Abstract
Most lymphoblastic lymphomas (LBLs) are regarded as neoplasms of immature T cells because they express cytoplasmic CD3 and frequently carry T-cell receptor (TCR) gene rearrangements. Immature natural killer (NK) and T cells, however, have a common bipotent T/NK-cell precursor in the thymus, and NK cells also express cytoplasmic CD3. Thus, some LBLs could arise from immature NK cells. Mature NK cells express 2 CD94 transcripts: 1A, induced by interleukin 15 (IL-15), and 1B constitutively. Because immature NK cells require IL-15 for development, CD94 1A transcripts could be a marker of NK-LBL. To test this hypothesis, we used laser capture microdissection to isolate IL-15 receptor α+ lymphoid cells from the thymus and showed that these cells contained CD94 1A transcripts. We then assessed for CD94 transcripts in 21 cases of LBL that were cytoplasmic CD3+, nuclear terminal deoxynucleotidyl transferase positive (TdT+), and CD56-, consistent with either the T-cell or NK-cell lineage. We found that 7 LBLs expressed CD94 1A transcripts without TCR gene rearrangements, suggesting NK-cell lineage. Patients with NK-LBL were younger than patients with T-LBL (15 years versus 33 years; P = .11) and had a better 2-year survival (100% versus 27%; P < .01). These results improve the current classification of LBL and contribute to our understanding of NK-cell differentiation.
Introduction
Precursor lymphoblastic lymphoma/leukemia (LBL) frequently presents as a mediastinal mass in young patients. Immunophenotypically, most neoplasms are positive for cytoplasmic CD3 and nuclear terminal deoxynucleotidyl transferase (TdT) and carry T-cell receptor (TCR) rearrangements.1 Thus, most LBL cases are thought to be of T lineage and represent lymphomas of immature T cells from the thymus. However, it is difficult to distinguish immature T cells from immature natural killer (NK) cells because both are derived from bipotent T/NK precursors in the thymus and share similar immunophenotypes.2-5 Cytoplasmic CD3 can be found in both immature T and NK cells.6 Similarly, nuclear TdT is expressed by lymphoid precursors of various lineages and is not specific for T cells.7 In addition, TCR gene rearrangement, despite being a good marker for T cells, can be found in cases of acute myeloid leukemia and precursor B-cell lymphoblastic leukemia because of the phenomenon of lineage promiscuity.8 These traditional criteria, therefore, do not exclude the possibility that lymphoblastic lymphoma/leukemia of the NK lineage exists.9 Although CD56 (neural-cell adhesion molecule; NCAM) has been recognized as a sensitive marker for NK cells, particularly mature NK cells,10 CD56 is not specific and can be expressed by neoplastic myeloid, lymphoid, plasmacytoid dendritic,11-13 and plasma cells. Other than CD56, CD161 and CD44 could be used as markers for NK14-17 and T cells,18,19 respectively, but their sensitivity and specificity in distinguishing NK-LBLs from T-LBLs have yet to be tested. The lack of suitable markers for immature NK cells thereby makes it difficult to identify NK-cell LBL.
For the NK-cell lineage to diverge from the T-cell lineage, interleukin 15 (IL-15) and transcription factor ID2 are essential. By contrast, IL-2 and transcription factor E2A are important for T-cell development.20-26 ID2 and E2A are highly homologous, and both belong to the helix-loop-helix family of transcription factors. ID2 is a dominant-negative inhibitor of E2A and promotes NK-cell development at the expense of T-cell development.27,28 Likewise, IL-2 and IL-15 are highly homologous, and their receptors are heterotrimers that share the same β and γ chains but differ in the α chains.29 The IL-2 receptor α (IL-2Rα) is also known as CD25, whereas the IL-15 receptor α (IL-15Rα) does not yet have a cluster designation (CD). Accordingly, NK-cell precursors most likely express IL-15Rα and ID2, and T-cell precursors are positive for CD25 and E2A.
T-cell precursors undergo TCR gene rearrangement and become immature T cells that are committed to the T-cell lineage.30 Expansion and survival of immature T cells depend on another transcription factor, TCF-1 (T-cell factor 1),31,32 which is also present in a subset of mature NK cells.33 For NK-cell precursors, their subsequent development and lineage commitment to immature NK cells is unclear. It is known only that mature human NK cells use both the C-type lectinlike CD94 and killer immunoglobulin-like receptors (KIRs) as antigen receptors.34-36 Recently, it was found that CD94 has both a distal 1A promoter and a proximal 1B promoter.37,38 These promoters generate transcripts with identical coding regions. In mature NK cells, the 1B promoter is constitutively active and produces the predominant transcript, whereas the 1A promoter is activated only by IL-15. The finding of an IL-15–regulated CD94 1A promoter raises the possibility that CD94 1A might be the predominant form expressed in immature NK cells.
In this study, we used laser capture microdissection to isolate IL-15Rα+ lymphoid cells from the thymus, and we confirmed that these immature NK cells are indeed CD94 1A+. We also studied a group of LBL cases that were positive for cytoplasmic CD3 and nuclear TdT and distinguished 2 immunophenotypic groups. One group that we propose is of immature NK-cell origin is characterized by a CD94 1A+/CD56-/ID2- immunophenotype and lacks TCR gene rearrangements. In contrast, the second group had a CD94 1A- immunophenotype and carries TCR gene rearrangements supporting T-cell lineage. The identification of a subset of LBL cases of immature NK-cell origin has both clinical and immunologic implications.
Materials and methods
Tissue samples
The study was approved by the institutional review board of National Taiwan University Hospital and informed consent was obtained according to the Declaration of Helsinki. Twenty-one LBL samples were collected. Diagnoses were made by histologic and immunophenotypic examination of lymph nodes or mediastinal biopsy specimens in the Pathology Department of the National Taiwan University Hospital between 1993 and 2001. All diagnoses were established on pretreatment biopsy specimens at the time the patients presented initially. Histologically, all specimens showed a characteristic diffuse infiltration of lymphoblasts positive for cytoplasmic CD3 and nuclear TdT and were negative for CD56 and CD20. All neoplasms were negative for Epstein-Barr virus (EBV)–encoded small RNA (EBER); thus, they were not associated with EBV infection. Clinical data were obtained from the medical records.
For comparison, other specimens were examined, including 10 cases of extranodal nasal-type NK/T-cell lymphoma as defined according to the criteria specified in the World Health Organization (WHO) classification; 6 samples of the thymus were obtained from thymectomy specimens of 2 adults who had myasthenia gravis and from autopsy specimens of 4 infants who died of prematurity; 3 spleens obtained were removed for cirrhosis and gastric cancer; 2 samples of endometrium were obtained from curettage specimens for dysfunctional uterine bleeding; 1 tonsil was excised for chronic tonsillitis; and 4 peripheral-blood specimens were obtained from volunteer healthy donors.
Also studied were 4 samples of umbilical cord blood obtained from healthy newborns according to a protocol approved by the institutional review committee. Umbilical cord mononuclear cells were isolated by density gradient centrifugation with use of Ficoll-Paque at a density of 1.077. CD34+ progenitor cells were then purified by an immunomagnetic cell separation system using multisort microbeads conjugated to a monoclonal mouse anti–human CD34 antibody (QBENDD/10; Miltenyi Biotech, Auburn, CA). The beads were dissociated from the purified CD34+ progenitors after enzymatic release. The CD34+ progenitor cells were further incubated with a mixture of biotin-conjugated antibodies against lineage (Lin) markers, including CD2 (clone LT2), CD3 (clone UCHT-1), CD14 (clone UCHM-1), CD16 (clone LNK16), CD19 (clone LT19), CD33 (clone WM53), CD41 (clone PM6/248), CD56 (clone MEM188), and glycophorin (clone AC107.3). All antibodies were obtained from Serotec (Raleigh, NC), except for CD3 from BD Biosciences (San Jose, CA) and glycophorin from Miltenyi Biotech. The CD34+Lin+ progenitors and CD34+Lin- progenitors were separated by streptavidin-conjugated microbeads. Flow cytometry was used to check the purity of the cells. For flow cytometry, the CD34+Lin- progenitors or CD34+Lin+ progenitors were labeled with a phycoerythrin-cyanin 5 (PC5)–conjugated CD34 antibody (clone 581; Coulter Immunotech, Marseille, France).
CD94 transcripts by RT-PCR
The sequences of the primers used for reverse transcription–polymerase chain reaction (RT-PCR) analyses are listed in Table 1. The table also provides the GenBank accession number of the CD94 cDNA, the positions of the primers, and the sizes of the PCR products. The “RS” in Table 1 is a random sequence, 5′-TGACAAACTGTGTTCACTAGC-3′, used for increased PCR specificity and incorporation of fluorescent labels.
Primers for CD94 and CD161*
GenBank . | Forward . | 5′-3′ . | Exon . | Reverse . | 5′-3′ . | Exon . | Size, bp . |
|---|---|---|---|---|---|---|---|
| CD94 1A | 5′-AGC-TCA-TCA-AGT-TCA-AGA-GAC-3′ | 40-60 | 1A | 5′-RS TTT-TCA-GAA-AGA-ACA-GCG-TTG-3′ | 150-130 | 1 | 136 |
| AY227806 | |||||||
| CD94 1B | 5′-ATT-TCT-TCA-TAC-TCA-ACT-TTC-3′ | 138-158 | 1B | 5′-RS ACC-TCC-ACA-GAG-TGG-TCT-TAA-3′ | 291-271 | 2 | 179 |
| NM002262 | |||||||
| CD161 | 5′-CTT-AGC-TGT-GCT-GGG-ATT-ATT-CTC-3′ | 187-210 | 2 | 5′-RS-CTG-CAT-TTT-TCT-ATT-GAT-GAT-TT-3′ | 284-262 | 3 | 123 |
| NM002258 | |||||||
| β2M | 5′-CTT-TGT-CAC-AGC-CCA-AGA-TAG-3′ | 377-397 | 2 | 5′-RS-CAG-AAT-TTG-GAA-TTC-ATC-CAA-3′ | 477-457 | 4 | 126 |
| NM004048 |
GenBank . | Forward . | 5′-3′ . | Exon . | Reverse . | 5′-3′ . | Exon . | Size, bp . |
|---|---|---|---|---|---|---|---|
| CD94 1A | 5′-AGC-TCA-TCA-AGT-TCA-AGA-GAC-3′ | 40-60 | 1A | 5′-RS TTT-TCA-GAA-AGA-ACA-GCG-TTG-3′ | 150-130 | 1 | 136 |
| AY227806 | |||||||
| CD94 1B | 5′-ATT-TCT-TCA-TAC-TCA-ACT-TTC-3′ | 138-158 | 1B | 5′-RS ACC-TCC-ACA-GAG-TGG-TCT-TAA-3′ | 291-271 | 2 | 179 |
| NM002262 | |||||||
| CD161 | 5′-CTT-AGC-TGT-GCT-GGG-ATT-ATT-CTC-3′ | 187-210 | 2 | 5′-RS-CTG-CAT-TTT-TCT-ATT-GAT-GAT-TT-3′ | 284-262 | 3 | 123 |
| NM002258 | |||||||
| β2M | 5′-CTT-TGT-CAC-AGC-CCA-AGA-TAG-3′ | 377-397 | 2 | 5′-RS-CAG-AAT-TTG-GAA-TTC-ATC-CAA-3′ | 477-457 | 4 | 126 |
| NM004048 |
RS indicates 5′-TGA-CAA-ACT-GTG-TTC-ACT-AGC-3′; β2M, β2-microglobulin.
Size of the RT-PCR product in base pairs (bp)
Reverse transcription was performed in a 20-μL mixture at 40°C for 50 minutes. The mixture included 0.1 μg RNA purified from formalin-fixed, paraffin-embedded tissue blocks, 0.5 μM antisense primer, 200 U reverse transcriptase, 50 mM Tris (tris(hydroxymethyl)aminomethane)–HCl at pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT (dithiothreitol), and 200 μMof each dNTP (deoxyribonucleoside triphosphate). The cDNA from 2 μL reverse transcription mixture was amplified with the forward primer and a fluorescence-labeled primer of the random sequence 5′-tet-CTTCTGACAAACTGTGTTCACTAGC-3′ in a standard PCR reaction.
PCR was done in a 20-μL mixture, including the DNA template, 0.3 μM of each primer, 15 mM Tris-HCl at pH 8.0, 1.5 mM MgCl2, 50 mM KCl, 200 μM of each dNTP, and 1 U Taq polymerase. The reaction mixture was subjected to 35 cycles of PCR. Each cycle consisted of denaturation at 94°C for 45 seconds, annealing at 45°C for 45 seconds, and extension at 72°C for 45 seconds.
At the end of 35 cycles, a portion of the PCR products was separated by a high-resolution electrophoretic system (ABI377 with GeneScan software (Perkin-Elmer, Foster City, CA). The sizes of the RT-PCR products of CD94 are listed in Table 1. A reaction for β2M was run as a positive control, and a reaction under the same conditions except for the omission of reverse transcriptase was run as a negative control. The RT-PCR is deemed positive if its peak is higher than 10% of β2M.
Killer immunoglobulin-like receptor repertoire analysis
This analysis was done according to a protocol developed in our laboratory.39 Briefly, KIRs can be categorized in 1 of 3 splicing groups, KIR2DL4, KIR2D, or KIR3D. Three pairs of group-specific primers were designed and used in RT-PCR reactions to evaluate the presence or absence of the KIR transcripts. The primers were 5′-GCTGAGAGAGAAGGTTCTCATAT-3′ and 5′-CACTCCCCCACTGGGTGGTCGGC-3′ for KIR 2DL4, 5′-TGGGTGGGCCAGGAGGAAGGTTT-3′ and 5′-CATGGCGTGTGTTGGGTTCTTCTTG-3′ for KIR2D, and 5′-TGGGTGGGCCAGGAGGAAGGTTT-3′ and 5′-CACTCCCCCCACTGGGTGGTCGGC-3′ for KIR3D.
T-cell–receptor γ rearrangement
Standard PCR was performed to assess for TCR γ chain gene rearrangements according to a protocol developed in our laboratory.40 A monoclonal TCR rearrangement is considered to be present if a dominant clone, making up greater than 30% of the total clones, is found.
CD161 transcripts by RT-PCR
RT-PCR for CD161 was performed using the primers listed in Table 1. The reaction conditions were identical to those for CD94. The RT-PCR result is considered to be positive if the amplified peak is greater than 20% of β2M.
Immunohistochemistry
Immunohistochemical stains for CD44, E2A, ID2, and TCF-1 were performed on formalin-fixed, paraffin-embedded tissue sections of all 21 LBL cases. The antibodies used were CD44 (clone 2C5; R&D Systems, Minneapolis, MN), E2A (polyclonal; Santa Cruz Biotech, Santa Cruz, CA), ID2 (polyclonal; Santa Cruz Biotech), and TCF-1 (7H3; Exalpha, Watertown, MA). The staining was done using conditions recommended by the vendor.
Lineage assignment of LBLs according to CD94 1A transcripts and TCR gene rearrangement
The 21 LBLs were divided into 3 groups, according to the patterns of CD94 transcripts and TCR gene rearrangement. The T-cell lineage was characterized by the presence of monoclonal TCR gene rearrangement. The NK-cell lineage was characterized by the presence of CD94 1A transcripts without monoclonal TCR gene rearrangement. The uncommitted cases had neither a monoclonal TCR gene rearrangement nor CD94 1A transcripts.
Comparison between LBLs of T-cell and NK-cell lineages
The uncommitted group was excluded from this comparison because of the small sample size (n = 3). Two-sample comparisons between the LBLs of T-cell and NK-cell lineages were performed with the binomial test for categorical data and the nonparametric Mann-Whitney U test for continuous data.
Correlation with 2-year survival
The 21 LBLs were divided into 2 groups according to the 2-year survival. Two-sample comparisons were done with the binomial test for categorical data and with the nonparametric Mann-Whitney U test for continuous data.
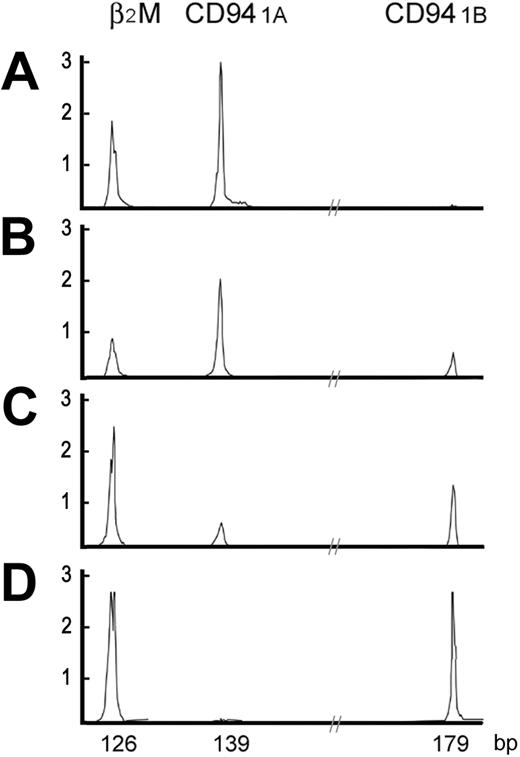
High-resolution electrophoretic tracings of mature and immature CD94 patterns. In A-D, 2 distinct patterns of CD94 transcripts are shown. The immature CD94 1A pattern, either CD94 1A alone or CD94 1A >> CD94 1B, was found in the thymus or LBLs. The mature CD94 1B pattern, either CD94 1B alone or CD94 1A << 1B, was found in peripheral tissues or extranodal nasal-type NK/T-cell lymphomas. X-axis is in base pairs; Y-axis, intensity of the fluorescence.
High-resolution electrophoretic tracings of mature and immature CD94 patterns. In A-D, 2 distinct patterns of CD94 transcripts are shown. The immature CD94 1A pattern, either CD94 1A alone or CD94 1A >> CD94 1B, was found in the thymus or LBLs. The mature CD94 1B pattern, either CD94 1B alone or CD94 1A << 1B, was found in peripheral tissues or extranodal nasal-type NK/T-cell lymphomas. X-axis is in base pairs; Y-axis, intensity of the fluorescence.
Isolation of IL-15Rα+ immature NK cells from thymus by reverse laser capture microdissection
IL-15Rα+ lymphoid cells, stained by a polyclonal goat antibody (R&D Systems), were isolated from 5-μm frozen sections of the thymus of infants who died of prematurity. In conventional laser capture microdissection (LCM), the laser hits the target cells directly, thereby causing disruption of the cells, loss of their histopathology, and damage to the nucleic acids. To circumvent these difficulties, we used a reverse LCM (rLCM) that removed IL-15Rα- cells from the slides. This approach preserved the original histopathology of the intact IL-15Rα+ cells and allowed better recovery of RNA.
Results
Immature NK cells from thymus are predominantly CD94 1A+, but mature NK cells from peripheral tissues or extranodal nasal-type NK/T-cell lymphomas are predominantly CD94 1B+
We evaluated the distribution patterns of CD94 transcripts in benign and neoplastic tissues known to have either mature or immature NK cells, such as umbilical cord blood, peripheral blood, tonsil, spleen, endometrium, thymus, and extranodal nasal-type NK/T-cell lymphomas (Table 2).
Tissue distribution of CD94 transcripts
. | Age, y . | Sex . | Tissue . | Diagnosis . | CD94 . |
|---|---|---|---|---|---|
| CD34 progenitors and thymus | |||||
| 1 | 0* | M | CD34+Lin+UC progenitors | Healthy newborn | Neg |
| 2 | 0* | M | CD34+Lin-UC progenitors | Same case above | Neg |
| 3 | 0* | F | CD34+Lin+UC progenitors | Healthy newborn | Neg |
| 4 | 0* | F | CD34+Lin-UC progenitors | Same case above | Neg |
| 5 | 1* | F | Thymus | Premature infant (GA, 29 wk) | 1A |
| 6 | 1* | F | Thymus | Premature infant (GA, 37 wk) | 1A |
| 7 | 1* | M | Thymus | Premature infant (GA, 27 wk) | 1A |
| 8 | 1* | M | Thymus | Premature infant (GA, 27 wk) | 1A |
| 9 | 40 | F | Thymus | MG with LH | 1A |
| 10 | 17 | F | Thymus | MG with LH | 1A>>1B |
| Peripheral tissues | |||||
| 1 | 44 | M | Spleen | Cirrhosis with hypersplenism and pancytopenia | 1A |
| 2 | 32 | F | Endometrium | Reactive endometrium | 1B |
| 3 | 75 | M | Spleen | Gastric cancer without splenic involvement | 1B |
| 4 | 45 | F | Endometrium | Reactive endometrium | 1B>>1A |
| 5 | 74 | M | Spleen | Cirrhosis with splenomegaly | 1B>>1A |
| 6 | 65 | M | Tonsil | Chronic tonsillitis | 1B>>1A |
| 7 | 43 | M | PB | Healthy volunteer | 1B>>1A |
| 8 | 24 | M | PB | Healthy volunteer | 1B>>1A |
| 9 | 28 | F | PB | Healthy volunteer | 1B>>1A |
| 10 | 27 | F | PB | Healthy volunteer | 1B>>1A |
| ENNLs | |||||
| 1 | 60 | M | Nasal cavity | ENNL | Neg |
| 2 | 65 | M | Nasal cavity | ENNL | Neg |
| 3 | 45 | F | Nasal cavity | ENNL | Neg |
| 4 | 61 | M | Nasal cavity | ENNL | 1A |
| 5 | 42 | M | Nasal cavity | ENNL | 1B>>1A |
| 6 | 62 | M | Nasal cavity | ENNL | 1B>>1A |
| 7 | 55 | M | Nasal cavity | ENNL | 1B>>1A |
| 8 | 39 | F | Nasal cavity | ENNL | 1B>>1A |
| 9 | 69 | M | Nasal cavity | ENNL | 1B>>1A |
| 10 | 38 | M | Nasal cavity | ENNL | 1B>>1A |
. | Age, y . | Sex . | Tissue . | Diagnosis . | CD94 . |
|---|---|---|---|---|---|
| CD34 progenitors and thymus | |||||
| 1 | 0* | M | CD34+Lin+UC progenitors | Healthy newborn | Neg |
| 2 | 0* | M | CD34+Lin-UC progenitors | Same case above | Neg |
| 3 | 0* | F | CD34+Lin+UC progenitors | Healthy newborn | Neg |
| 4 | 0* | F | CD34+Lin-UC progenitors | Same case above | Neg |
| 5 | 1* | F | Thymus | Premature infant (GA, 29 wk) | 1A |
| 6 | 1* | F | Thymus | Premature infant (GA, 37 wk) | 1A |
| 7 | 1* | M | Thymus | Premature infant (GA, 27 wk) | 1A |
| 8 | 1* | M | Thymus | Premature infant (GA, 27 wk) | 1A |
| 9 | 40 | F | Thymus | MG with LH | 1A |
| 10 | 17 | F | Thymus | MG with LH | 1A>>1B |
| Peripheral tissues | |||||
| 1 | 44 | M | Spleen | Cirrhosis with hypersplenism and pancytopenia | 1A |
| 2 | 32 | F | Endometrium | Reactive endometrium | 1B |
| 3 | 75 | M | Spleen | Gastric cancer without splenic involvement | 1B |
| 4 | 45 | F | Endometrium | Reactive endometrium | 1B>>1A |
| 5 | 74 | M | Spleen | Cirrhosis with splenomegaly | 1B>>1A |
| 6 | 65 | M | Tonsil | Chronic tonsillitis | 1B>>1A |
| 7 | 43 | M | PB | Healthy volunteer | 1B>>1A |
| 8 | 24 | M | PB | Healthy volunteer | 1B>>1A |
| 9 | 28 | F | PB | Healthy volunteer | 1B>>1A |
| 10 | 27 | F | PB | Healthy volunteer | 1B>>1A |
| ENNLs | |||||
| 1 | 60 | M | Nasal cavity | ENNL | Neg |
| 2 | 65 | M | Nasal cavity | ENNL | Neg |
| 3 | 45 | F | Nasal cavity | ENNL | Neg |
| 4 | 61 | M | Nasal cavity | ENNL | 1A |
| 5 | 42 | M | Nasal cavity | ENNL | 1B>>1A |
| 6 | 62 | M | Nasal cavity | ENNL | 1B>>1A |
| 7 | 55 | M | Nasal cavity | ENNL | 1B>>1A |
| 8 | 39 | F | Nasal cavity | ENNL | 1B>>1A |
| 9 | 69 | M | Nasal cavity | ENNL | 1B>>1A |
| 10 | 38 | M | Nasal cavity | ENNL | 1B>>1A |
UC indicates umbilical cord; Neg, negative; GA, gestational age; MG with LH, myasthenia gravis with lymphoid hyperplasia; PB, peripheral blood; ENNL, extranodal nasal-type NK/T-cell lymphoma.
Age in days
Umbilical cord progenitor cells, either CD34+Lin+ or CD34+Lin-, had neither CD94 1A nor CD94 1B transcripts. In 10 samples from peripheral blood, tonsil, spleen, and endometrium, had predominantly CD94 1B transcripts, including 2 samples with only CD94 1B transcripts, and 7 samples in which CD94 1B transcripts were much more numerous than CD94 1A. Only 1 of these samples, a spleen from a patient with cirrhosis with hypersplenism and pancytopenia, showed only CD94 1A transcripts. In 10 extranodal nasal-type NK/T-cell lymphomas assessed, CD94 transcripts were identified in 7 neoplasms, and in 6 tumors CD94 1B transcripts were more numerous than CD94 1A transcripts. By contrast, all 6 samples of thymus had predominantly CD94 1A transcripts, including 5 with only CD94 1A transcripts and 1 in which CD94 1A transcripts were more numerous than CD94 1B transcripts. Representative patterns of CD94 transcripts were shown in Figure 1.
These data suggest that immature NK cells use predominantly CD94 1A transcripts, whereas mature NK cells use predominantly CD94 1B transcripts. This pattern also indicates that CD94 1A transcripts can be used to identify LBLs of the NK lineage.
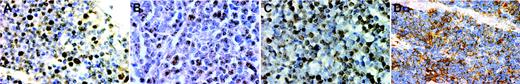
Immunohistochemistry for E2A and ID2 from LBLs and thymus of premature infants. Note the nuclear E2A stain in both the thymus (A) and a T-LBL (B). For ID2, the staining is nuclear in the normal thymus (C) but cytoplasmic in a case of T-LBL (D). (A-D, × 400, immunohistochemistry using formalin-fixed, paraffin-embedded tissue sections and hematoxylin counterstain). Images were visualized under an Olympus BX60 microscope equipped with a Plan-Apo 40×/0.95 objective lens, and were captured with a DP70 digital camera (Olympus, Tokyo, Japan).
Immunohistochemistry for E2A and ID2 from LBLs and thymus of premature infants. Note the nuclear E2A stain in both the thymus (A) and a T-LBL (B). For ID2, the staining is nuclear in the normal thymus (C) but cytoplasmic in a case of T-LBL (D). (A-D, × 400, immunohistochemistry using formalin-fixed, paraffin-embedded tissue sections and hematoxylin counterstain). Images were visualized under an Olympus BX60 microscope equipped with a Plan-Apo 40×/0.95 objective lens, and were captured with a DP70 digital camera (Olympus, Tokyo, Japan).
Lineage assignment of LBLs according to antigen-receptor patterns: transcripts of CD94 1A, but not KIRs, identify an NK-LBL
We used the presence of TCR gene rearrangements or expression of CD94 1A transcripts as criteria for T-cell or NK-cell lineage commitment, respectively (Table 3). According to this scheme, 11 LBLs carried monoclonal TCR gene rearrangements and were classified as T-LBL, and 7 LBLs had CD94 1A transcripts and were classified as NK-LBL. The remaining 3 cases had neither TCR gene rearrangement nor CD94 1A transcripts and were considered to be of uncommitted lineage. The T-LBLs were negative for CD94 1A and frequently expressed cytoplasmic ID2, whereas the NK-LBLs were positive for CD94 1A and negative for ID2.
Pathologic and clinical data for lymphoblastic lymphomas
. | Sex . | Age, y . | CD161 . | CD44 . | E2A . | ID2 . | TCF . | TCR . | 1A . | 1B . | KIR 2DL4 . | KIR 2D . | KIR 3D . | LDH . | Blast, % . | WBC . | HB . | PLT . | Med mass . | Stage . | ALL-like therapy* . | Survival, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uncommitted | ||||||||||||||||||||||
| 1 | F | 52 | – | + | – | C+ | – | – | – | – | – | – | – | 1298 | 70 | 22.1 | 10.4 | 32 | – | IV | No | 30 |
| 2 | F | 53 | – | – | + | – | – | – | – | – | – | – | – | NA | 0 | 7.2 | 15 | 194 | + | II | No | 120† |
| 3 | F | 29 | + | – | + | C+ | – | – | – | – | – | – | – | 419 | 30 | 10.7 | 12.1 | 434 | – | IV | No | 14 |
| T lineage | ||||||||||||||||||||||
| 4 | F | 7 | – | + | ND | – | – | + | – | – | – | – | – | 494 | 0 | 7.1 | 12.4 | 255 | + | III | Yes | 12 |
| 5 | M | 23 | + | – | + | C+ | – | + | – | – | – | – | – | 586 | 0 | 1.6 | 10.9 | 103 | – | IV | No | 21 |
| 6 | F | 69 | – | – | – | C+ | – | + | – | – | – | – | – | 373 | 0 | 7.6 | 3.5 | 52 | – | III | No | 1 |
| 7 | M | 33 | – | – | + | C+ | + | + | – | – | – | – | – | 2530 | 60 | 19.9 | 8.6 | 276 | – | IV | No | 18 |
| 8 | M | 43 | – | – | – | – | ND | + | – | – | – | – | – | 1970 | 57 | 4.7 | 3.5 | 44 | + | IV | No | 7 |
| 9 | M | 18 | – | – | + | C+ | + | + | – | – | – | – | – | 2340 | 0 | 5.6 | 11 | 190 | + | III | No | 1 |
| 10 | M | 29 | – | – | – | – | + | + | – | – | – | – | + | 577 | 0 | 8.7 | 13.9 | 280 | + | III | No | 14 |
| 11 | M | 28 | + | + | + | – | + | + | – | – | – | – | + | 289 | 6 | 7.4 | 16.1 | 166 | + | IV | No | 33 |
| 12 | M | 9 | + | + | – | C+ | + | + | – | – | – | – | + | 540 | NA | 2.8 | 8.2 | 125 | – | IV | Yes | 45 |
| 13 | M | 39 | – | + | ND | ND | ND | + | – | – | – | + | + | 908 | 0 | 5.6 | 14.1 | 313 | – | IV | No | 7 |
| 14 | M | 15 | + | – | + | C+ | – | + | – | – | – | – | + | 210 | 0 | 7.6 | 14.3 | 334 | – | III | Yes | 102† |
| NK lineage | ||||||||||||||||||||||
| 15 | M | 3 | – | – | + | – | + | – | + | + | – | – | + | 1900 | 0 | 7.5 | 3.6 | 10 | + | III | Yes | 34† |
| 16 | F | 19 | + | + | – | – | + | – | + | – | + | – | + | 247 | 12 | 9.1 | 13.3 | 129 | – | IV | No | 78† |
| 17 | F | 12 | + | – | – | – | + | – | + | – | – | – | + | NA | 0 | 7.2 | 13.5 | 271 | + | III | Yes | 105† |
| 18 | M | 27 | + | – | – | – | ND | – | + | – | – | + | – | 1407 | 0 | 6.1 | 13.3 | 232 | + | II | No | 108† |
| 19 | M | 12 | + | + | – | – | – | – | + | – | – | – | – | 372 | 0 | 5.2 | 13 | 238 | – | IV | Yes | 108† |
| 20 | M | 15 | + | + | – | – | + | – | + | – | – | – | – | 2353 | 23 | 15.3 | 12.2 | 92 | – | IV | Yes | 50† |
| 21 | F | 28 | + | + | + | – | + | – | + | + | – | – | – | 318 | 3 | 6.6 | 10.6 | 266 | – | IV | No | 45† |
. | Sex . | Age, y . | CD161 . | CD44 . | E2A . | ID2 . | TCF . | TCR . | 1A . | 1B . | KIR 2DL4 . | KIR 2D . | KIR 3D . | LDH . | Blast, % . | WBC . | HB . | PLT . | Med mass . | Stage . | ALL-like therapy* . | Survival, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uncommitted | ||||||||||||||||||||||
| 1 | F | 52 | – | + | – | C+ | – | – | – | – | – | – | – | 1298 | 70 | 22.1 | 10.4 | 32 | – | IV | No | 30 |
| 2 | F | 53 | – | – | + | – | – | – | – | – | – | – | – | NA | 0 | 7.2 | 15 | 194 | + | II | No | 120† |
| 3 | F | 29 | + | – | + | C+ | – | – | – | – | – | – | – | 419 | 30 | 10.7 | 12.1 | 434 | – | IV | No | 14 |
| T lineage | ||||||||||||||||||||||
| 4 | F | 7 | – | + | ND | – | – | + | – | – | – | – | – | 494 | 0 | 7.1 | 12.4 | 255 | + | III | Yes | 12 |
| 5 | M | 23 | + | – | + | C+ | – | + | – | – | – | – | – | 586 | 0 | 1.6 | 10.9 | 103 | – | IV | No | 21 |
| 6 | F | 69 | – | – | – | C+ | – | + | – | – | – | – | – | 373 | 0 | 7.6 | 3.5 | 52 | – | III | No | 1 |
| 7 | M | 33 | – | – | + | C+ | + | + | – | – | – | – | – | 2530 | 60 | 19.9 | 8.6 | 276 | – | IV | No | 18 |
| 8 | M | 43 | – | – | – | – | ND | + | – | – | – | – | – | 1970 | 57 | 4.7 | 3.5 | 44 | + | IV | No | 7 |
| 9 | M | 18 | – | – | + | C+ | + | + | – | – | – | – | – | 2340 | 0 | 5.6 | 11 | 190 | + | III | No | 1 |
| 10 | M | 29 | – | – | – | – | + | + | – | – | – | – | + | 577 | 0 | 8.7 | 13.9 | 280 | + | III | No | 14 |
| 11 | M | 28 | + | + | + | – | + | + | – | – | – | – | + | 289 | 6 | 7.4 | 16.1 | 166 | + | IV | No | 33 |
| 12 | M | 9 | + | + | – | C+ | + | + | – | – | – | – | + | 540 | NA | 2.8 | 8.2 | 125 | – | IV | Yes | 45 |
| 13 | M | 39 | – | + | ND | ND | ND | + | – | – | – | + | + | 908 | 0 | 5.6 | 14.1 | 313 | – | IV | No | 7 |
| 14 | M | 15 | + | – | + | C+ | – | + | – | – | – | – | + | 210 | 0 | 7.6 | 14.3 | 334 | – | III | Yes | 102† |
| NK lineage | ||||||||||||||||||||||
| 15 | M | 3 | – | – | + | – | + | – | + | + | – | – | + | 1900 | 0 | 7.5 | 3.6 | 10 | + | III | Yes | 34† |
| 16 | F | 19 | + | + | – | – | + | – | + | – | + | – | + | 247 | 12 | 9.1 | 13.3 | 129 | – | IV | No | 78† |
| 17 | F | 12 | + | – | – | – | + | – | + | – | – | – | + | NA | 0 | 7.2 | 13.5 | 271 | + | III | Yes | 105† |
| 18 | M | 27 | + | – | – | – | ND | – | + | – | – | + | – | 1407 | 0 | 6.1 | 13.3 | 232 | + | II | No | 108† |
| 19 | M | 12 | + | + | – | – | – | – | + | – | – | – | – | 372 | 0 | 5.2 | 13 | 238 | – | IV | Yes | 108† |
| 20 | M | 15 | + | + | – | – | + | – | + | – | – | – | – | 2353 | 23 | 15.3 | 12.2 | 92 | – | IV | Yes | 50† |
| 21 | F | 28 | + | + | + | – | + | – | + | + | – | – | – | 318 | 3 | 6.6 | 10.6 | 266 | – | IV | No | 45† |
1A indicates CD94 1A; 1B, CD94 1B; LDH, lactate dehydrogenase, IU/L; Blast, percentage of blasts in peripheral blood; WBC, white blood cell count, 103/μL; HB, hemoglobin level, g/dL; PLT, platelet count, 103/μL; Med mass, mediastinal mass; ALL, acute lymphoblastic leukemia; C, cytoplasmic; NA, not available; ND, not determined
Patients were treated with either ALL-like or CHOP (cyclophosphamide/doxorubicin/vincristine/prednisone)-based regimens
The case was censored
The immunohistochemical patterns of ID2 and E2A are shown in Figure 2. E2A exhibited nuclear staining in both normal thymus and T-LBL (Figure 2A-B). For ID2, there is nuclear staining in the normal thymus but cytoplasmic staining in T-LBL (Figure 2C-D).
As shown in Table 4, except for CD161, which had borderline significance at P = .05, the presence or absence of CD44, E2A, TCF-1, and KIRs was not lineage specific and did not correlate with the lineage assignment based on the presence of CD94 1A transcripts. In particular, the differences in the distributions of KIR2DL4, KIR2D, and KIR3D between the T- and NK-cell lineages, useful in lineage assignment in mature NK-cell lymphomas, were not significant statistically.
Lineage assignment according to CD94 1A and TCR rearrangement
. | Uncommitted . | T . | NK . | P . |
|---|---|---|---|---|
| No. of cases | 3 | 11 | 7 | |
| Median age, y | 52 | 33 | 15 | .11 |
| Sex, male:female | 0:3 | 9:2 | 4:3 | .28 |
| CD161, n/total | 1/3 | 4/11 | 6/7 | .05 |
| CD44, n/total | 1/3 | 4/11 | 4/7 | .39 |
| E2A, n/total | 2/3 | 5/9 | 2/7 | .27 |
| c-ID2, n/total | 2/3 | 6/10 | 0/7 | .02 |
| TCF-1, n/total | 0/3 | 5/9 | 5/6 | .28 |
| KIR 2DL4, n/total | 0/3 | 0/11 | 1/7 | .21 |
| KIR 2D, n/total | 0/3 | 1/11 | 1/7 | .73 |
| KIR 3D, n/total | 0/3 | 5/11 | 3/7 | .91 |
| LDH, IU/L | 859 | 586 | 890 | .49 |
| Blasts in blood, n | 2/3 | 3/10 | 3/7 | .59 |
| WBC count, 103/μL | 10.7 | 7.1 | 7.2 | .49 |
| HB level, g/dL | 12.1 | 11.0 | 13.0 | .89 |
| PLT count, 103/μL | 194 | 190 | 232 | .55 |
| Mediastinal mass, n/total | 1/3 | 5/11 | 3/7 | .91 |
| Stage IV, n/total | 2/3 | 6/11 | 4/7 | .91 |
| ALL-like regimens*, n/total | 0/3 | 3/11 | 4/7 | .22 |
| 2-y survival, n/total | 2/3 | 3/11 | 7/7 | .008 |
. | Uncommitted . | T . | NK . | P . |
|---|---|---|---|---|
| No. of cases | 3 | 11 | 7 | |
| Median age, y | 52 | 33 | 15 | .11 |
| Sex, male:female | 0:3 | 9:2 | 4:3 | .28 |
| CD161, n/total | 1/3 | 4/11 | 6/7 | .05 |
| CD44, n/total | 1/3 | 4/11 | 4/7 | .39 |
| E2A, n/total | 2/3 | 5/9 | 2/7 | .27 |
| c-ID2, n/total | 2/3 | 6/10 | 0/7 | .02 |
| TCF-1, n/total | 0/3 | 5/9 | 5/6 | .28 |
| KIR 2DL4, n/total | 0/3 | 0/11 | 1/7 | .21 |
| KIR 2D, n/total | 0/3 | 1/11 | 1/7 | .73 |
| KIR 3D, n/total | 0/3 | 5/11 | 3/7 | .91 |
| LDH, IU/L | 859 | 586 | 890 | .49 |
| Blasts in blood, n | 2/3 | 3/10 | 3/7 | .59 |
| WBC count, 103/μL | 10.7 | 7.1 | 7.2 | .49 |
| HB level, g/dL | 12.1 | 11.0 | 13.0 | .89 |
| PLT count, 103/μL | 194 | 190 | 232 | .55 |
| Mediastinal mass, n/total | 1/3 | 5/11 | 3/7 | .91 |
| Stage IV, n/total | 2/3 | 6/11 | 4/7 | .91 |
| ALL-like regimens*, n/total | 0/3 | 3/11 | 4/7 | .22 |
| 2-y survival, n/total | 2/3 | 3/11 | 7/7 | .008 |
c-ID2 indicates cytoplasmic ID2.
Patients were treated with either ALL-like or CHOP-based regimens
NK-LBL had a better 2-year survival rate than did T-LBL
Patients with NK-LBLs (CD94 1A positive) had a better prognosis than did those with T-LBLs (2-year survival, 100% versus 27%; P < .01; Table 4). NK-LBL patients also tended to be younger (15 years versus 33 years; P = .11; Table 4), but, because of the small sample size, a firm conclusion cannot be drawn regarding the age difference. Lineage assignment based on antigen receptors, using the presence of CD94 1A or TCR gene rearrangement, therefore, had both biologic and clinical significance.
As a confirmatory test (Table 5), we divided the patients with LBL into 2 groups, depending on whether the patients survived beyond 2 years. Among various clinical and biologic parameters tested, we found that 2-year survival correlated most strongly with presence of CD94 1A transcripts and absence of TCR gene rearrangement (P = .01).
Two-year survival correlates with CD94 1A and TCR rearrangement
. | Died before 2 y . | Alive after 2 y . | P . |
|---|---|---|---|
| No. of cases | 9 | 12 | |
| Median age, y | 29 | 17 | .11 |
| Sex, n male:female | 6:3 | 7:5 | .70 |
| CD161, n/total | 2/9 | 9/12 | .02 |
| CD44, n/total | 2/9 | 7/12 | .11 |
| E2A, n/total | 4/7 | 5/12 | .52 |
| c-ID2, n/total | 5/8 | 3/12 | .11 |
| TCF-1, n/total | 3/7 | 7/11 | .39 |
| CD94 1A, n/total | 0/9 | 7/12 | .01 |
| CD94 1B, n/total | 0/9 | 2/12 | .21 |
| KIR 2DL4, n/total | 0/9 | 1/12 | .38 |
| KIR 2D, n/total | 1/9 | 1/12 | .81 |
| KIR 3D, n/total | 2/9 | 6/12 | .20 |
| T-cell receptor, n/total | 8/9 | 3/12 | .01 |
| LDH, IU/L | 586 | 456 | .19 |
| Blasts in blood, n/total | 3/9 | 5/11 | .59 |
| WBC count, 103/μL | 7.1 | 7.3 | .64 |
| HB level, g/dL | 11 | 13.2 | .22 |
| PLT count, 103/μL | 255 | 194 | .35 |
| Mediastinal mass, n/total | 4/9 | 5/12 | .90 |
| Stage IV, n/total | 5/9 | 7/12 | .89 |
| ALL-like regimens, n/total | 1/9 | 6/12 | .07 |
. | Died before 2 y . | Alive after 2 y . | P . |
|---|---|---|---|
| No. of cases | 9 | 12 | |
| Median age, y | 29 | 17 | .11 |
| Sex, n male:female | 6:3 | 7:5 | .70 |
| CD161, n/total | 2/9 | 9/12 | .02 |
| CD44, n/total | 2/9 | 7/12 | .11 |
| E2A, n/total | 4/7 | 5/12 | .52 |
| c-ID2, n/total | 5/8 | 3/12 | .11 |
| TCF-1, n/total | 3/7 | 7/11 | .39 |
| CD94 1A, n/total | 0/9 | 7/12 | .01 |
| CD94 1B, n/total | 0/9 | 2/12 | .21 |
| KIR 2DL4, n/total | 0/9 | 1/12 | .38 |
| KIR 2D, n/total | 1/9 | 1/12 | .81 |
| KIR 3D, n/total | 2/9 | 6/12 | .20 |
| T-cell receptor, n/total | 8/9 | 3/12 | .01 |
| LDH, IU/L | 586 | 456 | .19 |
| Blasts in blood, n/total | 3/9 | 5/11 | .59 |
| WBC count, 103/μL | 7.1 | 7.3 | .64 |
| HB level, g/dL | 11 | 13.2 | .22 |
| PLT count, 103/μL | 255 | 194 | .35 |
| Mediastinal mass, n/total | 4/9 | 5/12 | .90 |
| Stage IV, n/total | 5/9 | 7/12 | .89 |
| ALL-like regimens, n/total | 1/9 | 6/12 | .07 |
CD94 1A transcripts could be detected in immature IL-15Rα+ lymphoid cells from thymus
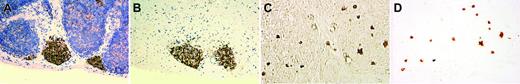
The existence of a bipotent T/NK precursor in the thymus is well known but has not been well characterized histologically. Immunohistochemistry on the thymus from an infant of 29-weeks' gestational age who died of prematurity showed either focal aggregates of IL-15Rα+ lymphoid cells (Figure 3A-B) or scattered interstitial staining (Figure 3C-D). We isolated these cells by rLCM and found that these cells had a CD94 1A transcript pattern similar to that shown in Figure 1A.
IL-15Rα+ immature lymphoid cells isolated from thymus of a premature infant by reverse laser capture microdissection. The figure shows a nodular pattern of IL-15Rα+ lymphoid cells in the thymus before and after rLCM (A-B, with hematoxylin counterstain), and an interstitial pattern of IL-15Rα+ lymphoid cells in the thymus before and after rLCM (C-D, phase-contrast view without hematoxylin counterstain). Note the well-preserved histopathology of IL-15Rα+ lymphoid cells isolated from a 5-μm frozen section of thymus with use of rLCM. Image acquisition was performed as described for Figure 2.
IL-15Rα+ immature lymphoid cells isolated from thymus of a premature infant by reverse laser capture microdissection. The figure shows a nodular pattern of IL-15Rα+ lymphoid cells in the thymus before and after rLCM (A-B, with hematoxylin counterstain), and an interstitial pattern of IL-15Rα+ lymphoid cells in the thymus before and after rLCM (C-D, phase-contrast view without hematoxylin counterstain). Note the well-preserved histopathology of IL-15Rα+ lymphoid cells isolated from a 5-μm frozen section of thymus with use of rLCM. Image acquisition was performed as described for Figure 2.
Discussion
In this study, we have attempted to improve the classification of LBLs by recognizing a subset of neoplasms that are of immature NK-cell lineage. We started with 21 LBLs, all of them positive for cytoplasmic CD3 and nuclear TdT, and negative for CD56. Because CD3 is part of the TCR complex and TdT is the enzyme that creates junctional diversity during TCR gene rearrangement, it is not surprising that immature T-cell or T/NK-cell precursors express cytoplasmic CD3 and TdT. It should be noted, however, that neither cytoplasmic CD3 nor TdT implies commitment to T-cell lineage. For example, immature or fetal NK cells also express cytoplasmic CD3,6 and TdT can be detected in the earliest Lin-/Sca-1+/ILR7-/C-Kithi/TdT+ common lymphoid precursors.7 Therefore, the cytoplasmic CD3 and nuclear TdT immunophenotype is not lineage specific.
With the use of CD94 1A expression and TCR gene rearrangement, our data support a reassessment of lineage assignment of LBLs, based on recent advances in the developmental biology of T and NK cells, both of which are derived from a common T/NK-cell thymic precursor. For the T-cell lineage,30 development precedes from a bipotent T/NK-cell precursor through T-cell precursor double negative (DN) stages 1 through 4, to CD4/CD8 double-positive immature T cells, and finally to CD4 or CD8 single-positive mature T cells. The process of lineage commitment takes place in CD44+/CD25+ T-cell precursors at stages DN2/DN3, characterized by TCRB gene rearrangement that is promoted by the transcription factor E2A. For the NK-cell lineage, development depends on IL-1520,21 and the transcription factor ID2.22,23 One of the earliest surface markers associated with the NK-cell lineage is CD161, but the intermediate stages preceding mature NK cells are still unknown. It is known only that mature NK cells use the CD94/NKG2 family and KIRs in humans as antigen receptors, but details on the regulation and acquisition of these receptors are limited.
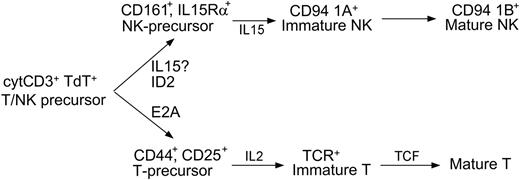
In accordance with these data, we present a model for NK-cell development that can be integrated into the classification of LBLs (Figure 4). In this 3-stage model, the bipotent T/NK-cell precursor develops into a unipotent precursor, then to immature stages, and finally to mature cells. CD94 1A transcripts and TCR rearrangements determine lineage commitment and have clinical significance. The presence of CD94 1A without monoclonal TCR gene rearrangement identifies NK-LBL, most of which are negative for cytoplasmic ID2
The absence of ID2 in NK-LBLs and the presence of cytoplasmic ID2 in T-LBLs is of interest. One might expect expression of nuclear ID2 to be associated with NK-LBL, and E2A to be associated with the T-LBL, because these molecules are required for NK and T-cell development, respectively.27,28 However, quite unexpectedly, we found that cytoplasmic ID2 was associated with T-LBL, and E2A was not lineage specific. How the loss of ID2 leads to NK-LBL is unknown, but it is known that redistribution of ID2 from the nucleus to the cytoplasm is associated with differentiation of murine hematopoietic cells,42 and that loss of E2A can cause T-lymphoblastic leukemia.43-45 Thus, the biologic significance of cytoplasmic ID2 is worthy of further investigation.
As summarized in Table 6, markers associated with precursors, such as CD161 and CD44, or those associated with mature cells, such as TCF-1 and KIRs, are not useful clinically. The lack of lineage specificity for CD161 is not totally unexpected, because CD161 is present in subsets of T cells, including those residing in the intestines.49 Markers associated with mature T or NK cells, such as TCF-1 and KIRs,31-33,36 are not useful for delineating T- or NK-LBLs that are derived from immature lymphoblasts. Because KIRs are related to T-cell receptors, and both belong to the immunoglobulin superfamily, one might suppose that the 2 evolutionarily conserved receptor families could be used as criteria for lineage commitment in 2 developmentally related cellular lineages. We found, however, that KIRs are not useful in lineage assignment for LBLs (Table 4). Because KIRs are found only in human NK cells, lineage commitment probably does not depend on KIRs. This finding is also consistent with a previous report that CD94 expression precedes KIRs, and that lineage commitment is independent of KIRs.46
Summary of candidate lineage markers
Marker . | Earliest detectable . | Role in T/NK development . | Role in mature T/NK lymphoma . | Role in lineage or prognosis of NK-LBL vs T-LBL . |
|---|---|---|---|---|
| CD161 | Before commitment | Present early during the development of NK-cell precursors, its expression persists into mature NK cells.16 | Not available | Less useful than CD94 1A |
| Also present in subsets of T cells, for example, in IFN-γ–producing intestinal T cells.49 | ||||
| CD44 | Before commitment | One of the earliest markers on T-cell precursors at the DN1 stage of development18 | Often lost in ENNLs but present in peripheral T-cell lymphoma40 | Not useful |
| ID2 and E2A | Before commitment | Transcription factors of the helix-loop-helix family; E2A induces the rearrangement of TCR; ID2 is a dominant-negative inhibitor of E2A and is essential for NK development.22-28 | Loss of E2A leads to T-cell leukemia43-45 | ID2 lost in NK-LBL |
| KIRs | After commitment | 3 alternative splicing groups, KIR2DL4, KIR2D, and KIR3D, found on human NK cells only.36 Expression of KIRs is preceded by CD94.46 | A restricted KIR repertoire found in ENNLs39,40 | Not useful |
| TCF-1 | After commitment | A transcription factor with functions in the maintenance, survival, and expansion of T cells31-33 | Loss of TCF-1 in a subset of peripheral T-cell lymphomas48 | Not useful |
| A different isoform found in mature human NK cells, also known to induce Ly49 NK-cell antigen receptors in the mouse47 |
Marker . | Earliest detectable . | Role in T/NK development . | Role in mature T/NK lymphoma . | Role in lineage or prognosis of NK-LBL vs T-LBL . |
|---|---|---|---|---|
| CD161 | Before commitment | Present early during the development of NK-cell precursors, its expression persists into mature NK cells.16 | Not available | Less useful than CD94 1A |
| Also present in subsets of T cells, for example, in IFN-γ–producing intestinal T cells.49 | ||||
| CD44 | Before commitment | One of the earliest markers on T-cell precursors at the DN1 stage of development18 | Often lost in ENNLs but present in peripheral T-cell lymphoma40 | Not useful |
| ID2 and E2A | Before commitment | Transcription factors of the helix-loop-helix family; E2A induces the rearrangement of TCR; ID2 is a dominant-negative inhibitor of E2A and is essential for NK development.22-28 | Loss of E2A leads to T-cell leukemia43-45 | ID2 lost in NK-LBL |
| KIRs | After commitment | 3 alternative splicing groups, KIR2DL4, KIR2D, and KIR3D, found on human NK cells only.36 Expression of KIRs is preceded by CD94.46 | A restricted KIR repertoire found in ENNLs39,40 | Not useful |
| TCF-1 | After commitment | A transcription factor with functions in the maintenance, survival, and expansion of T cells31-33 | Loss of TCF-1 in a subset of peripheral T-cell lymphomas48 | Not useful |
| A different isoform found in mature human NK cells, also known to induce Ly49 NK-cell antigen receptors in the mouse47 |
LBL expressing NK-cell–associated antigens was first recognized in 1987.50 By screening 38 cases of LBL, the investigators found 6 cases expressing the NK-associated antigens CD16 (Leu11b) and CD57 (Leu7). As a group, these cases of so-called “NK-LBL” were TdT+, CD2 (T11)+, CD4 (Leu3A)+, CD16+, and
CD57+, and they had a worse prognosis than that of the T-LBL. However, the true nature of these neoplasms was never further addressed at the molecular level. Subsequently, CD56 became a popular marker of NK cells even though CD56 is not specific and can be expressed by many other neoplasms. This has led to confusion in the literature regarding neoplasms of NK-cell lineage. For example, the entity described in the WHO classification as blastic NK-cell lymphoma51 is, in fact, not a neoplasm of NK cells and most likely is derived from type II plasmacytoid dendritic cells.13 For these reasons, we did not consider the absence of CD56 expression in LBLs as evidence excluding NK-cell lineage, prompting us to search for other markers useful for distinguishing immature NK from T cells.
A 3-stage model of NK development. The cytoplasmic CD3+ nuclear TdT+ bipotent T/NK-cell precursor may develop into either immature T cells under the actions of E2A and IL-2, or immature NK cells under the actions of ID2 and IL-15. Lineage commitment is fully established when immature T cells complete TCR gene rearrangement, or, as our data support, immature NK cells express CD94 1A transcripts. The model shows where the cytokines and transcription factors are critical, but does not exclude their possible roles at other developmental stages. For example, the assignment of ID2 before IL-15 is based on previous data23 and a recent review.41 According to this model, the loss of ID2 in NK-LBLs is due to abnormal differentiation. Whether the expression of IL-15Rα or, equivalently, the role of IL-15, might precede that of ID2 in the development of normal NK cells, requires further investigation.
A 3-stage model of NK development. The cytoplasmic CD3+ nuclear TdT+ bipotent T/NK-cell precursor may develop into either immature T cells under the actions of E2A and IL-2, or immature NK cells under the actions of ID2 and IL-15. Lineage commitment is fully established when immature T cells complete TCR gene rearrangement, or, as our data support, immature NK cells express CD94 1A transcripts. The model shows where the cytokines and transcription factors are critical, but does not exclude their possible roles at other developmental stages. For example, the assignment of ID2 before IL-15 is based on previous data23 and a recent review.41 According to this model, the loss of ID2 in NK-LBLs is due to abnormal differentiation. Whether the expression of IL-15Rα or, equivalently, the role of IL-15, might precede that of ID2 in the development of normal NK cells, requires further investigation.
In conclusion, we propose that LBLs can be divided into those of NK- and T-cell lineage using CD94 1A and TCR gene rearrangement. Among 21 cases of LBL that were positive for cytoplasmic CD3 and nuclear TdT and negative for CD56, 7 cases expressed CD94 1A transcripts and did not carry monoclonal TCR gene rearrangements supporting immature NK-cell lineage. These NK-LBLs occurred in younger patients who had a better prognosis than patients with T-LBL after appropriate chemotherapy. The use of CD94 1A, instead of CD56, as a marker of NK-cell lineage reflects recent advances in NK-cell biology and emphasizes the importance of complete lineage commitment in the biology of NK cells. On the basis of the identification of a true NK-LBL, we present a revised model of NK-cell development. This model not only stresses the importance of lineage commitment for clinical prognosis, but it also implies further constraints on the ultimate model of T/NK-cell development.
Prepublished online as Blood First Edition Paper, July 26, 2005; DOI 10.1182/blood-2005-02-0519.
Supported by grants from the National Taiwan University Hospital and the National Science Council, Taiwan (NTUH 94-S001 and NSC93-2320-B002-077) (C.-W.L.), (NSC-93-2314-B-002-005 and NSC-93-3112-B-041-Y) (S.-M.H.).
C.-W.L. proposed that CD94 1A transcripts might be used by immature NK cells and wrote the initial manuscript. T.-Y.L. and K.-T.W. conducted most of the experiments. S.-U.C. collected the cord blood and isolated the progenitor cells. L.J.M. reviewed the data and participated in the writing of the manuscript. S.-M.H. supervised the whole project.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mr C.C. Lu for the artwork, and the National Taiwan University Center for Genomic Medicine for technical support of laser capture microdissection.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal