Abstract
Immunoglobulin kappa (IGK) and immunoglobulin lambda (IGL) light chain repertoire was analyzed in 276 chronic lymphocytic leukemia (CLL) cases and compared with the relevant repertoires from normal, autoreactive, and neoplastic cells. Twenty-one functional IGKV genes were used in IGKV-J rearrangements of 179 kappa-CLL cases; the most frequent genes were IGKV3-20(A27), IGKV1-39/1D-39(O2/O12), IGKV1-5(L12), IGKV4-1(B3), and IGKV2-30(A17); 90 (50.3%) of 179 IGK sequences were mutated (similarity < 98%). Twenty functional IGLV genes were used in IGLV-J rearrangements of 97 lambda-CLL cases; the most frequent genes were IGLV3-21(VL2-14), IGLV2-8(VL1-2), and IGLV2-14(VL1-4); 44 of 97 IGL sequences (45.4%) were mutated. Subsets with “CLL-biased” homologous complementarity-determining region 3 (CDR3) were identified: (1) IGKV2-30-IGKJ2, 7 sequences with homologous kappa CDR3 (KCDR3), 5 of 7 associated with homologous IGHV4-34 heavy chains; (2) IGKV1-39/1D-39-IGKJ1/4, 4 unmutated sequences with homologous KCDR3, 2 of 4 associated with homologous IGHV4-39 heavy chains; (3) IGKV1-5-IGKJ1/3, 4 sequences with homologous KCDR3, 2 of 4 associated with unmutated nonhomologous IGHV4-39 heavy chains; (4) IGLV1-44-IGLJ2/3, 2 sequences with homologous lambda CDR3 (LCDR3), associated with homologous IGHV4-b heavy chains; and (5) IGLV3-21-IGLJ2/3, 9 sequences with homologous LCDR3, 3 of 9 associated with homologous IGHV3-21 heavy chains. The existence of subsets that comprise given IGKV-J/IGLV-J domains associated with IGHV-D-J domains that display homologous CDR3 provides further evidence for the role of antigen in CLL pathogenesis.
Introduction
Immunoglobulin kappa (IGK) V-J rearrangements in normal human B cells exhibit biased gene usage despite substantial germ-line-encoded diversity.1,2 Some genes are overrepresented in the nonexpressed repertoire, perhaps as a result of recombinational bias, whereas others are preferentially used in the expressed repertoire due to positive selection.1,2 Similar strong biases in the use of individual immunoglobulin lambda (IGL) V genes have been described in the expressed human repertoire.3-5 These observations have been attributed to several, non-mutually exclusive mechanisms, such as pairing of heavy and light chains, positive or negative selective processes, and possibly receptor editing processes. In this context, while the possibility of preferential pairing of specific immunoglobulin heavy (IGHV) and light gene products cannot be excluded, no evidence for pairing of specific IGHV or IGKV/IGLV subgroups was observed in either naive or memory B cells.6,7 The random pairing indicates that no particular limitations are imposed upon the expressed IG repertoire by specific favored interactions at heavy/light chain pairing.
Restricted light chain diversity has also been reported in the fetus8,9 ; furthermore, identical IGLV-J junctions were observed in the productive fetal repertoire at a greater extent compared with the adult repertoire. Based on these findings, it has been proposed that the highly restricted early light chain repertoire is strongly influenced by intrinsic developmental signals, exposure to self-antigens, as well as the need to effectively counteract ubiquitous bacterial pathogens.8,9 In the setting of autoimmunity, given the important influence of light chains in autoantigen binding, it has been suggested that the expressed light chain repertoire is shaped by secondary rearrangements, perhaps triggered upon chronic stimulation of the immune system by autoantigen.10-18
The expressed IGHV gene repertoire of B chronic lymphocytic leukemia (CLL) cells differs from that of normal peripheral blood CD5+ B cells.19-22 The most frequent IGHV genes in CLL are IGHV1-69, IGHV3-07, IGHV3-23, and IGHV4-34, although their relative frequency in different cohorts varies.19-26 Mutation analysis of IGHV genes has enabled a subdivision of CLL in 2 subsets, with and without somatic mutations, associated, respectively, with an indolent or a more aggressive clinical course.27-30 Nevertheless, regardless of IGHV mutation status, recent data suggest that all CLL cells resemble antigen-experienced and activated B cells.19,21,31 The importance of antigen in B-CLL evolution is underscored by the recent identification of subsets of CLL cases with “stereotyped” antigen-binding sites that could recognize individual, discrete antigens or classes of structurally similar epitopes.25,26,32-36
The study of IG repertoire in CLL has focused mainly on IGHV-D-J genes. There is considerably less information available for IGKV-J and IGLV-J genes; furthermore, in several reports, light chain data are provided for selected subgroups of CLL patients with particular molecular or clinical features. In the present study, we analyzed the expressed IGK/IGL rearrangements in a series of 276 consecutive, unselected patients with typical CLL, aiming at gaining insight into individual gene frequencies; the configuration and specific features of IGKV-J and IGLV-J junctions; possible biases in heavy/light chain associations; evidence for antigenic influences; and differences as well as similarities in the expressed IG light chain repertoire between B-CLL versus normal versus autoreactive B cells.
Patients, materials, and methods
Patient samples
Peripheral blood samples were collected from 276 typical, unselected CLL cases (CD5+, CD19+, CD20+, CD23+, low expression of surface immunoglobulin [sIg]). A case was considered to be kappa or lambda expressing if the ratio of kappa to lambda expression on CD19+ cells was more than 6 or less than 0.3, respectively. Based on the above definitions, 179 of 276 cases expressed kappa and 97 of 276 expressed lambda light chain. In all cases, the tumor load was at least 70%.
All patients met the diagnostic criteria of the National Cancer Institute-Working Group.37 Included in the analysis were 169 males and 107 females with a median age of 65 years (range, 26-88 years). Most patients were at early clinical stages by Rai/Binet classification systems (Rai stage/number of patients: 0/138, I/52, II/60, III/13, IV/13; Binet stage/number of patients: A/188, B/59, C/29). The median follow-up time from diagnosis was 41 months (range, 1-256 months). Written informed consent was obtained at study entry. The study was approved by the local ethics review committee of each institution.
PCR amplification of IG rearrangements and sequence analysis
Total cellular RNA isolation, cDNA preparation (after DNAse treatment), and reverse transcriptase-polymerase chain reaction (RT-PCR) amplification of IGHV-D-J, IGKV-J, and IGLV-J rearrangements were performed with primers specific for the framework region 1 (FR1) and the IGJ genes, as previously described.26
Direct sequencing of RT-PCR products was performed as described in Stamatopoulos et al.38 Sequence data were analyzed using the International ImMunoGeneTics information system (IMGT, http://imgt.cines.fr; initiator and coordinator: Marie-Paule Lefranc, Montpellier, France)39,40 and, more particularly, the IMGT/V-QUEST41 and IMGT/Junction Analysis42 tools. IGH, IGK, and IGL gene names from IMGT43-46 (for review, see Lefranc and Lefranc47 ) were approved in 1999 by the Human Genome Organisation (HUGO) Nomenclature Committee (HGNC)48 and entered in IMGT/GENE-database (DB),49 LocusLink National Center for Biotechnology Information (NCBI) in 1999 and in Entrez Gene NCBI. To facilitate direct reference and comparison with other studies, in the case of IGKV and IGLV genes, when a gene is first mentioned in the text, its name after the Zachau (IGKV) or Kawasaki (IGLV) nomenclature is provided in parentheses. All sequences reported in this study can be found in the GenBank, European Molecular Biology Laboratory (EMBL), and Laboratoire d'Immunogenetique Moleculaire (LIGM-DB) databases under accession numbers DQ098689 to DQ098828 and DQ100615 to DQ101183.
IG sequences of the present study were compared with cDNA IG sequences from multiple myeloma (MM) patients followed at our institutions as well as all IGKV-J and IGLV-J cDNA sequences retrieved from the IMGT/LIGM-DB sequence database (http://imgt.cines.fr/cgi-bin/IMGTlect.jv?)40 based on their accession numbers as reported in their respective publications. In this way, CLL sequences from our study were aligned and compared with (1) 43 IGKV-J and 43 IGLV-J cDNA MM sequences from our collection; (2) 677 IGKV-J and 368 IGLV-J sequences from normal and autoreactive cells, respectively (public databases); and (3) 122 IGKV-J and 47 IGLV-J cDNA sequences from CLL patients retrieved from public databases. All sequences from public databases used for comparisons in this study are presented in Table S1, available on the Blood website (see the Supplemental Table link at the top of the online article).
Somatic mutation distribution analysis in mutated sequences was conducted by the multinomial distribution model.50
Statistical analysis
Descriptive statistics were used for the presentation of data in terms of frequency distributions (discrete variables) and mean and median values (quantitative variables). Progression-free survival (PFS) was measured from enrollment to disease progression, and overall survival (OS) was measured from enrollment to death or last follow-up. PFS and OS curves were plotted using the Kaplan-Meier method. Bivariate differences in survival distributions were studied with the use of log-rank test. Multivariate Cox regression models were implemented for the study of the simultaneous effect of factors on survival outcomes taking into account the relative effect of remaining parameters. For the selection of best regression model, the automated forward model selection technique was implemented at a significance level of a = 5%. Hazard ratios of outcomes under study were calculated for each parameter estimate as well as 95% confidence intervals. All analyses were performed at a significance level of a = 5% with the statistical package SPSS 12.0 (SPSS, Chicago, IL).
Results
IGHV-D-J rearrangements
Productive IGHV-D-J rearrangements were successfully sequenced in 270 patients. Thirty-nine functional IGHV genes were identified; the most frequent genes were IGHV4-34 (31/270 sequences, 11.5%), IGHV3-23 (23/270, 8.5%), IGHV3-7 (21/270, 7.8%), IGHV1-69 (19/270, 7.0%), IGHV3-30 (18/270, 6.7%), and IGHV4-39 (16/270, 5.9%). The length of the complementarity-determining region 3 of the heavy chain (HCDR3) region length ranged from 6 to 32 amino acids (median, 16 amino acids). Using the 98% cut off for homology to germ line, 155 (57.4%) of 270 cases were considered as mutated, while the remainder (115/270, 42.6%) was considered as unmutated. The most frequent IGHV genes within the mutated subset were IGHV4-34 and IGHV3-7 (respectively, 27/31 and 19/21 rearrangements of each gene were mutated); IGHV1-69 was the most frequent gene in the unmutated group (16/19 IGHV1-69 rearrangements were unmutated).
IGKV/IGLV repertoire
Twenty-one functional IGKV genes were identified in 179 IGKV-J rearrangements; IGKV3-20 (A27) was the most frequent IGKV gene, followed by IGKV1-39/1D-39 (O2/O12), IGKV4-1 (B3), IGKV1-5 (L12), IGKV2-30 (A17), IGKV3-11 (L6), and IGKV1-8 (L9) (Table 1). Collectively, the aforementioned IGKV genes comprised 69.3% of all IGKV-J rearrangements. In total, 137 of 179 IGKV-J rearrangements used IGKV genes from the proximal cluster versus only 5 of 179 rearrangements that used genes from the distal cluster. In 37 of 179 cases, genes could not be distinguished as to cluster, since several genes from the J-distal cluster are identical to genes in the J-proximal cluster, and, therefore, members of the pairs cannot be distinguished (Table 1).
IGKV- and IGLV-rearranged genes in CLL cases of the present study
IGKV gene* . | No. (%) . |
|---|---|
| IGKV1-12 (L5) | 5 (2.79) |
| IGKV1-13/1D-13 (L4,L18) | 2 (1.12) |
| IGKV1-16 (L1) | 1 (0.56) |
| IGKV1-17 (A30) | 4 (2.23) |
| IGKV1-27 (A20) | 7 (3.91) |
| IGKV1-33/1D-33 (O18,O18a,O8) | 6 (3.35) |
| IGKV1-39/1D-39 (O12,O12a,O2) | 22 (12.29) |
| IGKV1-5 (L12,L12a) | 19 (10.61) |
| IGKV1-6 (L11) | 2 (1.12) |
| IGKV1-8 (L9) | 12 (6.70) |
| IGKV1-9 (L8) | 5 (2.79) |
| IGKV1D-16 (L15, L15a) | 1 (0.56) |
| IGKV2-24 (A23) | 2 (1.12) |
| IGKV2-28/2D-28 (A19,A3) | 7 (3.91) |
| IGKV2-30 (A17) | 13 (7.26) |
| IGKV2D-29 (A2a, A2c) | 3 (1.68) |
| IGKV2D-30 (A1) | 1 (0.56) |
| IGKV3-11 (L6, L6a) | 12 (6.70) |
| IGKV3-15 (L2) | 9 (5.03) |
| IGKV3-20 (A27,A27a) | 27 (15.08) |
| IGKV4-1 (B3) | 19 (10.61) |
| Total | 179 (100.00) |
| IGLV gene† | |
| IGLV1-36*01(Vλ1-11) | 1 (1.03) |
| IGLV1-40*01 (Vλ1-13) | 8 (8.25) |
| IGLV1-44*01 (Vλ1-16) | 7 (7.22) |
| IGLV1-51*01 (Vλ1-19) | 5 (5.15) |
| IGLV2-11*01 (Vλ1-3) | 4 (4.12) |
| IGLV2-14*01(Vλ1-4) | 10 (10.31) |
| IGLV2-23*02 (Vλ1-7) | 3 (3.09) |
| IGLV2-8*01 (Vλ1-2) | 14 (14.43) |
| IGLV3-1*01 (Vλ2-1) | 7 (7.22) |
| IGLV3-10*01(Vλ2-7) | 1 (1.03) |
| IGLV3-12*01(Vλ2-8) | 1 (1.03) |
| IGLV3-19*01 (Vλ2-13) | 3 (3.09) |
| IGLV3-21*01 (Vλ2-14) | 20 (20.62) |
| IGLV3-25*02(Vλ2-17) | 5 (5.15) |
| IGLV3-9*01(Vλ2-6) | 1 (1.03) |
| IGLV4-60*02(Vλ5-4) | 2 (2.06) |
| IGLV4-69*01 (Vλ5-6) | 1 (1.03) |
| IGLV6-57*01 (Vλ1-22) | 2 (2.06) |
| IGLV9-49*03(Vλ5-2) | 1 (1.03) |
| IGLV10-54*01(Vλ1-20) | 1 (1.03) |
| Total | 97 (100.00) |
IGKV gene* . | No. (%) . |
|---|---|
| IGKV1-12 (L5) | 5 (2.79) |
| IGKV1-13/1D-13 (L4,L18) | 2 (1.12) |
| IGKV1-16 (L1) | 1 (0.56) |
| IGKV1-17 (A30) | 4 (2.23) |
| IGKV1-27 (A20) | 7 (3.91) |
| IGKV1-33/1D-33 (O18,O18a,O8) | 6 (3.35) |
| IGKV1-39/1D-39 (O12,O12a,O2) | 22 (12.29) |
| IGKV1-5 (L12,L12a) | 19 (10.61) |
| IGKV1-6 (L11) | 2 (1.12) |
| IGKV1-8 (L9) | 12 (6.70) |
| IGKV1-9 (L8) | 5 (2.79) |
| IGKV1D-16 (L15, L15a) | 1 (0.56) |
| IGKV2-24 (A23) | 2 (1.12) |
| IGKV2-28/2D-28 (A19,A3) | 7 (3.91) |
| IGKV2-30 (A17) | 13 (7.26) |
| IGKV2D-29 (A2a, A2c) | 3 (1.68) |
| IGKV2D-30 (A1) | 1 (0.56) |
| IGKV3-11 (L6, L6a) | 12 (6.70) |
| IGKV3-15 (L2) | 9 (5.03) |
| IGKV3-20 (A27,A27a) | 27 (15.08) |
| IGKV4-1 (B3) | 19 (10.61) |
| Total | 179 (100.00) |
| IGLV gene† | |
| IGLV1-36*01(Vλ1-11) | 1 (1.03) |
| IGLV1-40*01 (Vλ1-13) | 8 (8.25) |
| IGLV1-44*01 (Vλ1-16) | 7 (7.22) |
| IGLV1-51*01 (Vλ1-19) | 5 (5.15) |
| IGLV2-11*01 (Vλ1-3) | 4 (4.12) |
| IGLV2-14*01(Vλ1-4) | 10 (10.31) |
| IGLV2-23*02 (Vλ1-7) | 3 (3.09) |
| IGLV2-8*01 (Vλ1-2) | 14 (14.43) |
| IGLV3-1*01 (Vλ2-1) | 7 (7.22) |
| IGLV3-10*01(Vλ2-7) | 1 (1.03) |
| IGLV3-12*01(Vλ2-8) | 1 (1.03) |
| IGLV3-19*01 (Vλ2-13) | 3 (3.09) |
| IGLV3-21*01 (Vλ2-14) | 20 (20.62) |
| IGLV3-25*02(Vλ2-17) | 5 (5.15) |
| IGLV3-9*01(Vλ2-6) | 1 (1.03) |
| IGLV4-60*02(Vλ5-4) | 2 (2.06) |
| IGLV4-69*01 (Vλ5-6) | 1 (1.03) |
| IGLV6-57*01 (Vλ1-22) | 2 (2.06) |
| IGLV9-49*03(Vλ5-2) | 1 (1.03) |
| IGLV10-54*01(Vλ1-20) | 1 (1.03) |
| Total | 97 (100.00) |
Genes belonging to the same IGKV or IGLV subgroup are listed together.
For all IGKV genes, the gene name in parentheses indicates the Zachau nomenclature
For all IGLV genes, the gene name in parentheses indicates the Kawasaki nomenclature
Twenty functional IGLV genes were identified; IGLV3-21 (Vλ2-14) was the most frequent IGLV gene, followed by IGLV2-8 (Vλ1-2), IGLV2-14 (Vλ1-4), IGLV1-40 (Vλ1-13), IGLV3-1 (Vλ2-1), and IGLV1-44 (Vλ1-16) (Table 1). Collectively, the aforementioned IGLV genes comprised 68% of all IGLV-J rearrangements. In total, 69 of 97 λ-CLL cases carried IGLV genes from cluster A, while 22 of 97 and 6 of 97 cases used genes from clusters B and C, respectively.
Analysis of the kappa CDR3 (KCDR3) and lambda CDR3 (LCDR3) regions
Complete analysis of the CDR3 region was possible in 176 of 179 IGKV-J and all 97 IGLV-J rearrangements. In IGKV-J sequences, IGKJ2 was the most frequent gene (67/176 cases), followed in order by IGKJ1 (54/176 cases), IGKJ4 (31/176 cases), IGKJ3 (21/176 cases), and IGKJ5 (3/176 cases).
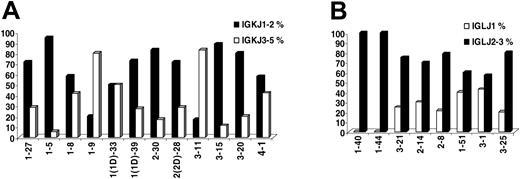
IGKJ3-5 gene usage was observed in 55 (31.24%) of 176 rearrangements. Important differences were observed among IGKV genes with regard to frequency of rearrangement to the IGKJ3-5 genes. Thus, the IGKV1-9 (L8) and IGKV3-11 genes were rearranged to the IGKJ3-5 genes in 80% or more of cases, contrasting the IGKV1-5 and IGKV3-15 (L2) genes with less than 20% frequency of rearrangement to the IGKJ3-5 genes (Figure 1). For most IGKV genes, similar patterns of recombination were observed in sequences from normal and autoreactive cells. “CLL-biased” patterns of recombination to IGKJ1-2 versus IGKJ3-5 were identified for some IGKV genes; thus, the IGKV3-20 and IGKV3-15 genes were much less frequently rearranged to the IGKJ3-5 genes in CLL (5/25 and 1/9 sequences, respectively) compared with either normal (49/110 and 12/34 sequences, respectively) or autoreactive (35/80 and 6/22 sequences, respectively) cells, while preferential rearrangement of the IGKV3-11 gene to the IGKJ3-5 genes was observed only in CLL (10/12 CLL vs 36/58 normal vs 13/29 autoreactive rearrangements).
In IGLV-J sequences, the IGLJ1 gene was used in 20 (20.6%) of 97 rearrangements, while the remainder (77/97, 79.4%) used the IGLJ2/3 genes. There were differences in IGLJ1 gene usage among individual IGLV genes (Figure 1); thus, no IGLV1-44 and IGLV1-40 rearrangements to the IGLJ1 gene were identified; on the other hand, 3 of 7 IGLV3-1, 2 of 5 IGLV1-51, and 5 of 20 IGLV3-21 rearrangements used the IGLJ1 gene. The aforementioned recombination patterns were CLL-biased for the IGLV1-40 and IGLV3-1 genes.
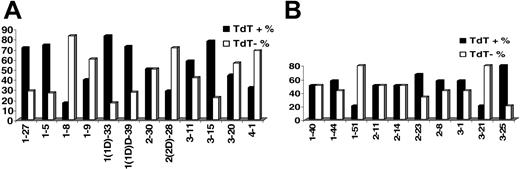
KCDR3 length ranged from 6 to 11 amino acids (median, 9 amino acids); LCDR3 length ranged from 8 to 13 amino acids (median, 11 amino acids). Details on terminal deoxyribonucleotidyl transferase (TdT), 5′- and 3′-exonuclease activities, as well as the presence of P nucleotides in IGKV-J and IGLV-J junctions are given in Table 2. Evidence for TdT activity was identified in 51.5% and 47.7% of IGKV-J and IGLV-J rearrangements, respectively. Nevertheless, TdT activity varied significantly among rearrangements of different IGKV or IGLV genes (Figure 2). Similar patterns of TdT activity were observed in normal and autoreactive cDNA sequences from public databases.
TdT activity, 5′- and 3′-exonuclease activities, and insertion of P nucleotides in IGKV-J and IGLV-J rearrangements in the present series
. | κ-CLL . | . | λ-CLL . | . | ||
|---|---|---|---|---|---|---|
. | Positivity, no. (%) . | Median (range) . | Positivity, no. (%) . | Median (range) . | ||
| TdT | 91 (51.5) | 3 (1-12) | 46 (47.7) | 3 (1-15) | ||
| P nucleotides | 18 (10.3) | 2 (1-3) | 4 (4.5) | 2 (2-3) | ||
| 5′-exonuclease | 132 (75.2) | 3 (1-16) | 66 (68.2) | 2 (1-10) | ||
| 3′-exonuclease | 116 (66.1) | 2 (1-8) | 71 (72.7) | 3 (1-7) | ||
. | κ-CLL . | . | λ-CLL . | . | ||
|---|---|---|---|---|---|---|
. | Positivity, no. (%) . | Median (range) . | Positivity, no. (%) . | Median (range) . | ||
| TdT | 91 (51.5) | 3 (1-12) | 46 (47.7) | 3 (1-15) | ||
| P nucleotides | 18 (10.3) | 2 (1-3) | 4 (4.5) | 2 (2-3) | ||
| 5′-exonuclease | 132 (75.2) | 3 (1-16) | 66 (68.2) | 2 (1-10) | ||
| 3′-exonuclease | 116 (66.1) | 2 (1-8) | 71 (72.7) | 3 (1-7) | ||
Somatic mutation analysis
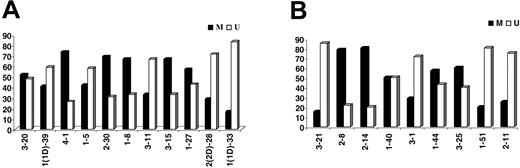
Eighty-nine (49.7%) of 179 IGKV-J rearrangements and 53 (54.6%) of 97 IGLV-J rearrangements were “unmutated” (≥ 98% homology to the closest germ-line gene); 58 of 89 “unmutated” IGKV-J and 28 of 53 IGLV “unmutated” sequences had 100% homology. Among “mutated” rearrangements, 27 of 90 IGKV and 11 of 44 IGLV sequences had less than 95% homology. The most frequent IGKV and IGLV genes within the mutated and unmutated subsets are shown in Tables 3-4. The distribution of individual IGKV or IGLV genes among mutated and unmutated rearrangements varied significantly (Figure 3). In the case of particular IGKV/IGLV genes (IGKV1-33/1D-33, IGKV2-30, IGLV3-1, IGLV3-21, and IGLV2-14), comparison with normal and autoreactive sequences revealed that mutation patterns observed in the present study were CLL-biased.
Most frequently used IGKV genes in mutated and unmutated IGKV-J rearrangements
Rank . | Mutated IGKV gene (%) . | Unmutated IGKV gene (%) . |
|---|---|---|
| 1 | IGKV4-1 (15.56) | IGKV3-20 (14.61) |
| 2 | IGKV3-20 (15.56) | IGKV1-39/1D-39 (14.61) |
| 3 | IGKV2-30 (10.00) | IGKV1-5 (12.36) |
| 4 | IGKV1-39/1D-39 (10.00) | IGKV3-11 (8.99) |
| 5 | IGKV1-5 (8.89) | IGKV4-1 (5.62) |
| 6 | IGKV1-8 (8.89) | IGKV2-28/2D-28 (5.62) |
| 7 | IGKV3-15 (6.67) | IGKV1-33/1D-33 (5.62) |
| 8 | IGKV3-11 (4.44) | IGKV1-8 (4.49) |
| 9 | IGKV1-27 (4.44) | IGKV2-30 (4.49) |
Rank . | Mutated IGKV gene (%) . | Unmutated IGKV gene (%) . |
|---|---|---|
| 1 | IGKV4-1 (15.56) | IGKV3-20 (14.61) |
| 2 | IGKV3-20 (15.56) | IGKV1-39/1D-39 (14.61) |
| 3 | IGKV2-30 (10.00) | IGKV1-5 (12.36) |
| 4 | IGKV1-39/1D-39 (10.00) | IGKV3-11 (8.99) |
| 5 | IGKV1-5 (8.89) | IGKV4-1 (5.62) |
| 6 | IGKV1-8 (8.89) | IGKV2-28/2D-28 (5.62) |
| 7 | IGKV3-15 (6.67) | IGKV1-33/1D-33 (5.62) |
| 8 | IGKV3-11 (4.44) | IGKV1-8 (4.49) |
| 9 | IGKV1-27 (4.44) | IGKV2-30 (4.49) |
Most frequently used IGLV genes in mutated and unmutated IGLV-J rearrangements
Rank . | Mutated IGLV gene (%) . | Unmutated IGLV gene (%) . |
|---|---|---|
| 1 | IGLV2-8 (25.00) | IGLV3-21 (32.08) |
| 2 | IGLV2-14 (18.18) | IGLV3-1 (9.43) |
| 3 | IGLV1-40 (9.09) | IGLV1-40 (7.55) |
| 4 | IGLV1-44 (9.09) | IGLV1-51 (7.55) |
| 5 | IGLV3-21 (6.82) | IGLV2-8 (5.66) |
| 6 | IGLV3-25 (6.82) | IGLV2-11 (5.66) |
| 7 | IGLV3-1 (4.55) | IGLV1-44 (5.66) |
| 8 | IGLV2-23 (4.55) | IGLV2-14 (3.77) |
Rank . | Mutated IGLV gene (%) . | Unmutated IGLV gene (%) . |
|---|---|---|
| 1 | IGLV2-8 (25.00) | IGLV3-21 (32.08) |
| 2 | IGLV2-14 (18.18) | IGLV3-1 (9.43) |
| 3 | IGLV1-40 (9.09) | IGLV1-40 (7.55) |
| 4 | IGLV1-44 (9.09) | IGLV1-51 (7.55) |
| 5 | IGLV3-21 (6.82) | IGLV2-8 (5.66) |
| 6 | IGLV3-25 (6.82) | IGLV2-11 (5.66) |
| 7 | IGLV3-1 (4.55) | IGLV1-44 (5.66) |
| 8 | IGLV2-23 (4.55) | IGLV2-14 (3.77) |
IG joining-region gene (IGJ) usage. (A) IGKJ1-IGKJ2 versus IGKJ3-IGKJ5 gene usage among individual rearranged IGKV genes. (B) IGLJ1 versus IGLJ2/IGLJ3 gene usage among individual rearranged IGLV genes.
IG joining-region gene (IGJ) usage. (A) IGKJ1-IGKJ2 versus IGKJ3-IGKJ5 gene usage among individual rearranged IGKV genes. (B) IGLJ1 versus IGLJ2/IGLJ3 gene usage among individual rearranged IGLV genes.
Parallel assessment of IGH and IGK/IGL mutation status was possible for 175 of 179 κ-CLL and 95 of 97 λ-CLL cases. In most cases, mutated IGHV rearrangements were associated with mutated IGK/IGL rearrangements and vice versa (Table 5). Fifteen of 21 IGH-mutated/IGK-unmutated κ-CLL and all 12 IGH-mutated/IGL-unmutated λ-CLL cases were found to carry IGKV- or IGLV-rearranged genes with a few somatic mutations (> 98% but < 100% homology to germ line); in the remaining cases, IGKV- or IGLV-rearranged genes had 100% homology to germ line. Three of 6 κ-CLL and all 4 λ-CLL cases carrying, respectively, unmutated IGH/mutated IGK or IGL genes were found to carry IGHV-rearranged genes with a few somatic mutations (> 98% but < 100% homology to germ line); in the remaining cases, IGHV-rearranged genes had 100% homology to germ line.
Parallel assesment of IGH and IGK/IGL mutation status in κ-CLL and λ-CLL cases of the present series
Type of rearrangements . | % . |
|---|---|
| κ-CLL | |
| IGKV-M + IGHV-M | 46.9 |
| IGKV-M + IGHV-U | 3.4 |
| IGKV-U + IGHV-U | 37.7 |
| IGKV-U + IGHV-M | 12.0 |
| λ-CLL | |
| IGLV-M + IGHV-M | 42.1 |
| IGLV-M + IGHV-U | 4.2 |
| IGLV-U + IGHV-U | 41.1 |
| IGLV-U + IGHV-M | 12.6 |
Type of rearrangements . | % . |
|---|---|
| κ-CLL | |
| IGKV-M + IGHV-M | 46.9 |
| IGKV-M + IGHV-U | 3.4 |
| IGKV-U + IGHV-U | 37.7 |
| IGKV-U + IGHV-M | 12.0 |
| λ-CLL | |
| IGLV-M + IGHV-M | 42.1 |
| IGLV-M + IGHV-U | 4.2 |
| IGLV-U + IGHV-U | 41.1 |
| IGLV-U + IGHV-M | 12.6 |
M indicates mutated; U, unmutated.
The vast majority of CDR-IMGT mutations were replacement (R) mutations, contrasting the situation in FR-IMGT, where a more even incidence of R and silent (S) mutations was observed (mean R/S ratios in, respectively, CDR-IMGT and FR-IMGT: IGKV, 2.6 and 1.5; IGLV, 3.7 and 1.6).
Homologous subsets
Each CLL-derived IGKV-J and IGLV sequence in our database was compared with every B-CLL and MM sequence of our collection as well as with IGKV-J and IGLV-J sequences from normal, autoreactive, and CLL cells in public cDNA gene databases (Table S1) using nucleotide and protein sequence IMGT. This analysis identified subsets of very similar sequences with closely homologous CDR3 regions (“homologous subsets”) composed of predominantly or exclusively CLL sequences (Table 6). Several of these subsets (subset 1: IGHV4-34/IGKV2-30; subset 2b: IGHV4-39/IGKV1-39-1D-39; subset 4: IGHV3-21/IGLV3-21) have been reported recently by several groups, including ours.26,33,35,36
Homologous subsets
. | Light chain . | . | . | . | Heavy chain . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | IGKV . | IGKJ . | HOM . | KCDR3 . | IGHV . | IGHD . | IGHJ . | HOM . | HCDR3 . | |||||||
| Set 1 | ||||||||||||||||
| P1422 | IGKV2-30 | IGKJ2 | 95, 7 | MQGTHW PYT | IGHV4-34 | IGHD5-12 | IGHJ6 | 91, 9 | ARGYADTPTFRRYYYYGMDV | |||||||
| P103 | IGKV2-30 | IGKJ2 | 97, 3 | ------P--- | IGHV4-34 | IGHD3-10 | IGHJ6 | 95, 9 | ----P---VV---------- | |||||||
| P907 | IGKV2-30 | IGKJ2 | 97 | ----Y- --- | IGHV4-34 | IGHD4-17 | IGHJ6 | 93, 2 | ----GTSA-TK--------- | |||||||
| P1626 | IGKV2-30 | IGKJ2 | 96, 2 | ------ --- | IGHV4-34 | IGHD4-17 | IGHJ6 | 94, 8 | -S-S ---T---------- | |||||||
| P1939 | IGKV2-30 | IGKJ2 | 96 | -------G-- | IGHV4-34 | IGHD5-5 | IGHJ6 | 94, 5 | V---P--AVVK-------E- | |||||||
| P711 | IGKV2-30 | IGKJ2 | 99, 3 | ------ --- | IGHV4-34 | IGHD6-19 | IGHJ4 | 97, 4 | ARGQWLDNY | |||||||
| P860 | IGKV2-30 | IGKJ2 | 96, 3 | ------ --- | IGHV4-30-4 | IGHD4-23 | IGHJ5 | 91 | VRDGNSN | |||||||
| Set 2a | ||||||||||||||||
| P1986 | IGKV1-39/1D-39 | IGKJ2 | 100 | QQSYSTPPYT | IGHV1-8 | IGHD6-13 | IGHJ6 | 100 | ARGLLSPYSSSWYWDFRHYYYGMDV | |||||||
| N1457 | IGKV1-39/1D-39 | IGKJ2 | 100 | ---------- | IGHV1-18 | IGHD6-19 | IGHJ4 | 99, 3 | ARKQWLPQYYFDY | |||||||
| P439 | IGKV1-39/1D-39 | IGKJ2 | 100 | ---------- | IGHV1-3 | IGHD6-19 | IGHJ4 | 100 | --W---VLG---- | |||||||
| Set 2b | ||||||||||||||||
| P1114 | IGKV1-39/1D-39 | IGKJ4 | 100 | QQSYSTPLT | IGHV4-39 | IGHD6-13 | IGHJ5 | 100 | ARRTGYSSSWYSTYNWFDP | |||||||
| P1615 | IGKV1-39/1D-39 | IGKJ1 | 100 | -------P- | IGHV4-39 | IGHD6-13 | IGHJ5 | 100 | -S-R------FNV-A---- | |||||||
| P191 | IGKV1-39/1D-39 | IGKJ4 | 100 | --------- | IGHV3-49 | IGHD3-3 | IGHJ4 | 100 | TRGPAEEYYDFWSGYYRLGGYY | |||||||
| N235 | IGKV1-39/1D-39 | IGKJ4 | 99, 6 | --------- | IGHV3-74 | IGHD3-9 | IGHJ4 | 100 | ARDLSGHYDILTGYYREIFDY | |||||||
| Set 3 | ||||||||||||||||
| N31 | IGKV1-5 | IGKJ1 | 100 | QQYNSYPWT | IGHV4-39 | IGHD3-16 | IGHJ3 | 100 | ARTLSYYDYVWANAFDI | |||||||
| N232 | IGKV1-5 | IGKJ1 | 100 | --------- | IGHV3-30 | IGHD2-15 | IGHJ6 | 100 | AKDSVSVVVVAATYLTPYYYYGMDV | |||||||
| N1622 | IGKV1-5 | IGKJ1 | 100 | --------- | IGHV4-39 | IGHD2-8 | IGHJ6 | 100 | ARGCRCTNGVCYVYYYYGMDV | |||||||
| P1289 | IGKV1-5 | IGKJ3 | 96, 4 | -------F- | IGHV4-59 | IGHD5-24 | IGHJ3 | 87 | GRGGNGYNYIQI | |||||||
| Set 4 | ||||||||||||||||
| P1531 | IGLV3-21 | IGLJ2/3 | 98, 3 | QVWDSGSDHPWV | IGHV3-21 | ND | IGHJ6 | 97, 3 | ARDANGMDV | |||||||
| P1321 | IGLV3-21 | IGLJ2/3 | 99 | ------------ | IGHV3-21 | IGHD1-1 | IGHJ6 | 98, 6 | ---Q-A--- | |||||||
| P326 | IGLV3-21 | IGLJ2/3 | 99, 7 | -----S------ | IGHV3-21 | IGHD1-14 | IGHJ6 | 98, 6 | -I-R----- | |||||||
| P173 | IGLV3-21 | IGLJ2/3 | 100 | ------------ | IGHV3-21 | IGHD3-10 | IGHJ4 | 99 | AREPLSVLLWFGESFGGGFDY | |||||||
| P458 | IGLV3-21 | IGLJ2/3 | 98 | ------------ | IGHV3-11 | IGHD5-24 | IGHJ4 | 97, 7 | ARDRGQQPFDY | |||||||
| P786 | IGLV3-21 | IGLJ2/3 | 99 | ------------ | IGHV3-23 | IGHD3-22 | IGHJ4 | 97 | AKDWRGESYYDSSGCIDY | |||||||
| N1533 | IGLV3-21 | IGLJ2/3 | 100 | -----S----V- | IGHV1-69 | IGHD3-22 | IGHJ4 | 100 | AYYYYDSSGYFRY | |||||||
| P1132 | IGLV3-21 | IGLJ2/3 | 99 | -----S------ | IGHV4-39 | IGHD3-22 | IGHJ4 | 98, 3 | ARAPHDSSGYDAYVWDY | |||||||
| N1707 | IGLV3-21 | IGLJ2/3 | 98, 9 | -----S------ | IGHV3-23 | IGHD3-22 | IGHJ1 | 96, 2 | AKDYDSSGANFQH | |||||||
| Set 5 | ||||||||||||||||
| P460 | IGKLV1-44 | IGLJ2/3 | 99 | AAWDDSLNGPV | IGHV4-b | IGHD6-13 | IGHJ5 | 97, 3 | ARIRWGSSWYDCFDP | |||||||
| P1558 | IGKLV1-44 | IGLJ2/3 | 100 | ---------Q- | IGHV4-b | IGHD6-19 | IGHJ4 | 100 | --A-YS-V---Y--Y | |||||||
. | Light chain . | . | . | . | Heavy chain . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | IGKV . | IGKJ . | HOM . | KCDR3 . | IGHV . | IGHD . | IGHJ . | HOM . | HCDR3 . | |||||||
| Set 1 | ||||||||||||||||
| P1422 | IGKV2-30 | IGKJ2 | 95, 7 | MQGTHW PYT | IGHV4-34 | IGHD5-12 | IGHJ6 | 91, 9 | ARGYADTPTFRRYYYYGMDV | |||||||
| P103 | IGKV2-30 | IGKJ2 | 97, 3 | ------P--- | IGHV4-34 | IGHD3-10 | IGHJ6 | 95, 9 | ----P---VV---------- | |||||||
| P907 | IGKV2-30 | IGKJ2 | 97 | ----Y- --- | IGHV4-34 | IGHD4-17 | IGHJ6 | 93, 2 | ----GTSA-TK--------- | |||||||
| P1626 | IGKV2-30 | IGKJ2 | 96, 2 | ------ --- | IGHV4-34 | IGHD4-17 | IGHJ6 | 94, 8 | -S-S ---T---------- | |||||||
| P1939 | IGKV2-30 | IGKJ2 | 96 | -------G-- | IGHV4-34 | IGHD5-5 | IGHJ6 | 94, 5 | V---P--AVVK-------E- | |||||||
| P711 | IGKV2-30 | IGKJ2 | 99, 3 | ------ --- | IGHV4-34 | IGHD6-19 | IGHJ4 | 97, 4 | ARGQWLDNY | |||||||
| P860 | IGKV2-30 | IGKJ2 | 96, 3 | ------ --- | IGHV4-30-4 | IGHD4-23 | IGHJ5 | 91 | VRDGNSN | |||||||
| Set 2a | ||||||||||||||||
| P1986 | IGKV1-39/1D-39 | IGKJ2 | 100 | QQSYSTPPYT | IGHV1-8 | IGHD6-13 | IGHJ6 | 100 | ARGLLSPYSSSWYWDFRHYYYGMDV | |||||||
| N1457 | IGKV1-39/1D-39 | IGKJ2 | 100 | ---------- | IGHV1-18 | IGHD6-19 | IGHJ4 | 99, 3 | ARKQWLPQYYFDY | |||||||
| P439 | IGKV1-39/1D-39 | IGKJ2 | 100 | ---------- | IGHV1-3 | IGHD6-19 | IGHJ4 | 100 | --W---VLG---- | |||||||
| Set 2b | ||||||||||||||||
| P1114 | IGKV1-39/1D-39 | IGKJ4 | 100 | QQSYSTPLT | IGHV4-39 | IGHD6-13 | IGHJ5 | 100 | ARRTGYSSSWYSTYNWFDP | |||||||
| P1615 | IGKV1-39/1D-39 | IGKJ1 | 100 | -------P- | IGHV4-39 | IGHD6-13 | IGHJ5 | 100 | -S-R------FNV-A---- | |||||||
| P191 | IGKV1-39/1D-39 | IGKJ4 | 100 | --------- | IGHV3-49 | IGHD3-3 | IGHJ4 | 100 | TRGPAEEYYDFWSGYYRLGGYY | |||||||
| N235 | IGKV1-39/1D-39 | IGKJ4 | 99, 6 | --------- | IGHV3-74 | IGHD3-9 | IGHJ4 | 100 | ARDLSGHYDILTGYYREIFDY | |||||||
| Set 3 | ||||||||||||||||
| N31 | IGKV1-5 | IGKJ1 | 100 | QQYNSYPWT | IGHV4-39 | IGHD3-16 | IGHJ3 | 100 | ARTLSYYDYVWANAFDI | |||||||
| N232 | IGKV1-5 | IGKJ1 | 100 | --------- | IGHV3-30 | IGHD2-15 | IGHJ6 | 100 | AKDSVSVVVVAATYLTPYYYYGMDV | |||||||
| N1622 | IGKV1-5 | IGKJ1 | 100 | --------- | IGHV4-39 | IGHD2-8 | IGHJ6 | 100 | ARGCRCTNGVCYVYYYYGMDV | |||||||
| P1289 | IGKV1-5 | IGKJ3 | 96, 4 | -------F- | IGHV4-59 | IGHD5-24 | IGHJ3 | 87 | GRGGNGYNYIQI | |||||||
| Set 4 | ||||||||||||||||
| P1531 | IGLV3-21 | IGLJ2/3 | 98, 3 | QVWDSGSDHPWV | IGHV3-21 | ND | IGHJ6 | 97, 3 | ARDANGMDV | |||||||
| P1321 | IGLV3-21 | IGLJ2/3 | 99 | ------------ | IGHV3-21 | IGHD1-1 | IGHJ6 | 98, 6 | ---Q-A--- | |||||||
| P326 | IGLV3-21 | IGLJ2/3 | 99, 7 | -----S------ | IGHV3-21 | IGHD1-14 | IGHJ6 | 98, 6 | -I-R----- | |||||||
| P173 | IGLV3-21 | IGLJ2/3 | 100 | ------------ | IGHV3-21 | IGHD3-10 | IGHJ4 | 99 | AREPLSVLLWFGESFGGGFDY | |||||||
| P458 | IGLV3-21 | IGLJ2/3 | 98 | ------------ | IGHV3-11 | IGHD5-24 | IGHJ4 | 97, 7 | ARDRGQQPFDY | |||||||
| P786 | IGLV3-21 | IGLJ2/3 | 99 | ------------ | IGHV3-23 | IGHD3-22 | IGHJ4 | 97 | AKDWRGESYYDSSGCIDY | |||||||
| N1533 | IGLV3-21 | IGLJ2/3 | 100 | -----S----V- | IGHV1-69 | IGHD3-22 | IGHJ4 | 100 | AYYYYDSSGYFRY | |||||||
| P1132 | IGLV3-21 | IGLJ2/3 | 99 | -----S------ | IGHV4-39 | IGHD3-22 | IGHJ4 | 98, 3 | ARAPHDSSGYDAYVWDY | |||||||
| N1707 | IGLV3-21 | IGLJ2/3 | 98, 9 | -----S------ | IGHV3-23 | IGHD3-22 | IGHJ1 | 96, 2 | AKDYDSSGANFQH | |||||||
| Set 5 | ||||||||||||||||
| P460 | IGKLV1-44 | IGLJ2/3 | 99 | AAWDDSLNGPV | IGHV4-b | IGHD6-13 | IGHJ5 | 97, 3 | ARIRWGSSWYDCFDP | |||||||
| P1558 | IGKLV1-44 | IGLJ2/3 | 100 | ---------Q- | IGHV4-b | IGHD6-19 | IGHJ4 | 100 | --A-YS-V---Y--Y | |||||||
Three novel subsets were identified in the present study (subsets 2a, 3, and 5 in Table 6). Two of 3 showed close, CLL-biased homology for light chains only. Specifically, subset 2a included 3 unmutated IGKV1-39/1D-39-IGKJ2 CLL rearrangements from our series with identical, CLL-biased, acidic, 10-amino acid-long KCDR3 (QQSYSTPPYT), all associated with unmutated IGHV1 heavy chains (HCs); an identical KCDR3 was identified in 3 IGKV1-39/1D-39 public CLL sequences (AY575940, AY575943, AY575945). Subset 3 included 4 IGKV1-5-IGKJ1/3 CLL sequences from our series (3 unmutated) with an almost identical, CLL-biased, acidic KCDR3 (QQYNSYP[W/F]T); 2 of 4 were associated with unmutated IGHV4-39 HCs with acidic, nonhomologous HCDR3. Finally, subset 5 included 2 cases with IGHV4-b HCs (one IgG, 100% identical to germ line; the other IgM, 97.3% homologous) with similar HCDR3s of 15 amino acids, associated with unmutated IGLV1-44/IGLJ2/3 CLL rearrangements with 10 of 11 identical LCDR3 amino acids (AAWDDSLNG[P/Q]V). Both HCDR3s and LCDR3s of the 2 cases in subset 5 had identical, acidic isoelectric point (pI) values (6,0 for both HCDR3s; 3,56 for both LCDR3s) as well as very similar molecular weights.
TdT activity in (A) IGKV-J and (B) IGLV-J rearrangements among individual rearranged IGKV and IGLV genes.
TdT activity in (A) IGKV-J and (B) IGLV-J rearrangements among individual rearranged IGKV and IGLV genes.
Survival data
Median PFS and OS were 87.7 and 156.7 months, respectively (95% CI: 61.5-112.0 for PFS and 93.8-219.6 for OS). Using univariate analysis, significant parameters (P < .001) for PFS and OS were clinical stage at diagnosis (Rai/Binet) and IGH/IGK-IGL mutation status (Figure 4A-B); CD38 positivity was significant only for PFS. For bivariate and multivariate analysis, Rai stage was excluded from the model due to the collinearity with Binet clinical stage. Patients were grouped into 2 subgroups, Binet A and Binet B/C patients. Bivariate analysis between (1) clinical stage and IGH mutation status, (2) clinical stage and IGK/IGL mutation status, and (3) IGHV and IGK/IGL mutational profile revealed significant differences (P < .001) in PFS and OS for all combinations (Figure 4C-F). Multivariate Cox regression analysis (including all factors with significant associations) revealed that only IGH mutation status and clinical stage remained statistically significant variables for both PFS and OS.
Discussion
We analyzed the expressed IGK/IGL repertoire along with the expressed IGH repertoire in a series of 276 consecutive, unselected patients with typical CLL. The IGHV genes used by these leukemic clones derived from the IGHV1-6 subgroups with 6 genes (IGHV4-34, IGHV3-23, IGHV3-7, IGHV1-69, IGHV3-30, and IGHV4-39) representing approximately 50% of cases. The relative frequency at which these genes occur in different CLL series varies. Several reasons could account for these differences,19-26 including (1) different cohort size as well as type of medical facility (referral versus primary centers) where patients are evaluated and (2) geographic differences that could allow peculiar genetic or environmental elements to differently shape both the normal and the “leukemic” repertoire (as exemplified by the markedly higher frequency of the IGHV3-21 gene in Scandinavian vs Mediterranean CLL repertoires).24-26
Seven IGKV genes (IGKV3-20, IGKV1-39/1D-39, IGKV1-5, IGKV4-1, IGKV2-30, IGKV3-11, and IGKV1-8) were collectively used in 69.3% of the analyzed IGKV-J rearrangements. The IGKV1-39/1D-39, IGKV4-1, and IGKV2-30 genes were also frequent both in productive and nonproductive IGKV-J rearrangements in normal peripheral blood IgM+ B cells (DNA-based, single-cell PCR study).1 In that study, the IGKV3-20 and IGKV1-5 genes were overrepresented in the productive but not the nonproductive repertoire, prompting speculations that their biased appearance in the expressed repertoire could be strictly related to expression of a functional kappa chain.1 A number of genes identified in CLL IGKV-J rearrangements have been found as components of autoantibodies (eg, IGKV1-33/1D-33, IGKV1-39/1D-39, IGKV3-20), although this may merely reflect their overall frequency in the repertoire.1,2,7
In the present CLL series, similar to normal B cells, only 3 of the 10 functional IGLV subgroups (IGLV1 to IGLV3) were used to a significant extent; furthermore, this skewed usage is due mainly to biases in the use of individual IGLV genes. Of the 29 to 33 functional IGLV genes, 6 (IGLV3-21, IGLV2-8, IGLV2-14, IGLV1-40, IGLV3-1, and IGLV1-44) encoded 68% of the expressed CLL IGLV repertoire. Similar to normal cells,3-6 IGLV genes from cluster A (closest to the IGLJ-IGLC pairs) accounted for 71% of the CLL clones, genes of cluster B constituted 23%, and genes from cluster C (furthest from the IGLJ-IGLC pairs) accounted for only 6% of the expressed CLL IGLV repertoire. Although this could be interpreted as evidence for biased recombination based on proximity of the IGLV gene to the IGLJ-IGLC pairs, the use of individual IGLV genes within each cluster shows no such effect.
Comparison of the CLL IGKV and IGLV repertoires of the present series to published repertoires from normal cells1-9 disclosed similarities as well as differences regarding individual gene frequencies. Among IGKV genes, the IGKV1-8 and IGKV2-30 genes were significantly more frequent in the present series than in 2 cDNA-based studies of normal cells2,6 ; the opposite was observed for the IGKV3-15 gene. Among IGLV genes, the IGLV3-21 and IGLV2-8 genes were significantly more frequent in CLL than in cDNA-based studies of normal cells,3,4,6 while the IGLV2-14 gene was significantly more frequent in normal cells. Nonetheless, one should interpret these results with caution, given the lack of a truly representative “normal” database that could be used for reference; thus, significant differences in gene frequencies are observed between studies of normal cells,1-9 most probably related to the sources of sequences. In this context, cDNA-based repertoires could potentially be biased toward activated cells, which have greater amounts of mRNA than resting cells.2 On the other hand, DNA-based studies discriminating only between productive (ie, in-frame) and nonproductive (ie, out-of-frame) rearrangements do not permit an accurate assessment of the expressed repertoire. It is perhaps relevant that a significant proportion (∼30%-40%) of nonexpressed IGKV-J rearrangements either in normal lambda-expressing B cells51 or in lambda-expressing CLL is productive (in-frame); as previously reported by our group, individual IGKV genes are used in nonexpressed, in-frame IGKV-J rearrangements in CLL at markedly different frequencies from the frequencies reported herein.52 Furthermore, the cloning procedure adopted in some studies may have introduced biases; finally, differences between databases could also be attributed to different donor sources.
IGKJ and IGLJ gene distributions in CLL show similarities to normal B cells.1-9 Nevertheless, significant differences in IGKJ and IGLJ gene usage were observed for different IGKV- or IGLV-rearranged genes. This was more pronounced for IGKV-J rearrangements; as revealed by comparison with normal and autoreactive cDNA sequences, extreme patterns of recombination to IGKJ1-2 versus IGKJ3-5 genes were often CLL biased. Single-cell studies of normal human B cells51 have shown downstream IGKJ gene (IGKJ3-5) usage in 45% versus 64% in kappa- and lambda-expressing cells, respectively. In autoantibody transgenic mice, use of downstream IGKJ genes is a hallmark of autoreactive cells that escape deletion by secondary rearrangements that alter antigen receptor specificity.53-55
Our parallel analysis of IG heavy and light chain genes in CLL demonstrates considerable heterogeneity regarding mutation “load” in both IGHV and IGKV/IGLV genes, with very few or even no mutations in a substantial proportion of cases. Nevertheless, a low level of mutations could be functionally relevant; as previously shown, even single-base changes may be important for improving antigen recognition (eg, high-affinity anti-DNA antibodies in systemic lupus erythematous with minimal somatic hypermutation).56 Mutation distribution analysis after the multinomial model50 provided evidence for selection by classical T-dependent antigen in only a subgroup of mutated cases from our series; nevertheless, these results should be interpreted with caution, as all available statistical methods cannot provide concrete evidence for antigen selection. The existence of CLL cases discrepant for somatic mutation status among heavy and light chains implies that a complementarity imprint of antigen witnessed either by IGHV, IGKV, or IGLV sequences might constitute an important event in the pathogenesis of, at least, a proportion of CLL cases.
Percentages (%) of mutated (M) versus unmutated (U) sequences per individual (A) IGKV or (B) IGLV gene.
Percentages (%) of mutated (M) versus unmutated (U) sequences per individual (A) IGKV or (B) IGLV gene.
The pioneering studies by Damle et al27 and Hamblin et al28 revealed that CLL patients who express V genes with more than 98% homology to the closest germ-line gene follow more aggressive clinical courses and have strikingly shorter survival than patients with significant V gene mutations; this has since been confirmed in several CLL cohorts. In the present study, we analyzed the prognostic value of IGHV mutation status in parallel with IGKV/IGLV mutation status within the different stages of clinical classification systems (Rai/Binet). On univariate analysis, IGKV/IGLV mutation status along with clinical stage at diagnosis (Rai/Binet) and IGH mutation status were significantly associated with outcome. On bivariate analysis, IGK/IGL mutation status in combination with either clinical stage or IGH mutation status was significantly associated with outcome. Nevertheless, on multivariate analysis, only IGHV mutation status and clinical stage at diagnosis retained independent prognostic status; in keeping with a previous study,57 our analysis confirms that clinical classification and IG gene mutation status are independent prognostic variables and most likely provide complementary information.
In both heavy and light chains, CDR3 length and amino acid composition make major contributions to antigen specificity.58-61 A continuous increase in length occurs during fetal life until birth in mice and humans, due mainly to the relative absence of N regions in fetal genes.61-63 Mutated antibodies have been shown to carry shorter CDR3s than nonmutated antibodies in both mice and humans.63 Although amino acid sequence alone cannot predict whether an antibody will be self-reactive, long KCDR3 or LCDR3 regions have been associated with self-reactive or polyreactive antibodies.64-66 In the present series, skewing to longer CDR3 was observed for IGKV-J rearrangements using IGKV3 subgroup genes (48% of IGKV3 rearrangements had a KCDR3 of > 9 amino acids); this is in keeping with previous reports from normal (fetal and adult) B cells.1,2,8,9 The fact that in CLL, as in the normal repertoire, the IGKV3 subgroup is represented more often than would be expected from random usage (based on its germ-line complexity40 ) might suggest cellular selection of the longer CDR3 lengths in IGKV3 sIGs. Skewing to longer CDR3 was also observed for IGLV-J rearrangements using IGLV3 subgroup genes and, in particular, IGLV3-21 (39% of all IGLV3 rearrangements and 60% of IGLV3-21 rearrangements had an LCDR3 of > 11 amino acids; in this context, 9/13 public/CU IGLV3-21 rearrangements had a 12-amino acid-long LCDR3). Interestingly, only 1 of 8 IGLV3-21 multiple myeloma sequences from our group had more than 11-amino acid-long LCDR3.
Survival curves. Kaplan-Meier PFS and OS curves comparing (A-B) mutated and unmutated IGKV/IGLV rearrangements; (C-D) IGHV and IGKV/IGLV mutation profile with bivariate analysis; and (E-F) Binet stage (A versus BC) and IGKV/IGLV mutation profile with bivariate analysis.
Survival curves. Kaplan-Meier PFS and OS curves comparing (A-B) mutated and unmutated IGKV/IGLV rearrangements; (C-D) IGHV and IGKV/IGLV mutation profile with bivariate analysis; and (E-F) Binet stage (A versus BC) and IGKV/IGLV mutation profile with bivariate analysis.
The mature normal repertoire is dominated by a few germ-line genes with no evidence for preferential pairings of specific IG heavy/light chain genes or subgroups.6,7 No associations exist between heavy/light chain CDR3 lengths and sequences or between IGHV and IGHD and IGHJ genes. In contrast, as recently shown by several groups around the world,25,26,32-36 a unique feature of the CLL IG repertoire is the existence of subsets of cases with “stereotyped” B-cell receptors (BCRs, use of the same IGHV and IGKV/IGLV genes with unique, shared HCDR3 and light chain CDR3 motifs). This BCR restriction could be the consequence of random transformation of B cells with very limited BCR diversity or specific transformation of B cells selected by antigen from a BCR-restricted or BCR-heterogeneous subpopulation or both.25,26,32-36
Although CDR3 length and diversity are much more restricted in light chains than heavy chains, by alignment and comparison of our sequences to cDNA sequences from public databases from normal and autoreactive cells we were able to document the existence of subsets of IGKV-J or IGLV-J rearrangements with CLL-biased amino acid sequence, often but not always associated with strikingly similar heavy chains. The only exception to the CLL bias for the subsets of light chain homologous rearrangements described here concerns IGKV1-39/1D-39-rearranged sequences, where an identical KCDR3 region to the one reported for subset 2a of the present series was also identified in 5 IGKV1-39/1D-39 public sequences (AJ399870, AF306388, L12112, X98985, L12065), all deriving from cells with anti-thyroid peroxidase activity.12,67,68 With the exception of the IGHV4-34/IGKV2-30 subset (previously described in Messmer et al35 ), sequences of these sets lacked or had very few somatic mutations, even in isotype-switched cases (2 unmutated sIgG+ cases with homologous IGHV4-39/IGKV1-39/1D-39 BCRs, in a pattern already described from groups in New York33 and Sweden36 ). A large number of somatic mutations is not a prerequisite for efficient antigen recognition; as previously shown for IgG autoantibodies in lupus, minimally mutated antibodies with particular HCDR3 features (eg, predominance of basic residues) may recognize DNA with high affinity.56 Intriguingly, in the IGHV4-34/IGKV2-30 subset, although all 10 IGHV and IGKV sequences were mutated, only 1 IGHV4-34 rearrangement had high R/S mutation ratios in the CDRs, while the remainder exhibited an almost uniform distribution of R and S mutations over both FRs and CDRs. The IGVH4-34 gene encodes an antibody that recognizes autologous determinants on red blood cells, the I/i antigens.69 Mutated IGHV4-34 rearrangements with similar mutation distribution patterns to the ones found in the CLL cases of the IGHV4-34/IGKV2-30 subset have also been reported in studies of microdissected marginal zone normal B cells,70 single B cells from patients with lupus,71 and splenic marginal-zone lymphoma.38
CLL cells can frequently express IgM antibodies that display reactivity to self-proteins (eg, IgG, cardiolipin, actin, thyroglobulin) or DNA.72-76 A recent study on transgenic expression of a human IgMκ polyreactive rheumatoid factor from a patient with CLL demonstrated that expression of polyreactive autoantibodies can allow for development of B cells that are neither deleted nor rendered anergic but have a phenotype of antigen-experienced B cells that respond to nonspecific activation.77 The similarity between B cells producing natural autoantibodies and CLL B cells could mean that the process of positive selection of natural autoreactive B cells may carry a risk for malignant transformation.77,78 The results of the present study emphasize the close relationship between autoreactivity and CLL with similarities both in IGKV/IGLV repertoire and CDR3 formation, alluding to a different pathway of B-cell activation, not involving a classical germinal center (GC) reaction response to T-independent antigens.19-22,79 Finally, although the HCDR3 region is generally thought to play the key role in the formation of the antigen-binding site, our results point to the (complementary) importance of light chain CDR3 for antigen recognition by CLL BCRs. This is in keeping with results of previous reports in normal80 and autoreactive cells,81 with examples of antibodies creating a critical hydrogen bonding center through the use of specific light chain residues, thus providing the basis for a particular light chain sequence restriction for antibody specificity, while still allowing for extensive diversity in the heavy chain variable domain, including HCDR3.
Prepublished online as Blood First Edition Paper, August 2, 2005; DOI 10.1182/blood-2005-04-1511.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Prof Marie-Paule Lefranc and Dr Veronique Giudicelli (Laboratoire d'Immunogenetique Moleculaire, LIGM, Universite Montpellier II, UPR CNRS) for their support and generosity to share with us their insight on immunoglobulin gene analysis; Dr Paolo Ghia, Department of Oncology, Università Vita Salute-San Raffaele, Milano, Italy, and Dr Fred Davi, Laboratory of Hematology and University Paris 6, Hôpital Pitié-Salpètrière, Paris, France, for many stimulating talks and helpful comments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal