MPA and/or imatinib at the doses indicated for 24 and 72 hours, respectively. Apoptosis was measured by annexin-V and PI staining. The percentage of annexin V-negative and -positive cells is indicated on each histogram. The annexin V-positive peak shown in the histogram includes both the annexin V-positive/PI-negative, and the annexin V-positive/PI-positive populations. At least 3 experiments with multiple drug combinations were performed on each cell line. One representative experiment at a selected combination of doses is shown.
Combination index values for induction of apoptosis
Cell lines . | Drugs (μM) . | Cl . | Description . |
|---|---|---|---|
| 32D-p 185 | MPA (1) + imatinib (0.5) | 0.47 (± 0.13) | Synergism |
| MPA (1) + imatinib (1) | 0.28 (± 0.07) | Strong synergism | |
| MPA (2) + imatinib (0.5) | 0.34 (± 0.04) | Synergism | |
| MPA (2) + imatinib (1) | 0.21 (± 0.09) | Strong synergism | |
| K562 | MPA (1) + imatinib (0.25) | 0.04 (± 0.04) | Very strong synergism |
| MPA (2) + imatinib (0.5) | 0.04 (± 0.05) | Very strong synergism | |
| MPA (4) + imatinib (1) | 0.06 (± 0.08) | Very strong synergism |
Cell lines . | Drugs (μM) . | Cl . | Description . |
|---|---|---|---|
| 32D-p 185 | MPA (1) + imatinib (0.5) | 0.47 (± 0.13) | Synergism |
| MPA (1) + imatinib (1) | 0.28 (± 0.07) | Strong synergism | |
| MPA (2) + imatinib (0.5) | 0.34 (± 0.04) | Synergism | |
| MPA (2) + imatinib (1) | 0.21 (± 0.09) | Strong synergism | |
| K562 | MPA (1) + imatinib (0.25) | 0.04 (± 0.04) | Very strong synergism |
| MPA (2) + imatinib (0.5) | 0.04 (± 0.05) | Very strong synergism | |
| MPA (4) + imatinib (1) | 0.06 (± 0.08) | Very strong synergism |
Cl values were obtained by annexin V staining analyzed by flow cytometry. Data represent the mean ± SD of 3 independent experiments.
Specificity of induction of apoptosis for Bcr-Abl-positive cells
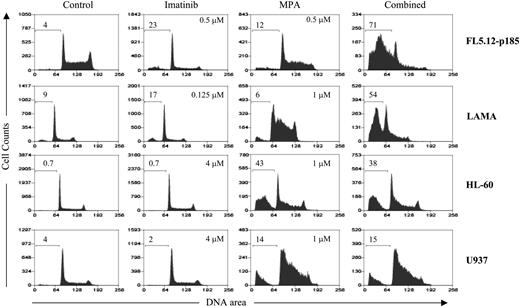
To determine whether the induction of apoptosis by combined treatment with MPA and imatinib is specific for Bcr-Abl-expressing cells, we treated vector control or Bcr-Abl-expressing 32D and FL5.12 cells with MPA and/or imatinib for 16 to 20 hours, and fragmented DNA was analyzed using the Cell Death Detection ELISA method.27 The level of apoptosis induced by the combination treatment in vector-control cells was similar to that of MPA alone, whereas the combination markedly enhanced apoptosis in Bcr-Abl-expressing cells (Figure 2). Apoptosis also was evaluated by the size of the sub-G1 peak obtained from cell-cycle analysis. Bcr-Abl-positive FL5.12 and LAMA84 cells and Bcr-Abl-negative HL-60 and U937 cells were analyzed by flow cytometry after single and combination drug treatment (Figure 3). In both FL5.12-p185 and LAMA cells, MPA and imatinib alone induced a small increase in the sub-G1 peak, whereas this peak was markedly increased by the combination treatment. In HL-60 and U937 cells, imatinib (4 μM) did not induce the apoptotic sub-G1 peak, nor did it enhance the MPA effect.
Effect of combination treatment on the induction of DNA fragmentation in Bcr-Abl-expressing cells. Both 32D (A) and FL5.12 (B) vector control cells (bars 1-4) or p185-expressing cells (bars 5-8) were either not treated (□) or treated with MPA alone (▪;1 μM for 32D, 0.5 μM for FL5.12) or imatinib alone (▦;1 μM for 32D, 0.5 μM for FL5.12 cells) or the combination of MPA and imatinib (▨) for 16 to 20 hours, and apoptosis was measured by the Cell Death Detection ELISA method. Data represent mean ± SD of 3 independent experiments. ELISA assays were performed in duplicate.
Effect of combination treatment on the induction of DNA fragmentation in Bcr-Abl-expressing cells. Both 32D (A) and FL5.12 (B) vector control cells (bars 1-4) or p185-expressing cells (bars 5-8) were either not treated (□) or treated with MPA alone (▪;1 μM for 32D, 0.5 μM for FL5.12) or imatinib alone (▦;1 μM for 32D, 0.5 μM for FL5.12 cells) or the combination of MPA and imatinib (▨) for 16 to 20 hours, and apoptosis was measured by the Cell Death Detection ELISA method. Data represent mean ± SD of 3 independent experiments. ELISA assays were performed in duplicate.
Induction of apoptosis in Bcr-Abl-positive and -negative cell lines using cell-cycle analysis. FL5.12-p185, LAMA84, HL-60, and U937 cells were treated with the indicated concentrations of MPA and/or imatinib for 24 hours (FL5.12-p185) and 48 hours (all other cell lines). The percent of sub-G1 (apoptotic) cells are shown on each histogram. At least 3 experiments with multiple drug combinations were performed on each cell line. One representative experiment at one selected dose combination is shown.
Induction of apoptosis in Bcr-Abl-positive and -negative cell lines using cell-cycle analysis. FL5.12-p185, LAMA84, HL-60, and U937 cells were treated with the indicated concentrations of MPA and/or imatinib for 24 hours (FL5.12-p185) and 48 hours (all other cell lines). The percent of sub-G1 (apoptotic) cells are shown on each histogram. At least 3 experiments with multiple drug combinations were performed on each cell line. One representative experiment at one selected dose combination is shown.
Synergistic activity in K562 cells resistant to imatinib
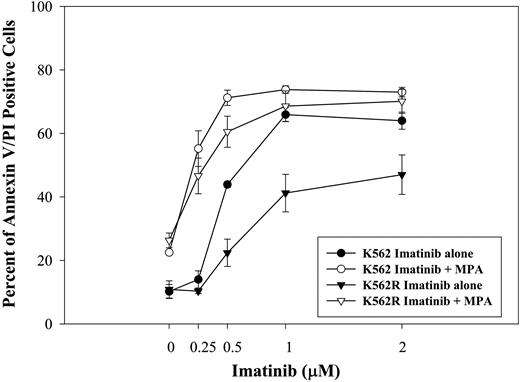
To determine whether MPA also enhances the response to imatinib in cells that demonstrate imatinib resistance, we tested the effect of the combination on induction of apoptosis using a K562-derivative cell line selected in the presence of imatinib (K562R40 ). As shown in Figure 4, K562R cells were significantly less sensitive to imatinib-induced apoptosis. K562 parental and K562R cells have similar sensitivity to MPA treatment. The combination of MPA and imatinib significantly enhanced the induction of apoptosis in both K562 parental and K562R cells.
Effect of combination treatment on imatinib-resistant cells. K562 and K562R cells were treated with imatinib at the indicated doses for 72 hours with or without MPA. Apoptosis was measured by annexin-V and PI staining. Imatinib treatment alone is indicated by the closed symbols. The controls in the absence of drug have 10.2% ± 2.2% and 10.9% ± 2.7% annexin V/PI-positive cells for K562 and K562R, respectively. Imatinib plus MPA (2 μM) is indicated by the open symbols. MPA alone induced 22.5% ± 0.6% and 26.3% ± 2.3% apoptosis for K562 and K562R, respectively. Data represent the mean ± SD of 3 independent experiments.
Effect of combination treatment on imatinib-resistant cells. K562 and K562R cells were treated with imatinib at the indicated doses for 72 hours with or without MPA. Apoptosis was measured by annexin-V and PI staining. Imatinib treatment alone is indicated by the closed symbols. The controls in the absence of drug have 10.2% ± 2.2% and 10.9% ± 2.7% annexin V/PI-positive cells for K562 and K562R, respectively. Imatinib plus MPA (2 μM) is indicated by the open symbols. MPA alone induced 22.5% ± 0.6% and 26.3% ± 2.3% apoptosis for K562 and K562R, respectively. Data represent the mean ± SD of 3 independent experiments.
Effect of imatinib and MPA on Stat5 phosphorylation
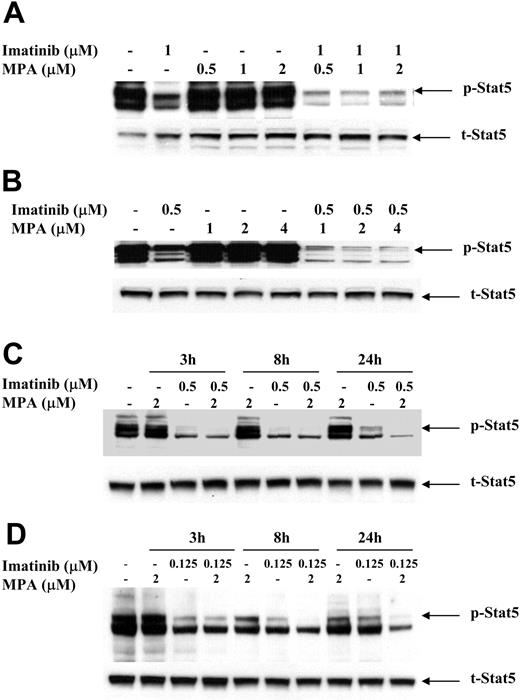
To study the possible mechanism(s) by which the combination of imatinib and MPA caused a synergistic effect on growth inhibition and apoptosis, we measured the expression of several downstream effectors of Bcr-Abl signaling that have been demonstrated to play important roles in Bcr-Abl transformation. The signal transducer and activator of transcription (Stat) 5 was highly phosphorylated in 32D-p185, FL5.12-p185 cells, and in both K562 and LAMA84 human cell lines as compared with vector-transfected 32D and FL5.12 parental cells and HL-60 and U937 cells (data not shown). To determine whether MPA and/or imatinib treatment decreases Stat5 phosphorylation, we treated 32D-p185 and FL5.12-p185 cell lines with either drug alone or in combination for 90 minutes. This time period is far shorter than the 4 to 8 hours required for caspase 3 activation in these 2 cell lines (data not shown). Although imatinib alone potently inhibited Stat5 phosphorylation (p-Stat5), the combination of imatinib and MPA further reduced the level of p-Stat5 (Figure 5A-B). Similar effects also are seen in K562 and LAMA cells, although at much later time points (between 8 and 24 hours; Figure 5C-D). The reduction of p-Stat5 levels by imatinib alone at 24 hours is less than the reduction at 3 and 8 hours, indicating that the effect of imatinib is relatively short. However, the combination of imatinib and MPA maintained lower levels of p-Stat5 at 24 hours. This prolongation of effect may have biologic significance in view of the 48 to 72 hours it takes for the combination to induce apoptosis in human cell lines. We also have demonstrated (data not shown) that Mcl-1 and Bcl-xL, both downstream targets of p-Stat5, are significantly reduced at these time points, underscoring the biologic consequences of reducing p-Stat5 levels.41,42
Effect of combination treatment on Stat5 phosphorylation. 32D-p185 (A), FL5.12-p185 (B), K562 (C), and LAMA84 (D) cells were treated with indicated concentrations of MPA and/or imatinib for 90 minutes (for 32D- and FL5.12-p185) or the indicated times for K562 and LAMA84 cells. Thirty μg of total cell lysate was loaded in each lane, and Western blot analysis was performed using an antibody specific for phospho-Stat5 (p-Stat5). Duplicate gels were probed with antibody specific for total Stat5 (t-Stat5). 32D-p185 and FL5.12-p185 results are representative of 4 and 3 independent experiments, respectively. K562 and LAMA84 results are representative of 2 independent experiments; treatment for 24 hours was repeated 3 times.
Effect of combination treatment on Stat5 phosphorylation. 32D-p185 (A), FL5.12-p185 (B), K562 (C), and LAMA84 (D) cells were treated with indicated concentrations of MPA and/or imatinib for 90 minutes (for 32D- and FL5.12-p185) or the indicated times for K562 and LAMA84 cells. Thirty μg of total cell lysate was loaded in each lane, and Western blot analysis was performed using an antibody specific for phospho-Stat5 (p-Stat5). Duplicate gels were probed with antibody specific for total Stat5 (t-Stat5). 32D-p185 and FL5.12-p185 results are representative of 4 and 3 independent experiments, respectively. K562 and LAMA84 results are representative of 2 independent experiments; treatment for 24 hours was repeated 3 times.
Effect of combination treatment on phospho-Jak2 and phospho-Src expression
We next examined upstream molecules that could phosphorylate Stat5. We failed to identify significant amounts of constitutively activated Janus kinase (Jak) 2 protein in our Bcr-Abl-expressing cell lines and did not detect any significant effect on phospho-Jak2 levels by either single or combination drug treatment (data not shown), suggesting that Jak2 is not responsible for the effect on Stat5 phosphorylation. This result is also consistent with the previous finding that inhibition of Jak2 does not affect Stat5 activity in Bcr-Abl-positive cells.43
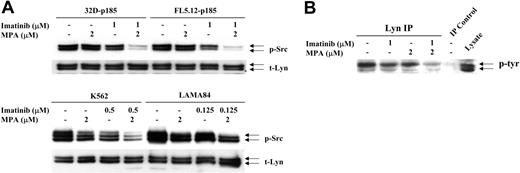
Increasing evidence has indicated that Stat proteins also can be activated through members of the Src gene family.44 In particular, 2 family members, Lyn and Hck, can phosphorylate Stat545,46 and have been implicated in Bcr-Abl-mediated transformation.38,47 We have found that levels of Src phosphorylation in 32D and FL5.12 cells were very high, independent of the expression of p185 Bcr-Abl, most likely due to the continuous IL-3 stimulation required for the growth of vector-control cells. However, Src is more highly phosphorylated in the human Bcr-Abl-positive cell lines, K562 and LAMA84 cells, than in HL-60 and U937 cells (data not shown). Although neither drug by itself significantly decreased p-Src levels, the combination dramatically reduced Src phosphorylation in 32D-p185 and FL5.12-p185 cells after 90 minutes (Figure 6A). Similar, although less dramatic, reductions were observed after 24 hours of combined treatment of K562 and LAMA cells (Figure 6A). The antibody used to detect p-Src (Y416) detects several activated Src family members, including Lyn (Y396, p53/p56) and Hck (Y390, p59). When the blots were probed with total Lyn, Hck, or Src antibodies, 2 bands detected by the Lyn antibody, p53 and p56, corresponded precisely to the 2 bands detected by p-Src antibody. In contrast, Hck-specific antibody demonstrated a single slightly higher band, the p59 protein, while Src-specific antibody also recognized a p60 protein. To confirm that the reduction in phosphorylation is that of Lyn, we performed immunoprecipitation using anti-Lyn antibody followed by Western blot analysis with a tyrosine phosphorylation-specific antibody (Figure 6B). These results clearly demonstrated that Lyn phosphorylation is reduced by the combination treatment.
Effect of combination treatment on Src and Lyn phosphorylation. (A) 32D-p185, FL5.12-p185, K562, and LAMA84 cells were treated with the indicated concentrations of MPA and/or imatinib for 90 minutes (for 32D- and FL5.12-p185 cells) and 24 hours (for K562 and LAMA84 cells). Total cell lysate (30 μg) was used in each lane. Western blots were first probed with anti-phospho-Src antibody. Membranes were washed and reprobed with an antibody to detect total levels of Lyn (t-Lyn). This result is representative of 3 independent experiments. (B) 32D-p185 cells were treated with individual drugs or the combination of drugs as indicated for 3 hours. Lysate was immunoprecipitated with anti-Lyn antibody and blotted with an antibody (4G10) specific for tyrosine phosphorylated proteins (p-tyr). Lysate precipitated with protein A beads in the absence of Lyn antibody (IP control), and total lysate are shown in the last 2 lanes.
Effect of combination treatment on Src and Lyn phosphorylation. (A) 32D-p185, FL5.12-p185, K562, and LAMA84 cells were treated with the indicated concentrations of MPA and/or imatinib for 90 minutes (for 32D- and FL5.12-p185 cells) and 24 hours (for K562 and LAMA84 cells). Total cell lysate (30 μg) was used in each lane. Western blots were first probed with anti-phospho-Src antibody. Membranes were washed and reprobed with an antibody to detect total levels of Lyn (t-Lyn). This result is representative of 3 independent experiments. (B) 32D-p185 cells were treated with individual drugs or the combination of drugs as indicated for 3 hours. Lysate was immunoprecipitated with anti-Lyn antibody and blotted with an antibody (4G10) specific for tyrosine phosphorylated proteins (p-tyr). Lysate precipitated with protein A beads in the absence of Lyn antibody (IP control), and total lysate are shown in the last 2 lanes.
Effect of combination treatment on mTOR pathway
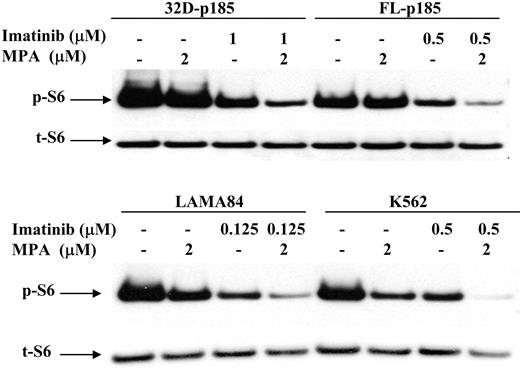
It has been shown that the mammalian target of rapamycin (mTOR) pathway is activated in Bcr-Abl-transformed cells,48 as indicated by hyperphosphorylation of p70 S6 kinase and S6 ribosomal proteins, as well as 4EBP1, resulting in enhanced protein translation and cell growth. We have shown previously that the p70 S6 kinase phosphorylation is down-regulated by MPA treatment in 32D cells.27 We therefore examined the S6 phosphorylation level following MPA and/or imatinib treatment in both murine and human Bcr-Abl-expressing cell lines. The level of phosphorylated S6 (p-S6) was slightly reduced by either MPA or imatinib treatment alone but was markedly reduced with combination of MPA and imatinib (Figure 7).
Effect of combination treatment on S6 phosphorylation. 32D-p185, FL5.12-p185, LAMA84, and K562 cells were treated with the indicated concentrations of MPA and/or imatinib for 3 hours (for 32D- and FL5.12-p185 cells) and 24 hours (for LAMA84 and K562 cells). Western blots were probed with antibody specific for phospho-S6 ribosomal protein (p-S6). Duplicate gels were probed with antibody specific for total S6 (t-S6). This is representative of 3 independent experiments.
Effect of combination treatment on S6 phosphorylation. 32D-p185, FL5.12-p185, LAMA84, and K562 cells were treated with the indicated concentrations of MPA and/or imatinib for 3 hours (for 32D- and FL5.12-p185 cells) and 24 hours (for LAMA84 and K562 cells). Western blots were probed with antibody specific for phospho-S6 ribosomal protein (p-S6). Duplicate gels were probed with antibody specific for total S6 (t-S6). This is representative of 3 independent experiments.
Discussion
Inhibition of the enzyme IMPDH and the consequent guanine nucleotide depletion have previously been shown to reduce blast counts in patients with CML in blast crisis,31-33 but the effect was transient and the toxicity of the drug precluded further investigation. Although inhibition of IMPDH also has been shown to result in the differentiation of leukemic cell lines such as HL-60 and K-56223,49 and to cause either cell-cycle arrest20,21 or apoptosis25-27 in other cell lines, the alterations in specific signaling pathways that result in these effects have not been fully elucidated. In previous work, we demonstrated that inhibition of the de novo synthesis of guanine nucleotides in IL-3-dependent cell lines resulted in apoptosis, while simultaneously causing the early down-regulation of both the mitogen-activated protein kinase (MEK/Erk) and the mTOR signaling pathways.27 These results and the demonstration of synergy between MPA and imatinib in Bcr-Abl-expressing cells led us to examine the effects of combined treatment on specific signaling molecules that have been demonstrated to be important in Bcr-Abl-mediated transformation.
The Stat proteins are very important in Bcr-Abl-mediated oncogenesis.50,51 These transcription factors are normally present in the cytoplasm of quiescent cells and are phosphorylated in response to stimulation by cytokines or growth factors resulting in dimerization and nuclear translocation and ultimately to the expression of specific target genes.52 Stat5 is the major Stat protein activated by Bcr-Abl in cell lines and in hematopoietic cells derived from patients with CML.43,53 Although an increase in Stat5 phosphorylation alone, using a dominant-active Stat5, is not sufficient to transform primary murine bone marrow cells, expression of a dominant-negative Stat5 reduces the efficiency of Bcr-Abl-mediated transformation and impairs leukemogenesis.54 It also has been shown that the antiapoptotic effect of Bcr-Abl is dependent upon Stat5 signaling to activate the Bcl-2 family members, Bcl-xL and Mcl-1.55,56 We have demonstrated that MPA treatment significantly enhanced imatinib's ability to decrease Stat5 phosphorylation and also reduced Mcl-1 and Bcl-xL expression. This effect was also enhanced at later time points in human cell lines, suggesting that the inhibitory effect of imatinib can be extended by treatment with MPA.
The Src family of tyrosine kinases are known to phosphorylate Stat proteins.44 The 2 Src family members, Lyn and Hck, are highly activated in Bcr-Abl-expressing cells38 as a results of direct physical interaction with Bcr-Abl.57,58 Expression of a kinase-defective mutant of Hck significantly down-regulated p-Stat546 and prevented transformation by Bcr-Abl,47 while inhibition of Lyn kinase expression using antisense oligonucleotides also significantly reduced leukemia cell growth.59,60 Our results have demonstrated that the combination of MPA and imatinib markedly reduces Lyn phosphorylation. Since overexpression of activated Lyn has been documented in later stages of CML and in Bcr-Abl-expressing cells that have developed resistant to imatinib,60 the additive effect of MPA on reducing p-Lyn may well be pivotal to its synergistic effect.
Although the PI3 kinase pathway is believed to be important for Bcr-Abl transformation and leukemia cell proliferation,61,62 we failed to observe any change in phospho-Akt level at either Thr 308 and Ser 473 sites following either MPA or imatinib alone or in combination. However, we did observe a dramatic down-regulation of the phosphorylated S6 ribosomal protein, a direct target of the mTOR/S6K1 pathway. It has been demonstrated previously that a Bcr-Abl mutant that failed to activate PI3K/Akt still can activate S6K1,62 indicating that the mTOR pathway may be activated independent of the PI3K/Akt pathway. The importance of the mTOR pathway in CML has become increasingly evident. This pathway acts as a sensor for the deprivation of essential nutrients such as amino acids, sources of nitrogen, or ATP63,64 and regulates the translation of a specific subset of mRNAs that contain polypyrimidine tracts at their 5′ ends.65 Both ribosomal protein S6, a substrate of S6K1, and 4EBP1 are constitutively phosphorylated in CML cells.48 A recent paper has documented synergy between the mTOR inhibitor, rapamycin, and imatinib in Bcr-Abl-expressing cell lines, as well as synergy of rapamycin with PKC412, an inhibitor of Flt-3 tyrosine kinase, in cell lines overexpressing Flt-3.66 The addition of a MEK inhibitor further potentiated the cytotoxicity. Clearly, pharmacological agents that inhibit multiple signaling pathways including mTOR have the potential to be more effective than single agents in inducing apoptosis in malignant cell lines.
Down-regulation of several signaling pathways caused by GTP depletion may enhance the effects of several other more specifically targeted chemotherapeutic agents. Our hypothesis is that guanine nucleotide depletion has a central sensing mechanism with multiple downstream effectors, ultimately resulting in growth arrest or apoptosis. Although this molecular sensor has not been definitively identified, we are exploring several small GTP binding proteins as candidates. One, the Rheb GTPase, was initially isolated as a Ras homolog enriched in brain67 and directly activates the mTOR pathway.68 Transient expression of a wild-type Rheb induces a dramatic increase in S6K phosphorylation and mobility shift of 4EBP1, and this high level of activity is consistent with its being predominantly GTP bound.68 A dominant-negative Rheb, defective in GTP binding, was found to block nutrient and serum-induced p70 S6K activation.69 We are currently testing the hypothesis that a reduction of intracellular GTP resulting from inhibition of de novo guanine nucleotide synthesis might decrease the level of GTP-bound Rheb, thus inhibiting the mTOR/S6K1 pathway.
Our data suggest that the IMPDH inhibitors may constitute a major addition to the armamentarium of drugs that target oncogenic receptor tyrosine kinases, and we are implementing a clinical protocol to test the combination of imatinib and MPA in patients with persistent positivity for the Bcr-Abl transcript on imatinib alone after 12 months. Our results demonstrating enhanced toxicity of the combination over either agent alone in cells that are partially resistant to imatinib on the basis of Lyn overexpression40 constitutes preliminary evidence that partial resistance to imatinib may be overcome by this approach. Although we have attempted to test this hypothesis in the leukemic cells from 2 cases of CML in blast crisis, the high degree of baseline apoptosis resulting from incubation of these cells ex vivo made interpretation of the results impossible.
Guanine nucleotides can be synthesized directly through the salvage of guanosine in vivo, thereby compensating for IMPDH inhibition. However, it has been demonstrated that imatinib inhibits nucleoside uptake through selective inhibition of nucleoside transporters.70 We have confirmed that imatinib treatment blocked the nucleoside transporter in a dose-dependent manner in leukemic blast cells from a CML patient (data not shown). Addition of guanosine to the culture medium did not reverse the synergistic effects of MPA and imatinib (data not shown). This observation enhances the probability that this combination will be effective in patients, since imatinib will inhibit purine salvage. The proposed clinical trial will be the optimal approach to definitively test our hypothesis and will incorporate the specific molecular end point of a more-than-1 log reduction in the Bcr-Abl transcript 6 months after the addition of MPA.
Prepublished online as Blood First Edition Paper, December 16, 2004; DOI 10.1182/blood-2004-10-3864.
Supported by National Institutes of Health grant CA 64193 (B.S.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Lee Graves for reading and critiquing the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal