A broad range of hematopoietic stem cells and progenitors reside within a fraction of umbilical cord blood (UCB) that exhibits low light scatter properties (SSClo) and high expression of aldehyde dehydrogenase (ALDHbr). Many SSClo ALDHbr cells coexpress CD34; however, other cells express either ALDH or CD34. To investigate the developmental potential of these cell subsets, purified ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg UCB cells were characterized within a variety of in vivo and in vitro assays. Primitive progenitors capable of multilineage development were monitored in long- and short-term repopulation assays performed on nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice, and in primary and secondary long-term culture assays. These progenitors were highly enriched within the ALDHbr CD34+ fraction. This cell fraction also enriched short-term myeloid progenitors that were detected in vitro. By comparison, ALDHneg CD34+ cells contained few primitive progenitors and had diminished short-term myeloid potential but exhibited enhanced short-term natural killer (NK) cell development in vitro. The ALDHbr CD34neg cells were not efficiently supported by any of the assays used. These studies suggested that in particular the expression of ALDH delineated distinct CD34+ stem cell and progenitor compartments. The differential expression of ALDH may provide a means to explore normal and malignant processes associated with myeloid and lymphoid development.
Introduction
Hematopoietic stem cells (HSCs) and progenitors maintain and replenish the blood in steady state and after blood or marrow transplantation.1,2 The HSCs themselves provide long-term hematopoietic repopulation of all of the blood cell lineages. Progenitor cells are more restricted in their ability to proliferate and in their capacity to generate multiple cell lineages.3,4 The CD34 cell surface antigen is expressed by human HSCs and progenitors and has been widely used to characterize these cells.5-13 However, CD34+ cells are heterogeneous,10-13 and recent evidence suggests that not all HSCs or progenitors express CD34 at all times.14-18 One promising complementary strategy for identifying and studying hematopoietic stem cells and progenitors is to rely on the expression of intracellular enzymes that may be important during development. One such enzyme is aldehyde dehydrogenase (ALDH). ALDH may play an important role in retinoid metabolism, and it appears to be relatively highly expressed in primitive hematopoietic cells in a number of species.19 Viable cells that express ALDH can be isolated by flow cytometry using fluorescent aldehyde substrates that freely diffuse across cell membranes.20,21 Polar fluorescent products accumulate when these dyes are oxidized in cells that express ALDH. Consequently, cells with high levels of ALDH activity stain more brightly than cells with lower ALDH expression and can be easily distinguished.
Proof of principle for this approach was first demonstrated in mice using a fluorescent ALDH substrate termed dansyl aminoacetaldehyde (DAAA).22 In those studies, the cells were first enriched from total bone marrow by counter-current elutriation (Fr25) and were then stained with DAAA to detect cells expressing high levels of ALDH (ALDHbr cells). Phenotypically, the murine Fr25 ALDHbr cells were CD34neg Sca-1neg cKitneg and were depleted of lineage-committed cells (linneg).22 These cells did not form colonies in vitro and did not establish splenic foci in short-term transplantation assays (splenic colony-forming units; CFUs-S). However, in long-term bone marrow transplantation assays, as few as 10 cells were sufficient to establish complete, multilineage hematopoietic repopulation. Thus, the Fr25 ALDHbr cells appeared to be a highly purified population of hematopoietic stem cells.
More recently, we developed an alternative fluorescent substrate for ALDH, termed BODIPY aminoacetaldehyde (BAAA) (Aldefluor; StemCo BioMedical, Durham, NC), to select human hematopoietic progenitors that express ALDH.21,23 In those studies, progenitors from human umbilical cord blood (UCB) and from human mobilized peripheral blood (mPB) were readily identified based on low orthogonal light scatter and high ALDH expression (SSClo ALDHbr cells).21,23 Hematopoietic progenitors within this cell fraction included clonogenic progenitors (CFUs) and cells with the capacity to initiate long-term culture (LTC). Subsequent studies indicated that SSClo ALDHbr UCB cells were also highly enriched for cells that engraft nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (SCID-repopulating cells [SRCs]).24-26 Thus, in contrast to the murine Fr25 ALDHbr HSCs, human cells that expressed ALDH exhibited a broad range of developmental potentials.
In all the previous studies on ALDHbr cells, it was noted that the SSClo ALDHbr UCB and mPB were enriched for CD34+ cells and for CD34+ CD38neg cells.21,23,25,26 However, though there was considerable overlap between cells that expressed CD34 and those that expressed ALDH, there were also cells that expressed one or the other determinant. In the current study, we sought to determine whether the differential expression of ALDH and CD34 could be used to define and segregate developmentally distinct progenitor populations. To pursue this, ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg hematopoietic cells were purified from SSClo UCB, and their developmental potential was tested in a variety of in vitro and in vivo assays. The most primitive hematopoietic progenitors were highly enriched within the ALDHbr CD34+ cell fraction. In addition, short-term myeloid progenitors were most abundant within the ALDHbr CD34+ cell subset. By comparison, the ALDHneg CD34+ cells contained few primitive progenitors and were reduced in short-term myeloid progenitors. However, when measured in short-term culture assays, the ALDHneg CD34+ cells contained higher frequencies of committed NK progenitors than did the ALDHbr CD34+ cells. The ALDHbr CD34neg cells were not consistently supported by any of the assays used, and their function remains undefined. Together these observations demonstrated that functionally distinct human hematopoietic progenitors could be resolved based on their relative expression of ALDH and CD34. In particular, these studies suggested that, within the CD34+ progenitor compartment, ALDH expression was significant for primitive multilineage progenitors and for the early myeloid compartment. In contrast, decreased ALDH expression was associated with enhanced short-term NK development by CD34+ progenitors.
Materials and methods
Reagents
BAAA (Aldefluor) was provided by StemCo Biomedical, Inc. Hydrocortisone hemisuccinate, verapamil, dimethylaminobenzaldehyde, Zn2SO4, Cu2SO4, Se2O3, and ascorbic acid were purchased from Sigma-Aldrich (St Louis, MO). 7-Amino actinomycin D (7AAD) was purchased from Molecular Probes (Eugene, OR). All cytokines were purchased from R&D Systems (Minneapolis, MN). All cell culture media and media supplements (2-mercaptoethanol [2-ME], pyruvate, glucose, Na2CO3, and HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]) were purchased from Invitrogen Life Technologies (Carlsbad, CA). Fetal calf serum (FCS) and equine serum were purchased from HyClone (Logan, UT).
ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells derived from UCB. UCB cells were labeled with BAAA, as described in “Materials and methods.” (A) Background fluorescence was established in the presence of DEAB. (B,E) SSClo ALDHneg and SSClo ALDHbr cell fractions were defined. (C-D) Both cell fractions contained CD34+ cells, although the SSClo ALDHbr fraction was highly enriched for CD34+ cells, to include CD34+ CD38neg cells (n = 8) (D). In most developmental studies, cells expressing lineage-specific antigens were excluded (n = 25). (F) CD34+ linneg cells were isolated from the SSClo ALDHneg. (G) CD34+ linneg and CD34neg linneg cells were isolated from SSClo ALDHbr UCB (H-M). To ensure purity, each cell fraction was sorted twice. Representative reanalyses depict twice-purified linneg SSClo ALDHneg CD34+ cells (H,K), linneg SSClo ALDHbr CD34neg cells (I,L), and linneg SSClo ALDHbr CD34+ cells (J,M).
ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells derived from UCB. UCB cells were labeled with BAAA, as described in “Materials and methods.” (A) Background fluorescence was established in the presence of DEAB. (B,E) SSClo ALDHneg and SSClo ALDHbr cell fractions were defined. (C-D) Both cell fractions contained CD34+ cells, although the SSClo ALDHbr fraction was highly enriched for CD34+ cells, to include CD34+ CD38neg cells (n = 8) (D). In most developmental studies, cells expressing lineage-specific antigens were excluded (n = 25). (F) CD34+ linneg cells were isolated from the SSClo ALDHneg. (G) CD34+ linneg and CD34neg linneg cells were isolated from SSClo ALDHbr UCB (H-M). To ensure purity, each cell fraction was sorted twice. Representative reanalyses depict twice-purified linneg SSClo ALDHneg CD34+ cells (H,K), linneg SSClo ALDHbr CD34neg cells (I,L), and linneg SSClo ALDHbr CD34+ cells (J,M).
UCB fractionation based on ALDH and CD34
UCBs were obtained from the Carolina Cord Blood Bank under protocols approved by the Institutional Review Board at Duke University Medical Center. UCB cells were stained with BAAA, as previously described.21 Briefly, cells were resuspended at 106 cells/mL in Iscove modified Dulbecco medium (IMDM) supplemented with 2% FCS (IMDM+2%) and equilibrated to 37°C. Cells were stained with 1 μM BAAA for 30 minutes in the presence of 50 μM verapamil (Sigma-Aldrich). Baseline fluorescence was established based on control cells stained with BAAA in the presence of 10 μM dimethylaminobenzaldehyde (DEAB), a potent inhibitor of ALDH.27 After 30 minutes, the cells were centrifuged, placed on ice, and stained with fluorescence-conjugated antibodies. These included monoclonal antibodies directed against CD3, CD19, CD56, CD235a (all phycoerythrin [PE] conjugated) and CD34 (allophycocyanin [APC] conjugated) (all from BD Pharmingen, San Jose, CA). Dead and dying cells were excluded by staining with 7AAD (1 μg/106 cells; Molecular Probes). The cells were washed and resuspended in chilled IMDM+2% containing 50 μM verapamil and were kept on ice before fluorescence-activated cell sorting.
Cell purification was performed using a FACS Vantage Cell Sorter (BD Biosciences). Samples were chilled throughout the cell sorting. During initial preparative sorts, the cells were captured in IMDM+2% containing 50 μM verapamil. For purity, the cells were immediately resorted into complete culture media that did not contain verapamil. UCB cells were fractionated to derive SSClo ALDHbr CD34+ cells, SSClo ALDHneg CD34+ cells, and SSClo ALDHbr CD34neg cells. In most studies, these cell fractions were derived from linneg UCB (Figure 1F-G; n = 25). In some studies, linneg UCB cells were enriched before sorting by depleting the PE-labeled CD3+, CD19+, CD56+, and CD235a+ cells using immunomagnetic beads (PE-specific EasySep beads; StemCell Technologies, Vancouver, BC, Canada). Residual lin+ (PE+) cells were excluded during the cell sorting. For some NOD/SCID reconstitution studies, control CD34+ cells were isolated from lineage-depleted UCB prepared using high-density magnetic enrichment (StemSep enrichment, human progenitor cocktail; StemCell Technologies).
NOD/SCID reconstitution
NOD/SCID (ltz-ltz) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Hematopoietic transplantations were performed according to protocols approved by the Institutional Animal Use and Care Committee at Duke University (Durham, NC). The mice underwent irradiation before transplantation (325 cGy). Each mouse underwent transplantation with purified cells derived from a single UCB. Cells were prepared in IMDM supplemented with 10% FCS and were transplanted by tail-vein injection. To prevent cell loss during the injections, carrier cells were added to purified cells before the transplantations. Carrier cells were total blood cells from irradiated NOD/SCID mice (106 cells per transplant; approximately 2 μL total blood).
NOD/SCID mice were killed 6 to 7 weeks after transplantation (short term) or 18 weeks or longer after transplantation (long term). Bone marrow cells were prepared in IMDM+2% and were concentrated to 107 cells/mL. Before antibody staining, murine Fc receptors were blocked by 15-minute incubation with an antibody specific for murine CD16 and CD32 (BD PharMingen). Dead and dying cells were excluded from analyses using 7AAD (Molecular Probes). The presence of human cells was detected with antibodies directed against CD45 and was further confirmed with antibodies directed against human class 1 HLA antigens (BD PharMingen). In addition, murine hematopoietic cells were excluded with antibodies directed against Ly5 (murine CD45; BD PharMingen). Human hematopoietic engraftment was defined by the presence of CD45+ Ly5neg cells (Figure 2A). Lineage-specific differentiation was determined using antibodies directed against human antigens (all BD PharMingen), including CD19 (B cell) and CD13 or CD15 (myeloid). Human progenitor cells were detected by their expression of CD34. In some animals, the total bone marrow cell preparations were also depleted of murine lineage-committed cells to enrich human cells (Figure 2B) (SpinSep isolations, murine progenitor enrichment cocktail; StemCell Technologies). Cells were stained with antibodies for 20 minutes at 4°C, washed with Dulbecco phosphate-buffered saline (PBS) supplemented with 2% FCS (PBS+2%), and fixed in PBS+2% containing 1% formaldehyde (methanol-free, EM grade; Polysciences, Warrington, PA).
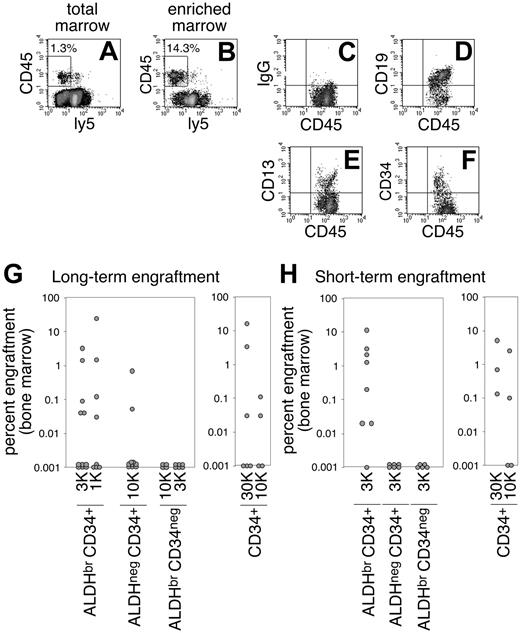
Human hematopoietic engraftment to NOD/SCID bone marrow. NOD/SCID mice received 1000 or 3000 transplanted SSClo ALDHbr CD34+ cells each. Comparisons were drawn to mice that received transplanted SSClo ALDHneg CD34+ or SSClo ALDHbr CD34neg cells (3000 or 10 000 cells per transplant). Transplantation controls included mice that underwent transplantation with 10 000 or 30 000 linneg CD34+ cells. (A) Human hematopoietic engraftment was assessed by the presence of CD45+ ly5neg (murine CD45neg) cells in NOD/SCID bone marrow. (B) To confirm engraftment, some marrows were enriched for human cells by depletion of lineage-committed murine cells using density-dependent negative selection. Percentages indicate the % CD45+ cells. (C) IgG indicates immunoglobulin G. Human hematopoietic engraftment was determined for (D) B-lymphoid, (E) myeloid, and (F) CD34+ progenitor cells. (A-F) Analyses performed at 20 to 21 weeks after transplantation of mice that underwent transplantation with 3000 SSClo ALDHbr CD34+ cells (A-B) or with 3000 linneg SSClo ALDHbr CD34+ cells (C-F). The percentage of long-term human hematopoietic engraftment for each cell fraction is depicted in panel G (as labeled; 18 to 21 weeks after transplantation; n = 6 UCB). The percentage of short-term human hematopoietic engraftment for each cell fraction is depicted in panel H (as labeled; 6 to 7 weeks after transplantation; n = 3 UCB). Each data point represents the percentage of human hematopoietic engraftment within a single mouse.
Human hematopoietic engraftment to NOD/SCID bone marrow. NOD/SCID mice received 1000 or 3000 transplanted SSClo ALDHbr CD34+ cells each. Comparisons were drawn to mice that received transplanted SSClo ALDHneg CD34+ or SSClo ALDHbr CD34neg cells (3000 or 10 000 cells per transplant). Transplantation controls included mice that underwent transplantation with 10 000 or 30 000 linneg CD34+ cells. (A) Human hematopoietic engraftment was assessed by the presence of CD45+ ly5neg (murine CD45neg) cells in NOD/SCID bone marrow. (B) To confirm engraftment, some marrows were enriched for human cells by depletion of lineage-committed murine cells using density-dependent negative selection. Percentages indicate the % CD45+ cells. (C) IgG indicates immunoglobulin G. Human hematopoietic engraftment was determined for (D) B-lymphoid, (E) myeloid, and (F) CD34+ progenitor cells. (A-F) Analyses performed at 20 to 21 weeks after transplantation of mice that underwent transplantation with 3000 SSClo ALDHbr CD34+ cells (A-B) or with 3000 linneg SSClo ALDHbr CD34+ cells (C-F). The percentage of long-term human hematopoietic engraftment for each cell fraction is depicted in panel G (as labeled; 18 to 21 weeks after transplantation; n = 6 UCB). The percentage of short-term human hematopoietic engraftment for each cell fraction is depicted in panel H (as labeled; 6 to 7 weeks after transplantation; n = 3 UCB). Each data point represents the percentage of human hematopoietic engraftment within a single mouse.
Hematopoietic progenitor colony assays
Hematopoietic progenitor colony assays were performed in MethoCult GF+ H4435 (StemCell Technologies). Myeloid colonies (more than 50 cells) were scored between 14 and 18 days.
Long-term culture assays
LTC assays were performed on AFT024 fetal liver stroma (kindly provided by Dr Ihor Lemischka, Princeton University, NJ).28 AFT024 stromas were maintained in DMEM/F12 medium supplemented with 10% FCS. For LTCs, the stromas were established in 24-well cell-culture dishes. Stromas, maintained at 33°C for normal passage, were irradiated (20 Gy) and transferred to 37°C before the establishment of LTCs. Primary and secondary LTCs were grown in MEMα supplemented with 12% FCS, 12% equine serum, 55 μM 2-mercaptoethanol (2-ME), 100 nM hydrocortisone, recombinant human interleukin-3 (IL-3; 5 ng/mL), and recombinant human macrophage inflammatory protein alpha-1 (MIP-1α) (100 pg/mL).29,30 Cultures were fed weekly by replacing half the medium in each culture. After 5 weeks, primary LTCs were harvested with trypsin-EDTA (ethylenediaminetetraacetic acid) to establish clonogenic assays in MethylCult GF+ H4435 (StemCell Technologies), to establish secondary LTCs on AFT024 stroma, and to establish secondary NK cell development assays on AFT024 stroma (see “Secondary NK assays”).
In some LTC assays, cell proliferation was monitored by fluorescence-activated cell sorter (FACS) analysis. For these analyses, a portion of each culture was concentrated to less than 100 μL, and the cells were stained with antibodies specific for human CD45, CD13, and CD34 (all BD PharMingen). Dead and dying cells were excluded with 7AAD (Molecular Probes). The cells were stained for 20 minutes at 4°C, washed in PBS+2%, and fixed in PBS+2% containing 1% formaldehyde (Polysciences). For accuracy, the CD45+7AADneg cells were enumerated relative to fixed numbers of 6-μm fluorescent reference beads that were added to the reactions (Polysciences).
Secondary NK assays
After primary LTCs, the potential for NK cell development was monitored by replating a fraction of the culture on irradiated AFT024 cells (20 Gy) in a DMEM/F12 medium supplemented with 10% FCS that was also modified in a manner similar to a media formulation previously described.31 This medium was adjusted to 20 mM HEPES, 2.5 g/L Na2CO3, 4.5 g/L glucose, 2 mM sodium pyruvate, 55 μM 2-ME, 5 μMZn2SO4, 100 nM Cu2SO4, 100 nM Se2O3, 100 nM ascorbic acid, and 0.3 μM biotin. NK cell development was encouraged with recombinant IL-2 (5 ng/mL), IL-7, IL-15, kit ligand, and Flt3 ligand (each at 10 ng/mL). Cultures were fed twice weekly by replacing half the medium in each culture.
After 5 to 6 weeks, the cultures were monitored for NK (CD56+) and myeloid (CD13+) cells by flow cytometry. Cultures were harvested with trypsin-EDTA and were stained with fluorescence-conjugated antibodies and 7AAD. The expansion of viable hematopoietic cells (CD45+7AADneg) was calculated relative to 6-μm fluorescent reference beads (Polysciences). Anti-CD45 (fluorescein isothiocyanate [FITC], clone J33) was purchased from Coulter-Immunotech (Miami, FL). All other antibodies were purchased from BD PharMingen.
Short-term assays for lymphoid development
Short-term culture assays for lymphoid development were monitored on STO murine embryonic fibroblasts (CRL-1503; American Type Culture Collection, Manassas, VA). For these assays, the STO cells were established as monolayers in 24-well dishes in DMEM/F12 media supplemented with 10% FCS. At confluence, the cells were irradiated (20 Gy). Purified cells were cultured on STO feeders in DMEM/F12 medium that was supplemented as described for secondary NK cultures. In short-term assays, NK cell development was encouraged with recombinant IL-3 (10 ng/mL), IL-7 (20 ng/mL), and IL-15 (10 ng/mL). After 21 days, NK cell development was monitored by flow cytometry. The cultures were harvested with trypsin-EDTA and were stained with fluorescence-conjugated antibodies and 7AAD. The expansion of viable hematopoietic cells (CD45+7AADneg) was calculated relative to 6-μm fluorescent reference beads. NK cells were monitored as CD56+ CD3neg cells. The total output of lymphoid cells was calculated as [(total cell expansion) × (percentage CD56+ cells)]. Myeloid cells were monitored as CD13+ cells. The total output of myeloid cells was calculated as [(total cell expansion) × (percentage CD13+ cells)]. The relative cell output was calculated as [(total output, ALDHneg CD34+ cells)/(total output, ALDHbr CD34+ cells)] or as [(total output, ALDHbr CD34+ cells)/(total output, ALDHneg CD34+ cells)]. Anti-CD45 (FITC, clone J33) was purchased from Coulter-Immunotech (Miami, FL). Anti-CD3 (PE) was purchased from Becton Dickinson. All other antibodies were purchased from BD PharMingen.
Statistical analyses
For most studies, the mean (± SD) and median values were presented. After development in vitro, the performances of linneg SSClo ALDHbr CD34+ and linneg SSClo ALDHneg CD34+ cells were compared in paired 1-tailed nonparametric analyses (Wilcoxon signed rank tests). Frequencies for NOD/SCID repopulating cells, for secondary LTCs, and for lymphoid progenitors were estimated by limiting dilution analyses using L-Calc (StemCell Technologies). For NOD/SCID studies, the frequencies of SRCs were calculated based on the mice that demonstrated multilineage engraftment, in which not more than 90% of human cells were CD19+. LTCs for secondary NK progenitors were considered negative when fewer than 10 CD56+ cells were generated per cell used to initiate the primary LTCs.
Results
Discrete ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells exist within UCB
The overall goal of this study was to determine whether distinct hematopoietic progenitors could be delineated within UCB based on their expression of ALDH and CD34. As had been previously noted,21 the SSClo ALDHbr UCB was highly enriched for CD34+ cells and contained 79.7% ± 16.8% of the SSClo CD34+ CD38neg cells (n = 8; Figure 1C-D [compare panels]). To characterize these cells, SSClo ALDHbr CD34+, SSClo ALDHneg CD34+, and SSClo ALDHbr CD34neg UCB cells were purified for functional analyses. Our previous studies had indicated that SSClo ALDHbr UCB contained few cells that expressed lineage-specific antigens.21 However, to ensure this, lineage-committed cells were excluded to yield a linneg population (depicted in Figure 1F-G). Each fraction contained small cells with minimal cytoplasm that were not morphologically distinguishable (data not shown). The SSClo ALDHbr cells represented 0.96% ± 0.5% of the UCB, and 50.9% ± 18.3% of these cells expressed CD34. Similarly, 0.9% ± 0.6% of the total UCB was linneg SSClo CD34+, and 47.9% ± 14.3% of these cells expressed ALDH. Absolute percentages of the linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg UCB cell subsets are summarized in Table 1. In addition, it was noted that CD34 expression was significantly more intense on linneg SSClo ALDHbr CD34+ cells than on linneg SSClo ALDHneg CD34+ cells (Table 1; Figure 1F-G). Together these observations suggested that discrete populations of cells could be defined based on their expression patterns of ALDH and CD34.
UCB cell fractionation
. | SSClo ALDHbr . | linneg SSClo CD34+ . | linneg SSClo ALDHbrCD34+ . | linneg SSClo ALDHnegCD34+ . | linneg SSClo ALDHbrCD34neg . |
|---|---|---|---|---|---|
| Total UCB, % | |||||
| Mean ± SD | 0.96 ± 0.5 | 0.90 ± 0.63 | 0.48 ± 0.29 | 0.27 ± 0.31 | 0.25 ± 0.26 |
| Median | 0.91 | 0.8 | 0.46 | 0.17 | 0.16 |
| Mean CD34 fluorescence | |||||
| Mean ± SD | — | 320.7 ± 154 | 391.3 ± 198.8 | 188.4 ± 107.5 | 5.69 ± 1.9 |
| Median | — | 305.5 | 369.7 | 146.4 | 5.46 |
. | SSClo ALDHbr . | linneg SSClo CD34+ . | linneg SSClo ALDHbrCD34+ . | linneg SSClo ALDHnegCD34+ . | linneg SSClo ALDHbrCD34neg . |
|---|---|---|---|---|---|
| Total UCB, % | |||||
| Mean ± SD | 0.96 ± 0.5 | 0.90 ± 0.63 | 0.48 ± 0.29 | 0.27 ± 0.31 | 0.25 ± 0.26 |
| Median | 0.91 | 0.8 | 0.46 | 0.17 | 0.16 |
| Mean CD34 fluorescence | |||||
| Mean ± SD | — | 320.7 ± 154 | 391.3 ± 198.8 | 188.4 ± 107.5 | 5.69 ± 1.9 |
| Median | — | 305.5 | 369.7 | 146.4 | 5.46 |
UCB cells were fractionated as depicted in Figure 1. Mean (± SD) and median percentages were calculated for linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg cell fractions (n = 25). Mean (± SD) and median mean fluorescence intensity (MFI) for CD34 expression were calculated for each cell fraction (n = 25). The intensity of CD34 expression was significantly different between the ALDHbr CD34+ and the ALDHneg CD34+ cell subsets (P < .0001). See also Figure 1F-G.
— indicates not determined.
NOD/SCID repopulating cells coexpress ALDH and CD34
To confirm that primitive progenitors could be distinguished on the basis of ALDH and CD34 expression, ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg UCB cells were transplanted into NOD/SCID mice. In 3 initial studies, these cell fractions were purified from SSClo UCB. In all subsequent studies, the cells were purified from linneg SSClo UCB, as depicted in Figure 1. Because no differences were observed between these studies, the results were pooled. Control mice underwent transplantation with CD34+ linneg cells. In all studies, human hematopoietic engraftment was monitored by the content of human CD45+ ly5neg present in NOD/SCID bone marrow (Figure 2A) and was routinely confirmed by their expression of class 1 HLA antigens (data not shown). When limiting cell doses were used, some mice achieved only limited (less than 0.1%) engraftment. Therefore, in some mice, low-level human hematopoietic engraftment was also confirmed by enriching the relative proportions of human cells by depleting lineage-committed murine cells (n = 7; Figure 2B). To test for multilineage engraftment, the NOD/SCID marrow was also stained for human B cells (Figure 2D), myeloid cells (Figure 2E), and CD34+ progenitors (Figure 2F).
Long-term human hematopoietic engraftment was reliably detected at varying levels in NOD/SCID mice in which 1000 to 3000 ALDHbr CD34+ cells were transplanted (Figure 2G). Human hematopoietic engraftment in these mice was 64.5% ± 30.7% B-lymphoid and 29.7% ± 34.7% myeloid cells. In addition, 5 of 9 mice maintained discernible CD34+ cells (9.16% ± 9.07%). SRCs were defined as cells that repopulated multiple human hematopoietic lineages in NOD/SCID mice. Limiting dilution analyses estimated the frequency of SRCs to be 1 in 4687 ALDHbr CD34+ cells (95% confidence interval [CI]: 1/2180-1/10 077). In control animals, the SRC frequency was estimated to be 1 in 31 486 CD34+ linneg cells (95% CI: 1/12 497-1/79 332). Therefore, the ALDHbr CD34+ cell fraction was enriched for SRC approximately 7-fold when compared with the CD34+ cell fraction. In contrast, the ALDHneg CD34+ cells exhibited very low levels of SRC activity. Only 2 of 10 mice that received 10 000 ALDHneg CD34+ cells had discernible human hematopoietic engraftment. With this cell fraction, estimates for the frequency of SRCs would not be accurate. However, limiting dilution analyses performed on these data suggested that SRCs were likely to represent less than 1 in 45 000 ALDHneg CD34+ cells. Higher cell transplant doses would be required to accurately resolve this issue. Finally, the ALDHbr CD34neg cells did not engraft NOD/SCID mice at any of the cell doses used (Figure 2G). These studies suggested that long-term multilineage NOD/SCID repopulating cells were enriched in cells that expressed both ALDH and CD34.
To continue these studies, short-term transplantation assays were performed using cell doses that approached the limiting dilution for the ALDHbr CD34+ cells. Purified linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, or linneg SSClo ALDHbr CD34neg cells were transplanted into NOD/SCID mice, and human hematopoietic engraftment was monitored at 6 to 7 weeks after transplantation (Figure 2H). Using 3000 cells per transplant, the ALDHbr CD34+ cells reliably engrafted NOD/SCID bone marrow, which recapitulated what had been observed in long-term hematopoietic transplantation assays. In contrast, the ALDHneg CD34+ and ALDHbr CD34neg cells did not achieve detectable human hematopoietic engraftment at these low cell doses, even in short-term assays. These data suggested that short-term NOD/SCID repopulating cells were enriched within the ALDHbr CD34+ cell fraction.
Progenitors that initiate LTCs coexpress ALDH and CD34
LTC assays were established as an alternative assay for primitive progenitors. These assays were used to test a potential for long-term multilineage growth without the more stringent requirement for cell homing that is present in NOD/SCID transplantation assays. linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg UCB cells were purified as in Figure 1. LTCs were initiated on AFT024 stroma using 50 to 500 cells/culture (n = 8).29,30 After 5 weeks, the LTCs were harvested, and the collected cells were monitored for total hematopoietic cell expansion, content of myeloid CFU, content of secondary LTC, and content of secondary NK progenitors. As with the primary LTCs, secondary LTCs were maintained in a standard Dexter-type medium supplemented with human IL-3 and MIP-1α. Secondary NK cultures were established without hydrocortisone in the presence of human Kit ligand, Flt3 ligand, IL-2, IL-7, and IL-15.
The distributions of cells that initiated LTCs closely paralleled the data for SRCs. In primary LTCs, the ALDHbr CD34+ cells expanded 4-fold more than the ALDHneg CD34+ cells and yielded approximately 60-fold more CFUs (Table 2). Similarly, the ALDHbr CD34+ cells yielded approximately 100-fold more CFUs after secondary LTCs when compared with ALDHneg CD34+ cells (Table 2). Limiting dilution analyses estimated that 1 in 43 ALDHbr CD34+ cells initiated secondary LTCs (95% CI: 1/18-1/103). In contrast, the ALDHneg CD34+ cells were nearly devoid of secondary LTCs, and only 3 of 8 ALDHneg CD34+ cell preparations initiated these cultures. Limiting dilution analyses estimated that 1 in 1130 ALDHneg CD34+ cells initiated secondary LTCs (95% CI: 1/364-1/3513). Finally, the ALDHbr CD34neg cell fraction failed to initiate primary or secondary LTCs.
Cell development in LTC
. | linneg CD34+ . | ALDHbr CD34+ . | ALDHneg CD34+ . | P . |
|---|---|---|---|---|
| Cell expansion in primary LTC, -fold | ||||
| Expansion* | .0625 | |||
| Mean ± SD | 4083 ± 1922 | 3664 ± 2120 | 827 ± 1142 | |
| Median | 3479 | 3350 | 285 | |
| Myeloid CFUs/100 initiating cells | ||||
| Primary LTC† | .004 | |||
| Mean ± SD | 260.9 ± 159 | 309.2 ± 180 | 4.85 ± 10.4 | |
| Median | 294.9 | 349.4 | 0 | |
| Secondary LTC† | .004 | |||
| Mean ± SD | 13.9 ± 8.5 | 54.6 ± 95.1 | 0.36 ± 0.56 | |
| Median | 9.74 | 25.9 | 0 |
. | linneg CD34+ . | ALDHbr CD34+ . | ALDHneg CD34+ . | P . |
|---|---|---|---|---|
| Cell expansion in primary LTC, -fold | ||||
| Expansion* | .0625 | |||
| Mean ± SD | 4083 ± 1922 | 3664 ± 2120 | 827 ± 1142 | |
| Median | 3479 | 3350 | 285 | |
| Myeloid CFUs/100 initiating cells | ||||
| Primary LTC† | .004 | |||
| Mean ± SD | 260.9 ± 159 | 309.2 ± 180 | 4.85 ± 10.4 | |
| Median | 294.9 | 349.4 | 0 | |
| Secondary LTC† | .004 | |||
| Mean ± SD | 13.9 ± 8.5 | 54.6 ± 95.1 | 0.36 ± 0.56 | |
| Median | 9.74 | 25.9 | 0 |
LTCs were established with linneg SSClo CD34+, linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg cells, as described in “Materials and methods.” Fifty to 500 cells were used to initiate the primary LTC. In some cultures, the total expansion of CD45+ cells was estimated by flow cytometry after 5 weeks.
Linneg SSClo ALDHbr CD34neg cells were not supported under these culture conditions. Statistical comparisons (P) were drawn between the linneg SSClo ALDHbr CD34+ and the linneg SSClo ALDHneg CD34+ cell fractions using paired, 1-tailed nonparametric analyses (Wilcoxon signed rank test).
Mean (±SD) and median frequencies for myeloid CFUs were normalized per 100 cells that were used to initiate the culture.
Secondary NK culture assays were established from primary LTC cultures and were used to measure long-term lymphoid development in vitro. In these assays, the linneg SSClo ALDHbr CD34+ cells consistently gave rise to high percentages of CD56+ cells in secondary cultures (Figure 3A-B). Because all these cultures contained NK cells, the frequencies of the long-term NK progenitor cells could not be estimated. However, under these same conditions, only half the primary LTCs that had been initiated with linneg SSClo ALDHneg CD34+ cells yielded any NK progeny in secondary cultures (Figure 3C-D). Limiting dilution analyses estimated that 1 in 709 linneg SSClo ALDHneg CD34+ cells initiated secondary NK cultures (95% CI: 1/258-1/1947). Collectively, these studies indicated that long-term multilineage growth in vitro was primarily associated with cells that expressed ALDH and CD34, similar to what had been observed in SCID repopulation studies.
Short-term myeloid progenitors are more frequent among ALDHbr CD34+ cells
The data for SRC and LTC activity suggested that primitive, multilineage progenitor cells were highly enriched within the ALDHbr CD34+ cell fraction. To determine whether this was also true for short-term myeloid progenitors, linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg UCB was monitored for clonogenic cells (CFUs) in hematopoietic progenitor colony assays (HPCAs). Both the ALDHneg CD34+ and the ALDHbr CD34+ cell fractions contained clonogenic myeloid progenitors (Table 3). However, myeloid CFUs were more than twice as frequent within the ALDHbr CD34+ subset as they were among the ALDHneg CD34+ cells. The ALDHbr CD34neg cells did not contain detectable clonogenic progenitors. These studies indicated that myeloid CFU activity was enriched in cells that coexpressed CD34 and ALDH but that some CFU activity remained in CD34+ cells that did not express ALDH. In contrast, no clonogenic myeloid progenitors were detected in cells that did not express CD34.
Clonogenic cell growth by ALDHbr CD34+ and ALDHneg CD34+ cells
. | ALDHbr CD34+ . | ALDHneg CD34+ . | ALDHbr CD34neg . | P . |
|---|---|---|---|---|
| Myeloid CFUs/100 initiating cells | ||||
| Mean ± SD | 26.8 ± 16.95 | 9.54 ± 11.8 | ND | .06 |
| Median | 24 | 3.75 | ND |
. | ALDHbr CD34+ . | ALDHneg CD34+ . | ALDHbr CD34neg . | P . |
|---|---|---|---|---|
| Myeloid CFUs/100 initiating cells | ||||
| Mean ± SD | 26.8 ± 16.95 | 9.54 ± 11.8 | ND | .06 |
| Median | 24 | 3.75 | ND |
linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg cells were isolated (Figure 1) and cultured in short-term myeloid clonogenic assays, as described in “Materials and methods” (n = 7). The total numbers of myeloid colonies was normalized per 100 cells that initiated each culture. Statistical comparisons (P) were drawn between the linneg SSClo ALDHbr CD34+ and the linneg SSClo ALDHneg CD34+ cell fractions using paired, 1-tailed nonparametric analyses (Wilcoxon signed rank test).
ND indicates none detected.
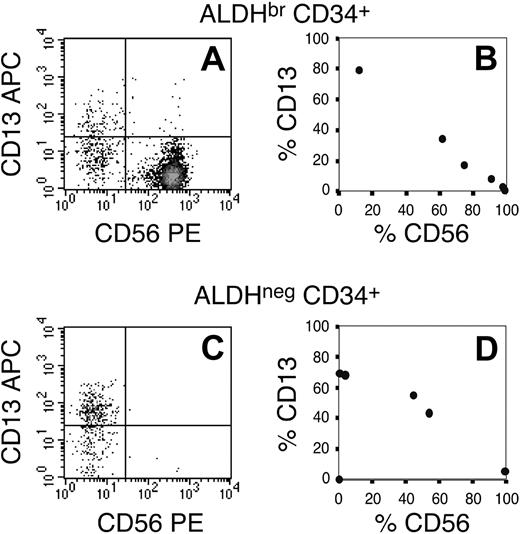
NK development in secondary LTC assays. Secondary NK cultures were established from primary LTCs that had been originally initiated with 50 to 500 cells per culture (n = 6). (A,C) Secondary cultures were analyzed for their content of CD56+ NK cells and CD13+ myeloid cells. (B,D) To represent the entire data set, the relative percentages of myeloid and lymphoid progeny were plotted for each culture, where each data point represents the average of duplicate cultures. (A-B) High percentages of CD56+ cells (more than 50%) were present in secondary cultures derived from primary LTCs originally established with linneg SSClo ALDHbr CD34+ cells. The single exception was from primary cultures that had been initiated with 50 cells/well. (C-D) In contrast, of the cultures established with linneg SSClo ALDHneg CD34+ cells, 2 secondary cultures gave rise to predominantly CD13+ myeloid progeny, and 1 secondary culture had no evident growth.
NK development in secondary LTC assays. Secondary NK cultures were established from primary LTCs that had been originally initiated with 50 to 500 cells per culture (n = 6). (A,C) Secondary cultures were analyzed for their content of CD56+ NK cells and CD13+ myeloid cells. (B,D) To represent the entire data set, the relative percentages of myeloid and lymphoid progeny were plotted for each culture, where each data point represents the average of duplicate cultures. (A-B) High percentages of CD56+ cells (more than 50%) were present in secondary cultures derived from primary LTCs originally established with linneg SSClo ALDHbr CD34+ cells. The single exception was from primary cultures that had been initiated with 50 cells/well. (C-D) In contrast, of the cultures established with linneg SSClo ALDHneg CD34+ cells, 2 secondary cultures gave rise to predominantly CD13+ myeloid progeny, and 1 secondary culture had no evident growth.
ALDHneg CD34+ cells contain short-term lymphoid progenitors
To complete these studies, we next sought to determine whether short-term lymphoid progenitors could be discriminated based on the expression of ALDH and CD34. Purified linneg SSClo ALDHbr CD34+, linneg SSClo ALDHneg CD34+, and linneg SSClo ALDHbr CD34neg cells were cultured on STO murine embryonic fibroblasts in the presence of recombinant human IL-3, IL-7, and IL-15 to encourage lymphoid progenitors to develop into NK cells. This culture system has been previously used to support the rapid emergence of NK cells from CD7+ CD34neg cells (R.W.S., unpublished observations, December 2003). After 21 days, the cultures were analyzed for their total expansion of hematopoietic cells (CD45+7AADneg) and for their relative frequencies of CD56+ and CD13+ cells.
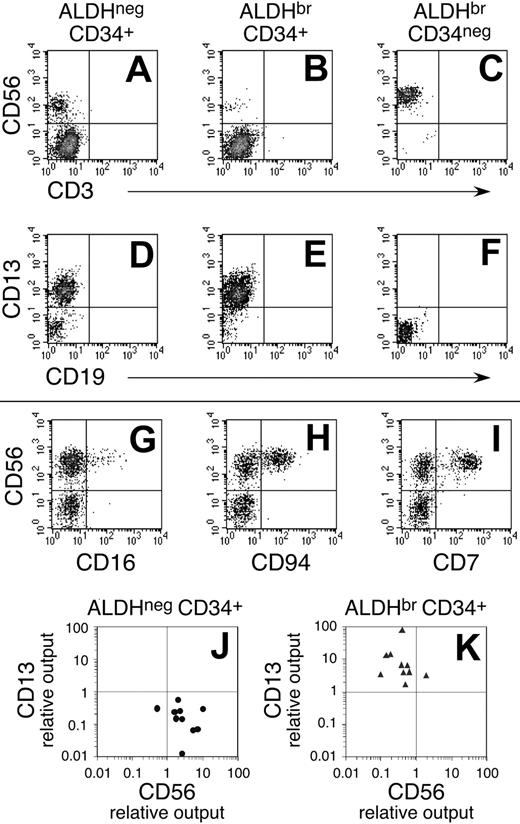
In these short-term cultures, the ALDHbr CD34+ cells expanded more efficiently than did the ALDHneg CD34+ cells. Both cell fractions gave rise to CD56+ progeny that, based on their phenotype, appeared to be NK cells. However, the ALDHneg CD34+ cells consistently gave rise to higher percentages of CD56+ lymphoid cells than did the ALDHbr CD34+ cells (Table 4; Figure 4A-B [compare panels]). Furthermore, even with lower total cell expansion, the ALDHneg CD34+ cells produced 3-fold more NK progeny than were observed in cultures initiated with ALDHbr CD34+ cells (Table 4). Indeed, in 9 of 10 cultures established, the ALDHneg CD34+ cells produced more CD56+ progeny (Figure 4J). This suggested that the ALDHneg CD34+ cell fraction was enriched for short-term NK progenitors relative to ALDHbr CD34+ progenitors. In contrast, the ALDHbr CD34+ cells predominantly gave rise to CD13+ myeloid cells (Table 4; Figure 4D-E [compare panels]). Taken together, the total output of myeloid progeny by ALDHbr CD34+ was approximately 7-fold greater than that of ALDHneg CD34+ cells (Table 4). This was consistent with their clonogenic potential as monitored in myeloid CFU assays (Table 3). Furthermore, relative to ALDHneg CD34+ cells, the ALDHbr CD34+ cell fraction always produced more CD13+ progeny (Figure 4K). Finally, of 9 culture assays performed, only 2 preparations of ALDHbr CD34neg cells proliferated to any degree. Furthermore, within those 2 assays, the cells expanded less than 10-fold and gave rise to progeny that were nearly entirely CD56+ (Figure 4C-F). Given that these events were sporadic, no conclusions were drawn as to the developmental potential of the ALDHbr CD34neg cells in these short-term assays.
Cell development in short-term cultures
. | ALDHneg CD34+ . | ALDHbr CD34+ . | ALDHbr CD34neg . | P . |
|---|---|---|---|---|
| Expansion, -fold | .001 | |||
| Mean ± SD | 233.7 ± 262 | 1185 ± 1352 | 5.41 ± 9.01* | |
| Median | 145.2 | 427.1 | 2.04* | |
| CD56+, % | .001 | |||
| Mean ± SD | 29.8 ± 28.2 | 2.52 ± 3.0 | 95.4 ± 3.95* | |
| Median | 19.4 | 1.97 | 95.4* | |
| CD56 output, no. cells | .014 | |||
| Mean ± SD | 80.6 ± 112 | 25.9 ± 32.4 | 14.8 ± 9.17* | |
| Median | 25 | 6.4 | 14.8* | |
| CD13+, % | .002 | |||
| Mean ± SD | 69.2 ± 24 | 90.7 ± 5.7 | 1.02 ± 0.6* | |
| Median | 75.2 | 91.3 | 1.02* | |
| CD13 output, no. cells | .001 | |||
| Mean ± SD | 152.5 ± 170.2 | 1085 ± 1275 | 0.28 ± 0.33* | |
| Median | 95.6 | 361.7 | 0.28* |
. | ALDHneg CD34+ . | ALDHbr CD34+ . | ALDHbr CD34neg . | P . |
|---|---|---|---|---|
| Expansion, -fold | .001 | |||
| Mean ± SD | 233.7 ± 262 | 1185 ± 1352 | 5.41 ± 9.01* | |
| Median | 145.2 | 427.1 | 2.04* | |
| CD56+, % | .001 | |||
| Mean ± SD | 29.8 ± 28.2 | 2.52 ± 3.0 | 95.4 ± 3.95* | |
| Median | 19.4 | 1.97 | 95.4* | |
| CD56 output, no. cells | .014 | |||
| Mean ± SD | 80.6 ± 112 | 25.9 ± 32.4 | 14.8 ± 9.17* | |
| Median | 25 | 6.4 | 14.8* | |
| CD13+, % | .002 | |||
| Mean ± SD | 69.2 ± 24 | 90.7 ± 5.7 | 1.02 ± 0.6* | |
| Median | 75.2 | 91.3 | 1.02* | |
| CD13 output, no. cells | .001 | |||
| Mean ± SD | 152.5 ± 170.2 | 1085 ± 1275 | 0.28 ± 0.33* | |
| Median | 95.6 | 361.7 | 0.28* |
ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells were isolated from SSClo linneg UCB (see Figure 1). Two hundred to 1000 purified cells were cultured on STO feeder stroma in the presence of IL-3, IL-7, and IL-15, as described in “Materials and methods.” Each culture was analyzed for its expansion of CD45+ cells and for its relative content of lymphoid (CD56+) or myeloid (CD13+) progeny (see Figure 4). The total output of lymphoid and myeloid cells was calculated per cell that initiated the culture. Mean (±SD) and median values were determined. Statistical comparisons (P) were drawn between the linneg SSClo ALDHbr CD34+ and the linneg SSClo ALDHneg CD34+ cell fractions using paired, 1-tailed nonparametric analyses (Wilcoxon signed rank test; n = 10).
Only 2 of 9 cultures initiated with ALDHbr CD34neg cells could be evaluated.
Cell development in short-term culture assays. ALDHneg CD34+, ALDHbr CD34+, and ALDHbr CD34neg cells were purified from linneg SSClo UCB, as depicted in Figure 1. Two hundred to 1000 purified cells were cultured on STO fibroblasts in the presence of IL-3, IL-7, and IL-15 (n = 10). Cultures initiated with ALDHneg CD34+ cells contained higher percentages of CD56+ lymphoid progeny than did the ALDHbr CD34+ cells (compare panels A and B; see Table 4). Similarly, the ALDHneg CD34+ cell fraction yielded lower percentages of CD13+ myeloid progeny (compare panels D and E). Only 2 cultures initiated with ALDHbr CD34neg cells had growth significant enough to evaluate lineage development, and these gave rise to CD56+ progeny (C,F). The CD56+ progeny of ALDHneg CD34+ (G-I) and ALDHbr CD34+ cells (data not shown) expressed other antigens consistent with NK cells. (J) Relative output of lymphoid cells was higher in cultures initiated with ALDHneg CD34+ cells when compared with paired cultures initiated with ALDHbr CD34+ cells. (K) Conversely, the relative cell output of myeloid cells was higher in cultures initiated with ALDHbr CD34+ cells when compared with paired cultures initiated with ALDHneg CD34+ cells. Estimations for relative cell output are described in “Materials and methods.” (J-K) Each data point represents an average from duplicate cultures.
Cell development in short-term culture assays. ALDHneg CD34+, ALDHbr CD34+, and ALDHbr CD34neg cells were purified from linneg SSClo UCB, as depicted in Figure 1. Two hundred to 1000 purified cells were cultured on STO fibroblasts in the presence of IL-3, IL-7, and IL-15 (n = 10). Cultures initiated with ALDHneg CD34+ cells contained higher percentages of CD56+ lymphoid progeny than did the ALDHbr CD34+ cells (compare panels A and B; see Table 4). Similarly, the ALDHneg CD34+ cell fraction yielded lower percentages of CD13+ myeloid progeny (compare panels D and E). Only 2 cultures initiated with ALDHbr CD34neg cells had growth significant enough to evaluate lineage development, and these gave rise to CD56+ progeny (C,F). The CD56+ progeny of ALDHneg CD34+ (G-I) and ALDHbr CD34+ cells (data not shown) expressed other antigens consistent with NK cells. (J) Relative output of lymphoid cells was higher in cultures initiated with ALDHneg CD34+ cells when compared with paired cultures initiated with ALDHbr CD34+ cells. (K) Conversely, the relative cell output of myeloid cells was higher in cultures initiated with ALDHbr CD34+ cells when compared with paired cultures initiated with ALDHneg CD34+ cells. Estimations for relative cell output are described in “Materials and methods.” (J-K) Each data point represents an average from duplicate cultures.
Discussion
We and others have previously demonstrated that the SSClo ALDHbr UCB was enriched for clonogenic progenitors, progenitors that initiate LTC and NOD/SCID repopulating cells.21,24-26 In all these studies there was substantial, but incomplete, overlap between cells that expressed ALDH and CD34. Therefore, the major focus for the current study was to determine whether the expression of ALDH and CD34 could be used to delineate functionally distinct progenitor cell subsets. To address this question, a variety of assays was used to explore the developmental potentials of 3 distinct and nonoverlapping cell populations: ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells.
In total our analyses suggested that substantial functional distinctions did exist between ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells. ALDHbr CD34+ cells had a high frequency of short- and long-term SRCs. In addition, this cell fraction was highly enriched for cells that initiated primary and secondary LTCs. In contrast, by these same assays, the ALDHneg CD34+ cell subset contained few primitive progenitors. Furthermore, the ALDHbr CD34neg cells did not repopulate NOD/SCID mice or initiate LTC. Thus, the ALDHbr CD34+ cells contained multilineage lymphomyeloid progenitors with an extensive capacity for proliferation.
ALDHbr CD34+ and ALDHneg CD34+ cells contained progenitor cells with more limited capacities that could be detected in short-term in vitro assays. ALDHbr CD34+ cells had more than a 2-fold higher content of clonogenic myeloid progenitors when compared with ALDHneg CD34+ cells. Furthermore, ALDHbr CD34+ cells expanded efficiently into myeloid progeny under conditions designed to primarily support NK cell growth. Under the same in vitro culture conditions, ALDHneg CD34+ cells consistently gave rise to more lymphoid progeny than ALDHbr CD34+ cells. Taken together, these studies suggested that, within the CD34+ cell compartment, ALDH is not maintained among short-lived NK progenitors. Finally, as in the studies that characterized primitive hematopoietic cells, the ALDHbr CD34neg cell fraction contained no consistent short-term myeloid or NK progenitor activity. Thus, though it is possible that this cell fraction may contain an NK progenitor, this phenomenon was too inconsistent to draw firm conclusions regarding the developmental potential of ALDHbr CD34neg cells.
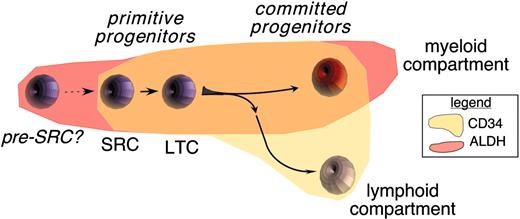
Collectively these observations suggest that the expression of ALDH and CD34 can be used to distinguish developmental stages of human hematopoiesis. As suggested by the model depicted in Figure 5, the most primitive hematopoietic cells expressed ALDH and CD34. CD34 expression and ALDH expression are maintained through early myeloid differentiation. However, during lymphoid differentiation, ALDH expression apparently diminishes before the loss of CD34 expression.
An important caveat must be considered with this model. It is possible that the most primitive hematopoietic cells failed to grow in the assay systems that were used. In our studies, the ALDHbr CD34neg UCB cells contained no SRC, LTC, or clonogenic myeloid progenitors and exhibited only minimal, sporadic growth in short-term NK cultures. However, murine ALDHbr CD34neg HSCs repopulate mice in long-term hematopoietic transplants with as few as 10 cells but do not contain clonogenic myeloid progenitors or more primitive multilineage CFUs-S.22 In addition, human CD34neg cells do not engraft NOD/SCID mice efficiently without direct injection to the bone marrow.14,17,32 Preliminary studies by Hess et al33 suggest that the ALDHbr CD34neg cells might represent a primitive progenitor. A related question is how these various progenitors might contribute to actual human engraftment and immune reconstitution after clinical transplantation. Our studies with long-term growth and NOD/SCID reconstitution suggest that the cells most likely to contribute to bone marrow reconstitution would coexpress CD34 and ALDH. The ALDHneg CD34+ cells had only limited capacity for long-term NOD/SCID reconstitution, and the lymphoid potential of these cells was elucidated only in short-term in vitro cultures. However, it is possible that some ALDHneg CD34+ cells may contribute to short-term immune reconstitution after transplantation. Future studies will more thoroughly compare the relative contributions of ALDHbr CD34+ and ALDHneg CD34+ cells toward short-term reconstitution in vivo. Last, our studies to date have only been performed using conditions that favor NK cell development. Future studies will also more thoroughly explore the relative capacities of ALDHbr CD34+, ALDHneg CD34+, and ALDHbr CD34neg cells to differentiate toward the B- and T-cell lineages in vitro.
Expression of ALDH and CD34 during early hematopoietic development. Our studies suggested that ALDHbr CD34+ cells contained high frequencies of primitive multilineage progenitors, as monitored in NOD/SCID repopulation assays and in LTC. CD34+ cells contained short-term progenitors for myeloid and lymphoid compartments; however, the expression of ALDH was maintained most strongly by myeloid progenitors. In contrast, ALDHneg CD34+ cells enriched short-term lymphoid progenitors, suggesting that ALDH may have a diminishing role during early lymphopoiesis. Finally, a population of ALDHbr CD34neg cells was not efficiently supported within any of the developmental assays used. Based on recent reports,33 this cell may represent a progenitor for the CD34+ compartment.
Expression of ALDH and CD34 during early hematopoietic development. Our studies suggested that ALDHbr CD34+ cells contained high frequencies of primitive multilineage progenitors, as monitored in NOD/SCID repopulation assays and in LTC. CD34+ cells contained short-term progenitors for myeloid and lymphoid compartments; however, the expression of ALDH was maintained most strongly by myeloid progenitors. In contrast, ALDHneg CD34+ cells enriched short-term lymphoid progenitors, suggesting that ALDH may have a diminishing role during early lymphopoiesis. Finally, a population of ALDHbr CD34neg cells was not efficiently supported within any of the developmental assays used. Based on recent reports,33 this cell may represent a progenitor for the CD34+ compartment.
Prepublished online as Blood First Edition Paper, March 24, 2005; DOI 10.1182/blood-2004-09-3652.
Supported by National Institutes of Health grant 1 PO1 HL67314.
Several of the authors (O.M.C., N.J.C., C.A.S.) serve on the Scientific Advisory Board for StemCo BioMedical, Inc. (Durham, NC), whose product (Aldefluor) was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mrs Adelyn Luther for her continuing generosity in support of this work. We also thank Lynn Martinek and Dr Michael Cook of the Duke University Cancer Center Flow Cytometry Core for their valuable expertise in cell sorting and Drs John Chute and Andrew Balber for their critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal