Abstract
To study natural killer (NK) cell–mediated antileukemic activity in chronic myelogenous leukemia (CML), we investigated the ability of HLA-matched and mismatched CD56+ cells to inhibit granulocyte macrophage–colony-forming unit (CFU-GM) formation by leukemic CD34+ cells. In 14 HLA-identical donor-recipient pairs, donor CD56+ cells inhibited CML CFU-GM comparably to effectors from 14 HLA-mismatched unrelated individuals (mean inhibition 42% ± 9% vs 39.5% ± 7% at a 10:1 effector-to-target (E/T) ratio), suggesting that killer inhibitory receptor (KIR) incompatibility was not essential for an antileukemic effect. Both CD56+CD3- (natural killer [NK]) and CD56+CD3+(NK-T) cells inhibited CFU-GM growth of CML but not normal CD34+ cells. A mechanism for this leukemia-specific cytotoxicity was suggested by the abnormal overexpression of major histocompatibility class I chain–related gene A or gene B (MICA/B) on CML CD34 cells and their ability to bind the NK activation ligand NKG2D. However, in vivo, CML cells may avoid NK-cell–mediated immune destruction by immune escape, shedding MICA into the plasma, thereby down-regulating NKG2D on CML CD56+ cells.
Introduction
Clinical results of allogeneic stem cell transplantation indicate that chronic myelogenous leukemia (CML) is particularly susceptible to an allogeneic graft-versus-leukemia (GVL) effect. The GVL effect is believed to be mediated in part by alloreacting T cells recognizing minor histocompatibility antigen disparities between donor and recipient,1 and in part by natural killer (NK) and NK-T cells.2-5 Both NK and NK-T cells express the CD56 antigen, a 140-kDa isoform of the neural-cell adhesion molecule (NCAM) and both cell types show major histocompatibility complex (MHC)–unrestricted cytotoxicity against tumor cells. Cytotoxicity is regulated by a balanced activation of natural cytotoxic receptors (NCRs)6-8 and anticytotoxic killer inhibitory receptors (KIRs).9-11 In humans, KIRs are composed of immunoglobulin-like receptors, which bind to specific HLA class I molecules.11 In particular, KIR2DL1 and KIR2DL2 bind 2 distinct subsets of HLA-I Cw alleles, whereas KIR3DL1 binds Bw4 allele.12,13 In humans, the only well-characterized NCR is NKG2D,14 whose ligands are the nonclassic MHC molecules major histocompatibility class I chain–related gene A or gene B (MICA/B)15 and the UL16 binding proteins (ULBPs).16,17
In the context of allogeneic stem cell transplantation, donor NK cells can exert antileukemic cytotoxicity through disparity between recipient MHC class and KIRs on donor NK cells (the missing self hypothesis),18,19 and in HLA-mismatched transplants, KIR disparity is associated with powerful GVL effects against myelogenous leukemia.18 NK-cell–mediated GVL effects have also been described in HLA-identical stem cell transplants for CML, but the mechanism for antileukemic cytotoxicity in a KIR-matched setting is not well understood.20,21 In mice, transfected NKG2D-activating ligands in syngeneic tumor cells are sufficient to generate a graft-versus-tumor effect, suggesting that NK-activating ligands on tumor cells can overcome KIR inhibitory signals.22 Since NK-cell–activating ligands MICA/B are also widely expressed by myeloid leukemia and melanoma cell lines,23,24 we hypothesized that GVL effects in CML might occur through this activation pathway in KIR-matched as well as KIR-mismatched donor recipient pairs. Here we present evidence that the strong cytotoxicity to CML progenitor cells of KIR-compatible CD56+ effector cells is mediated, at least in part, through NKG2D.
Patients, materials, and methods
Patients and healthy subjects
Lymphocytes and CD34 cells were obtained from patients with CML, their healthy stem-cell donors, and other family members. Subjects gave written informed consent for donating cell samples by venepuncture or leukapheresis. Donors gave permission for a proportion of their peripheral-blood stem-cell (PBSC) transplant product to be used for research. All samples were collected under institutional review board–approved National Heart Lung and Blood Institute (NHLBI) clinical research protocols. Collected cells were divided into aliquots and cryopreserved in liquid nitrogen in 10% dimethyl sulfoxide (DMSO) until use.
Antibodies and reagents
Fluoroscein isothiocyanate (FITC)–conjugated anti-CD56, anti-FcγRIII (CD16), anti-CD3, anti-KIR2DL1, anti-KIR3DL1, anti-CD94, anti–HLA class I antigen, phycoerythrin (PE)–conjugated anti-CD56, anti-CD16 mAb, matching isotype mouse mAb, Z-VAD-FMK, and Z-FA-FMK were purchased from BD Bioscience (San Jose, CA). PE-conjugated anti-CD34, anti-KIR2DL1, anti-KIR2DL2, and anti-NKG2A monoclonal antibodies (mAbs) were purchased from Beckman Coulter (Miami, FL). Magnetic bead–conjugated anti-CD34 and anti-CD56 mAbs and NK isolation kit “MiniMacs” magnet were purchased from Miltenyi Biotec (Auburn, CA). Allophycocyanin (APC)–conjugated anti-FAS (CD95) and PE-conjugated anti-FAS ligand (CD95L) mAbs were purchased from Caltag (Burlingame, CA). PE-conjugated and anti–human NKG2D mAb and a human recombinant NKG2D/Fc chimera were purchased from R&D Systems (Minneapolis, MN). Purified rabbit anti-MICA/B antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Interleukin-15 (IL-15), FLT3 ligand (FLT3-L), granulocyte macrophage–colony-stimulating factor (GM-CSF), and stem-cell factor (SCF) were purchased from Peprotech (Rocky Hill, NJ). Semisolid medium (MethoCult with or without recombinant cytokines) was purchased from Stem Cell Technologies (Vancouver, BC, Canada). Calcein AM was purchased from Molecular Probes (Eugene, OR).
Cell isolation, activation, and expansion
Isolation of total CD56+cells (NK-T and NK). Cryopreserved peripheral-blood mononuclear-cell (PBMCs) collections were thawed, mixed with anti-CD56 mAb magnetic beads, and passed through a magnetic column (Miltenyi Biotech, Auburn, CA). CD56+ cells retained in the column were then eluted.
Isolation of NK cells. Peripheral-blood NK cells were negatively selected from PBMCs by magnetic sorting using the Mini MACS NK isolation kit (Miltenyi Biotech). Positively and negatively selected CD56+ cells or NK cells were stimulated in vitro in RPMI 1640 medium supplemented with 10% fetal calf serum and 2 mM glutamine, hereafter referred to as complete medium. In experiments to measure NKG2D expression, cells were cultured in serum-free medium (X-VIVO 10; Cambrex, East Rutherford, NJ). CD56+CD3+ and CD3- subsets from PBMCs were obtained by labeling with a PE–anti-CD56 and an FITC–anti-CD3 mAb and electronic sorting using an EPICS ALTRA flow cytometer (Beckman Coulter). Cells were cultured in complete medium supplemented with IL-2 (200 U/mL) and IL-15 (50 ng/mL) until tested.
Isolation of CD34+cells. Patient's bone marrow or PBMCs were thawed, incubated with anti-CD34 mAb–conjugated magnetic beads (Miltenyi Biotech), and positively selected as described in the previous paragraph. CD34+ cells were positively selected from healthy donor G-CSF–mobilized peripheral-blood stem cells (PBSCs) using an Isolex 300i cell separator (Baxter, Irving, CA). In the same process, residual lymphocytes were removed by negative selection using a cocktail of anti-CD2, anti-CD6, and anti-CD7 mAbs to produce a stem-cell product with fewer than 0.001% CD3+ cells.
Acid stripping of membrane-bound HLA class I antigens
The removal of membrane-bound HLA- class I molecules from viable cells has been described.25 After thawing and washing once in complete medium, one million CD34+ cells from healthy subjects or patients with CML were incubated for 2 minutes at 4°C with a pH 3.0 buffer solution (citric acid–NA2PO4, 0.263 M and Na2HPO4 0.123 M, bovine serum albumin [BSA] 1%). The buffer was then neutralized by adding an excess of complete medium. HLA- class I antigen down-regulation was verified by flow cytometry using W6/32 mAb (FACS Caliber; BD Biosciences).
Colony-inhibition assay
CD34+ cells in complete medium were plated in duplicate in the presence or absence of effector cells at an effector-to-target (E/T) ratio of 10:1 in 96-well U-bottom plates. After 5 hours of incubation at 37°C, in 5% CO2, cells were transferred to 6-well plates containing methylcellulose (Methcult; Stem Cell Technologies) supplemented with GM-CSF, G-CSF, and SCF. Colonies (groups of 50 or more cells) were counted after 14 days. Results of duplicates target/effector combination were expressed as mean plus or minus standard deviation (SD).
Cytotoxicity assay
Replicates of 20 μL effector-cell suspension were serially diluted and incubated in 60-well (40 μL) plates (NalgeNunc International, Rochester, NY) for 30 minutes at room temperature. At the same time, 1 × 106 to 2 × 106 target cells were incubated in 1 mL complete medium supplemented with 10 μL Calcein-AM (Molecular Probes, Junction City, OR) for 30 minutes at 37°C, washed 4 times, and diluted to 1 × 105/mL. Subsequently, 10 μL target-cell suspension was added. Plates were then centrifuged and incubated at 37°C for 4 hours. A few minutes before scanning the plates, a fluorescent detector of 5 μL fluoro-quench was added to each well. The percent of lysis was calculated as follows: 1 - (mean test - mean blank) / (mean max - mean blank) × 100.
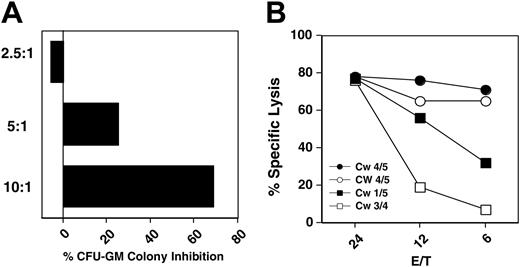
Antileukemia effect of autologous, HLA-matched and -mismatched CD56+ cells. (A) CML CFU-GM colony inhibition by IL-2–activated HLA-identical donor CD56+ cells at different effector-to-target ratios (patient 15). (B) Cytotoxicity (Calcein-AM release) of HLA-matched sibling transplant donor (•), recipient after transplantation (○), mother (▪), and father (□) CD56+ cells on CD34+-CML recipient cells (patient 3).
Antileukemia effect of autologous, HLA-matched and -mismatched CD56+ cells. (A) CML CFU-GM colony inhibition by IL-2–activated HLA-identical donor CD56+ cells at different effector-to-target ratios (patient 15). (B) Cytotoxicity (Calcein-AM release) of HLA-matched sibling transplant donor (•), recipient after transplantation (○), mother (▪), and father (□) CD56+ cells on CD34+-CML recipient cells (patient 3).
Flow cytometry analysis of NKG2D ligand expression on CD34+ cells
Normal and CML CD34+ cells were incubated on ice with rabbit or goat anti-MICA/B antibodies for 30 minutes. Goat or rabbit immunoglobulin G (IgG) were used as negative controls. After 2 washes, cell-surface–bound antibodies were detected using fluorescein-conjugated antibodies. The direct binding of NKG2D/Fc chimeric molecule was detected using a PE-conjugated anti-NKG2D mAb.
Detection of MICA in serum
The anti-MICA mAb, WW2G8 (IgG1) was generated in our laboratory (W.W., manuscript in preparation) using the methodology of Kohler and Milstein.26 The specificity of WW2G8 mAb was determined on bacteria lysate containing recombinant MICA or MICB by Western blot and on MICA+ or MICA- cell lines by cell enzyme-linked immunosorbent assay (ELISA) or flow cytometry analysis. Patient and normal sera were diluted 1:2 in phosphate-buffered saline (PBS). Quadruplicates of 100 μL diluted sera were coated overnight in 96-well U-bottom plates at 4°C. Wells were washed 4 times in PBS supplemented with 0.05% Tween 20 (PBS-T; Sigma, St Louis, MO). Subsequently, 100 μL biotynolated WW2G8 (1 μg/mL) in PBS, 1% BSA, was added to the wells. After a 2-hour incubation at room temperature, wells were washed 4 times with PBS-T. Then 100 μL horseradish peroxidase (HRP)–conjugated streptavidin (Pierce, Rockford, IL) was added to the wells for 1 hour at room temperature. Wells were washed 6 times with PBS-T. Finally, 100 μL TMB (3,3′,5,5′-tetramethylbenzidine) Microwell Peroxidase Substrate (KPL, Gaithersburg, MD) was added to the wells for 30 minutes at room temperature, in the dark. The substrate reaction was stopped with 50 μL of 1.8 M H2SO4. Within 1 to 2 hours after stop reaction, plates were read at a wavelength of 450 nm.
Results
Characteristics of study patients with CML
Table 1 lists the characteristics, HLA type, and predicted KIR compatibility of cells from patients with CML used as CD34+ leukemia targets of CD56+ effectors from autologous, HLA-identical sibling, related, or unrelated HLA-mismatched individuals. Serum from an additional 5 patients was examined for soluble MICA. Patients selected for individual experiments described below are indicated. All patient CD34+ CML cells were obtained from pretransplant collections. Most patients studied were in chronic phase with blood counts controlled by imatinib or hydroxyurea. There were 2 patients who had an atypical Ph-negative CML.
Characteristics of study patients with CML
Subject . | Time since diagnosis, mo . | WBCs × 109/L . | Diagnosis . | Age/sex . | HLA-Cw(Bw4+) . | HLA-C group . | Study performed . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 38 | CML-AP | 31/F | 04/07* (-) | 2/1 | 1 |
| 2 | 12 | 33.5 | CML-CP | 23/F | 05/07* (-) | 2/1 | 1,2 |
| 3 | 4 | 12 | CML-CP | 29/M | 04/05* (-) | 2/2 | 1,2,3 |
| 4 | 6 | 3.5 | CML-CP | 32/M | 03/07 (-) | 1/1 | 1,3,5 |
| 5 | 24 | 23 | CML-CP | 26/M | 04/15 (-) | 2/2 | 2,3,5 |
| 6 | 6 | 112 | CML-CP | 23/M | 07/16 (-) | 1/NA | 1,2 |
| 7 | 5 | 3 | CML-CP | 31/M | 07/17 (-) | 1/2 | 1,2 |
| 8 | 5 | 9.0 | CML-CP | 19/M | 04/04 (-) | 2/2 | 2 |
| 9 | 9 | 23 | CML-CP | 39/M | 06/07 (-) | 2/1 | 1,2 |
| 10 | 4 | 14 | CML-CP | 30/F | 7/ND (-) | 1 | 1,2 |
| 11 | 22 | 38 | CML-CP | 20/M | 04/05 (-) | 2/2 | 2,5 |
| 12 | 18 | 5.7 | CML-CP | 19/F | 04/05 (-) | 2/2 | 2 |
| 13 | 5 | 2.4 | CML-CP | 29/F | 03/08 (-) | 1/1 | 2 |
| 14 | 18 | 26 | CML-Ph- | 18/M | 02/12 | 2/1 | 1,2 |
| 15 | 9.5 | 6.8 | CML-CP | 18/F | 04/07 | 1/2 | 1,5 |
| 16 | 54 | 41.2 | CML-AP | 44/F | ND | ND | 4 |
| 17 | 6 | 5.6 | CML-CP | 28/M | ND | ND | 4 |
| 18 | 36 | 3.9 | CML-CP | 53/F | ND | ND | 4 |
| 19 | 31 | 31.7 | CML-CP | 39/F | ND | ND | 4 |
| 20 | 28 | 11.6 | CML-Ph- | 54/M | ND | ND | 4 |
| 21 | 32 | 2.5 | CML-CP | 28/F | 03/08 | 1/1 | 1,2 |
| 22 | 22 | 16.8 | CML-CP | 21/M | 06/15 (-) | 2/2 | 2 |
Subject . | Time since diagnosis, mo . | WBCs × 109/L . | Diagnosis . | Age/sex . | HLA-Cw(Bw4+) . | HLA-C group . | Study performed . |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 38 | CML-AP | 31/F | 04/07* (-) | 2/1 | 1 |
| 2 | 12 | 33.5 | CML-CP | 23/F | 05/07* (-) | 2/1 | 1,2 |
| 3 | 4 | 12 | CML-CP | 29/M | 04/05* (-) | 2/2 | 1,2,3 |
| 4 | 6 | 3.5 | CML-CP | 32/M | 03/07 (-) | 1/1 | 1,3,5 |
| 5 | 24 | 23 | CML-CP | 26/M | 04/15 (-) | 2/2 | 2,3,5 |
| 6 | 6 | 112 | CML-CP | 23/M | 07/16 (-) | 1/NA | 1,2 |
| 7 | 5 | 3 | CML-CP | 31/M | 07/17 (-) | 1/2 | 1,2 |
| 8 | 5 | 9.0 | CML-CP | 19/M | 04/04 (-) | 2/2 | 2 |
| 9 | 9 | 23 | CML-CP | 39/M | 06/07 (-) | 2/1 | 1,2 |
| 10 | 4 | 14 | CML-CP | 30/F | 7/ND (-) | 1 | 1,2 |
| 11 | 22 | 38 | CML-CP | 20/M | 04/05 (-) | 2/2 | 2,5 |
| 12 | 18 | 5.7 | CML-CP | 19/F | 04/05 (-) | 2/2 | 2 |
| 13 | 5 | 2.4 | CML-CP | 29/F | 03/08 (-) | 1/1 | 2 |
| 14 | 18 | 26 | CML-Ph- | 18/M | 02/12 | 2/1 | 1,2 |
| 15 | 9.5 | 6.8 | CML-CP | 18/F | 04/07 | 1/2 | 1,5 |
| 16 | 54 | 41.2 | CML-AP | 44/F | ND | ND | 4 |
| 17 | 6 | 5.6 | CML-CP | 28/M | ND | ND | 4 |
| 18 | 36 | 3.9 | CML-CP | 53/F | ND | ND | 4 |
| 19 | 31 | 31.7 | CML-CP | 39/F | ND | ND | 4 |
| 20 | 28 | 11.6 | CML-Ph- | 54/M | ND | ND | 4 |
| 21 | 32 | 2.5 | CML-CP | 28/F | 03/08 | 1/1 | 1,2 |
| 22 | 22 | 16.8 | CML-CP | 21/M | 06/15 (-) | 2/2 | 2 |
Due to the limited availability of the biologic material, the number of the assays performed for single patients vary.
CP indicates chronic phase; AP, accelerated phase; Ph-, Philadelphia chromosome; NA, not available; ND, not done; 1, CD34 CML for colony inhibition assays with matched and mismatched effectors with or without acid stripping; 2, CD34 CML cells for MICA/B and NKG2D labeling; 3, CD34 CML cells for cytotoxicity tests; 4, serum assay for MICA/B; 5, CD34 CML cells for proliferation assay.
Antileukemia effects of autologous, HLA-matched and -mismatched CD56+ effectors on CD34+ CML cells
When IL-2–stimulated HLA-matched CD56+ cells were cultured with recipient CD34+ CML cells (patient 15) CFU-GM colony formation was inhibited in dose-dependent manner (Figure 1).
Family study. To further investigate the antileukemia activity of HLA-matched CD56+ cells, we first evaluated HLA-Cw alleles and the expression of KIR2DL1, KIR2DL2, and KIR2DL3 in peripheral-blood lymphocytes (PBLs) of the family members of patient 3 (Table 2). Subsequently, the patient's CD34+ cells were cultured with HLA-matched and -mismatched CD56+ cells from family members in a 4-hour cytotoxicity assay. The patient and the HLA-identical stem-cell donor expressed HLA-Cw4 and Cw5 haplotypes (group 2 NK specificity), whereas the patient's parents expressed Cw1 and Cw5 or Cw3 and Cw4 haplotypes (groups 1 and 2 NK specificity) allowing a comparison of KIR-matched and -mismatched effectors on the same target. Surprisingly, HLA-matched CD56+ cells, obtained either directly from the donor or from the recipient following 100% donor engraftment, were more cytotoxic to the recipient CD34+ CML cells than mismatched CD56+ cells (Figure 1).
Phenotypic analysis of KIR expression on CD56+ cells of the family shown inFigure 1
. | . | . | % CD56+ . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | HLA-Cw . | HLA-C group . | KIR2DL1 . | KIR2DL2 . | KIR3DL1 . | KIR2DL2/KIR2DL1 . | |||
| Donor | 04/05 | 2/2 | 5 | 12 | 5 | 3.0/1 | |||
| Recipient | 04/05 | 2/2 | 4 | 11 | 7 | 2.8/1 | |||
| Father | 01/05 | 1/2 | 23 | 26 | 8 | 1.1/1 | |||
| Mother | 03/04 | 1/2 | 8 | 8 | 8 | 1.0/1 | |||
. | . | . | % CD56+ . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
. | HLA-Cw . | HLA-C group . | KIR2DL1 . | KIR2DL2 . | KIR3DL1 . | KIR2DL2/KIR2DL1 . | |||
| Donor | 04/05 | 2/2 | 5 | 12 | 5 | 3.0/1 | |||
| Recipient | 04/05 | 2/2 | 4 | 11 | 7 | 2.8/1 | |||
| Father | 01/05 | 1/2 | 23 | 26 | 8 | 1.1/1 | |||
| Mother | 03/04 | 1/2 | 8 | 8 | 8 | 1.0/1 | |||
HLA-Cw4 and Cw5 bind KIR2DL1; HLA-Cw 1 binds KIR2DL2; KIR3DL1 binds HLA-Bw4.
CFU-GM inhibition. The antileukemic activity of HLA-matched and -mismatched CD56+ LAK cells against CD34+ CML cells was then compared by the colony inhibition assay. In 14 paired comparisons of HLA-matched and -mismatched effectors against CML targets, there was no significant difference in the mean CFU-GM inhibition by HLA-matched (11 allogeneic, 3 autologous effectors), or HLA-mismatched CD56+ cells (Table 3), indicating that HLA-matched and -mismatched CD56+ cells had comparable antileukemic activity.
Colony-inhibition assays
Effector . | HLA match of E/T (n) . | N . | Target CD34+ . | % colony inhibition . | *P . |
|---|---|---|---|---|---|
| CD56+ donor, CD56+ autologous | Matched | 11,3 | CML | 42 ± 9 | NS |
| CD56+ donor | Mismatched | 14 | CML | 39 ± 7 | NS |
| CD56+ donor | Matched (11); mismatched (4) | 15 | CML class I–negative, acid stripped | 69.8 ± 7 | .01 |
| CD56+ donor | Matched (11); mismatched (4) | 15 | CML class I–positive | 39.6 ± 10 | .01 |
| CD56+CD3- (NK) | Auto (1), matched (1) | 3 | CML | 69.1 ± 12 | NS |
| CD56+ CD3+ (NKT) | Mismatched (1) | 3 | CML | 68 ± 16 | NS |
| CD56+ donor, HLA-C group homozygous | Matched (4) | 4 | CML | 55.5 ± 10 | NS |
| CD56+ donor, HLA-C group heterozygous | Matched (4) | 4 | CML | 53 ± 7 | NS |
Effector . | HLA match of E/T (n) . | N . | Target CD34+ . | % colony inhibition . | *P . |
|---|---|---|---|---|---|
| CD56+ donor, CD56+ autologous | Matched | 11,3 | CML | 42 ± 9 | NS |
| CD56+ donor | Mismatched | 14 | CML | 39 ± 7 | NS |
| CD56+ donor | Matched (11); mismatched (4) | 15 | CML class I–negative, acid stripped | 69.8 ± 7 | .01 |
| CD56+ donor | Matched (11); mismatched (4) | 15 | CML class I–positive | 39.6 ± 10 | .01 |
| CD56+CD3- (NK) | Auto (1), matched (1) | 3 | CML | 69.1 ± 12 | NS |
| CD56+ CD3+ (NKT) | Mismatched (1) | 3 | CML | 68 ± 16 | NS |
| CD56+ donor, HLA-C group homozygous | Matched (4) | 4 | CML | 55.5 ± 10 | NS |
| CD56+ donor, HLA-C group heterozygous | Matched (4) | 4 | CML | 53 ± 7 | NS |
Colony inhibition calculated as mean CFU-GM plated after 4-hour CD56+ incubation / mean CFU-GM after 4 hours without CD56+ cells. The CML CD34+ cell dose was selected to obtain 40 to 100 colonies/plate (mean of duplicate plates).
CD56+ indicates NK+NK-T cells sorted from leukapheresis product or PB; E/T, effector-to-target.
Paired t test
Effect of HLA class I antigen expression on antileukemia effect of HLA-matched CD56+ cells
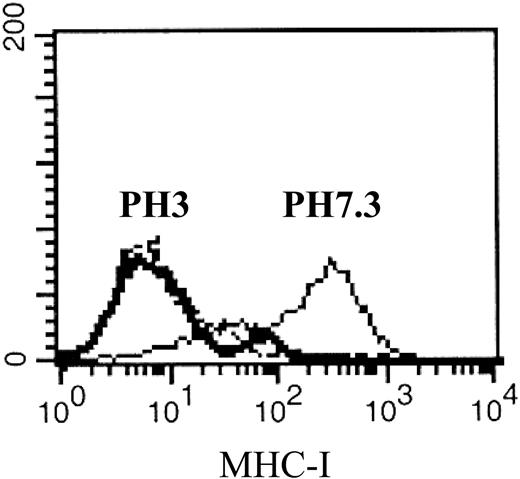
To evaluate the contribution of HLA class I antigens on CML CD34+ cells to CD56+-cell–mediated cytotoxicity, HLA class I antigen expression by CD34 cells was reduced by acid stripping. Most CML-cell samples lost surface HLA class I antigen expression within 2 minutes (without affecting CD34 expression; Figure 2). In 11 paired comparisons of colony inhibition by matched (n = 8) or mismatched (n = 3) CD56+ effectors against untreated or acid-stripped CML CD34+ cells, colony inhibition was significantly greater in the HLA-deficient targets (P = .01, Wilcoxon), indicating that target-cell cytotoxicity was in part blocked by KIR–HLA class I antigen interactions (Table 3). Since HLA class I antigens and KIRs are inherited separately, it is theoretically possible that CFU-GM inhibition could occur between HLA-identical effector-target pairs if individual NK clones express “orphan KIRs,” which do not bind to a cognate HLA class I ligand on the target. To test this possibility, the CFU-GM inhibitory effect of HLA-identical donors was compared in donor-recipient pairs either homozygous for HLA class I antigen group 1 or 2 (n = 4), or heterozygous (expressing both group 1 and group 2 HLA class I antigens; n = 4). We found no difference in the inhibition of recipient CFU-GM between HLA-C group heterozygous or homozygous effectors (Table 3). These results eliminate an important role for autologous antileukemic effects through an orphan KIR mechanism.
Antileukemia activity of CD56+ subsets
Peripheral-blood CD56+ cells include a major subset of CD56+CD3- NK cells and a minor subset of CD56+CD3+ NK-T cells. To characterize the antileukemia activity of these subpopulations, CD56+CD3- and CD56+CD3+ cells were flow-sorted from the HLA-matched donors and leukemia patient 7. CML CFU-GMs were comparably inhibited by NK (CD56+, CD3-) and NK-T cells (CD56+, CD3+) from either the autologous source or from the HLA-identical donor (Figure 3A-B).
Reduced HLA class I antigen expression on CD34+ CML cells following acid stripping. Cryopreserved bone marrow cells obtained from CML patient 2 were thawed and CD34+ cells were isolated and cultured in 1% BSA (solid line) or acid-stripped at pH 3.0 (bold solid line). An IgG1 isotype was used as a negative control (dashed line).
Reduced HLA class I antigen expression on CD34+ CML cells following acid stripping. Cryopreserved bone marrow cells obtained from CML patient 2 were thawed and CD34+ cells were isolated and cultured in 1% BSA (solid line) or acid-stripped at pH 3.0 (bold solid line). An IgG1 isotype was used as a negative control (dashed line).
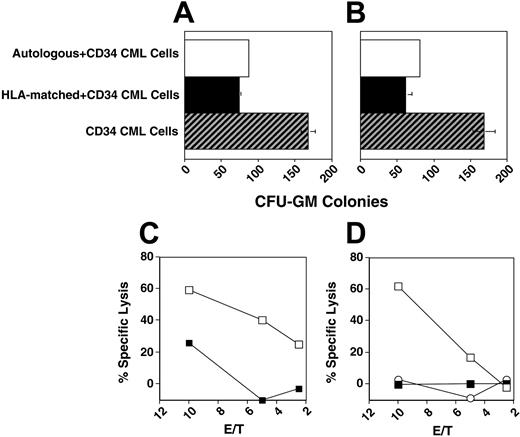
HLA-matched CD56+ effectors have leukemia-restricted CD34 cytotoxicity
To evaluate the specificity of the cytotoxic activity of HLA-matched CD56+ cells, we tested the effect of donor CD56+ cells in colony inhibition and direct cytotoxicity tests on the respective recipient CD34+ CML cells of patients 4 and 5 or on autologous CD34+ cells. The HLA-Cw alleles and KIR phenotype of HLA-matched CD56+ cells of patients 4 and 5 are shown in Table 4. Both HLA-matched donors showed cytotoxicity to patients' CD34+ CML cells but not to normal autologous CD34+ cells, suggesting that only CML CD34+ cells expressed NK-activating molecules capable of overcoming KIR inhibition (Figure 3). Similarly, sorted and irradiated CD56+ cells, as well as the CD56+-cell subpopulations KIR2DL1-KIR2DL2– and KIR2DL1-KIR2DL2+, obtained in 2 separate experiments, inhibited the proliferation of CD34+ CML cells of patient 11, but had minimal or no inhibition against autologous (nonleukemic) CD34+ cells (data not shown).
Phenotypic analysis of KIR expression on CD56+ cells of HLA-matched donors used as effector cells inFigure 3C-D
. | . | . | % CD56+ . | . | . | ||
|---|---|---|---|---|---|---|---|
| Donor . | HLA-Cw . | HLA-C group . | KIR2DL1 . | KIR2DL2 . | KIR2DL2/KIR2DL1 ratio . | ||
| Patient 4 | 03/07 | 1/1 | 7 | 10 | 1.4 | ||
| Patient 5 | 04/15 | 2/2 | 8 | 4.5 | 0.6 | ||
. | . | . | % CD56+ . | . | . | ||
|---|---|---|---|---|---|---|---|
| Donor . | HLA-Cw . | HLA-C group . | KIR2DL1 . | KIR2DL2 . | KIR2DL2/KIR2DL1 ratio . | ||
| Patient 4 | 03/07 | 1/1 | 7 | 10 | 1.4 | ||
| Patient 5 | 04/15 | 2/2 | 8 | 4.5 | 0.6 | ||
Expression of MICA/B on CD34+ CML cells
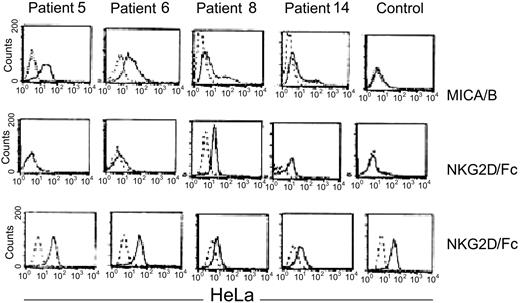
To explore the possibility that CML cells express NK-activating ligands, we studied the expression of the NKG2D ligand MICA/B on CML CD34+ cells. MICA/B was expressed by CD34+ cells in 10 of 12 CML samples studied. Of the 4 MICA/B-positive patients tested, 2 showed strong binding of NKG2D; 1 bound weakly and 1 did not bind, possibly indicating functional polymorphism in MICA/B alleles. In contrast, normal CD34+ cells neither expressed MICA/B nor bound the NKG2D/Fc chimeric molecule, whereas the MICA+ HeLa-cell–positive control bound NKG2D/Fc (Figure 4).
HLA-matched CD56+ cells show cytotoxicity restricted to leukemic CD34+ cells. CD56+ cells were cultured with either recipient CD34+ CML cells or autologous CD34+ cells. (A-B) Colony inhibition assay of electronically sorted autologous or HLA-matched NK or NK-T cells as indicated on patient 7 CD34+ CML cells. (C) Cytotoxicity assay of IL-2–stimulated HLA-matched donor CD56+ cells on CD34+ CML cells of patient 4 (□) or autologous normal CD34+ cells (▪). (D) Cytotoxicity assay of IL-2– and IL-15–stimulated donor HLA-matched CD56+ cells on CD34+ CML cells of patient (□) or autologous normal CD34+ cells (▪). IL-2–stimulated CD56-negative PBLs versus recipient CD34+ CML cells indicated by ○. Error bars indicate plus or minus standard deviation (SD).
HLA-matched CD56+ cells show cytotoxicity restricted to leukemic CD34+ cells. CD56+ cells were cultured with either recipient CD34+ CML cells or autologous CD34+ cells. (A-B) Colony inhibition assay of electronically sorted autologous or HLA-matched NK or NK-T cells as indicated on patient 7 CD34+ CML cells. (C) Cytotoxicity assay of IL-2–stimulated HLA-matched donor CD56+ cells on CD34+ CML cells of patient 4 (□) or autologous normal CD34+ cells (▪). (D) Cytotoxicity assay of IL-2– and IL-15–stimulated donor HLA-matched CD56+ cells on CD34+ CML cells of patient (□) or autologous normal CD34+ cells (▪). IL-2–stimulated CD56-negative PBLs versus recipient CD34+ CML cells indicated by ○. Error bars indicate plus or minus standard deviation (SD).
NKG2D expression
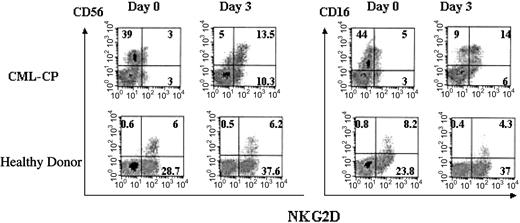
To address the paradox that CML cells evidently persist in vivo despite the susceptibility of CML cells to NKG2D-mediated cytotoxicity in vitro, we examined NKG2D expression by NK cells of patients with CML. Three of 4 patients with CML (patients 10, 12, and 13) showed reduced expression of NKG2D by CD56+CD3- and also CD56+CD3+ lymphocytes. In contrast, most healthy donor NK cells strongly expressed NKG2D. Reduced NKG2D expression appeared to be mediated by ligand-induced down-regulation, since expression by CML PBLs was restored after a 3-day culture in serum-free medium and IL-2 (Figure 5).
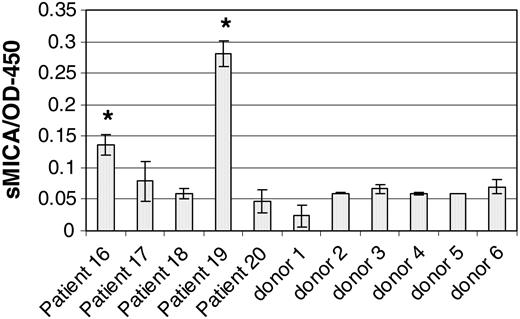
Shedding of MICA/B by CML cells and subsequent blocking of NKG2D on NK cells is another mechanism that might reduce in vivo autoreactivity against CML. We therefore assayed MICA in the serum of patients with CML and in healthy controls. Figure 6 shows that 2 of 5 patients with CML (patients 16 and 19) had detectable serum MICA and both had high leukocyte counts. In contrast, all of the normal sera tested had background levels of soluble MICA. This suggests that in vivo CML cells may escape NK-mediated cytotoxicity by shedding soluble MICA, thereby down-regulating NK cells by NKG2D blockade.
Expression of NKG2D ligands MICA/B on CD34 CML cells. From left to right: patients 5, 6, 8, and 14. Top panel: CD34+ CML cells and normal CD34+ cells were stained with polyclonal anti-MICA/B (solid line) or isotype antibodies (dashed line). Middle panel: direct binding of an NKG2D/Fc chimeric molecule to the CD34 CML cells from the indicated patients (solid lines). The bound NKG2D/Fc was detected by a PE-conjugated anti-NKG2D that was also used as a negative control (dashed lines). Lower panel: direct binding of NKG2D/Fc (solid line) to MICA+ HeLa cells or PE-conjugated anti-NKG2D used as negative control (dashed lines). HeLa cells were used as an internal positive control for each patient indicated in the middle panel (variability of HeLa staining may reflect batch or dose variation in NKG2D/Fc antibody).
Expression of NKG2D ligands MICA/B on CD34 CML cells. From left to right: patients 5, 6, 8, and 14. Top panel: CD34+ CML cells and normal CD34+ cells were stained with polyclonal anti-MICA/B (solid line) or isotype antibodies (dashed line). Middle panel: direct binding of an NKG2D/Fc chimeric molecule to the CD34 CML cells from the indicated patients (solid lines). The bound NKG2D/Fc was detected by a PE-conjugated anti-NKG2D that was also used as a negative control (dashed lines). Lower panel: direct binding of NKG2D/Fc (solid line) to MICA+ HeLa cells or PE-conjugated anti-NKG2D used as negative control (dashed lines). HeLa cells were used as an internal positive control for each patient indicated in the middle panel (variability of HeLa staining may reflect batch or dose variation in NKG2D/Fc antibody).
Mechanism of NKG2D-mediated cytotoxicity against CML CD34+ cells
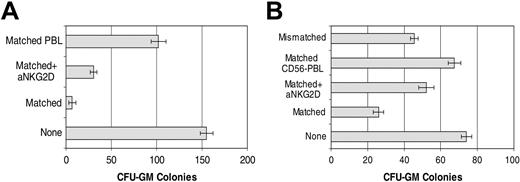
Cytotoxicity by CD56+ cells was compared in 2 selected patients according to positivity for MICA/B expression (patient 2) or high affinity for NKG2D/Fc chimeric molecule (patient 14) and, using HLA-matched and -mismatched (KIR incompatible) cells as effectors. To exclude a role for granule-independent cytotoxicity, experiments were carried out in the presence of Z-VAD-FMK, a general caspase inhibitor. CFU-GM colony formation in patients with CML exhibiting high NKG2D binding was clearly inhibited. Furthermore, this inhibition was partially reversed by an anti-NKG2D blocking antibody. These results suggest a role for NKG2D-ligand interaction in the regulation of HLA-matched CD56+-cell recognition of CML cells (Figure 7).
Discussion
Pioneering observations by Ruggeri and colleagues in the last few years have reawakened interest in the possibility of using NK-cell–based strategies to treat leukemia. These investigators have defined the KIR ligand disparities between recipient leukemia and donor NK cells required to elicit alloreactive cytotoxicity against leukemia cells.18 However, their results, derived from mismatched allogeneic stem cell transplantation studies, have to be reconciled with earlier observations that some leukemias are susceptible to autologous inhibition by NK cells, and that NK-cell susceptibility of leukemic cell lines is highly variable.27 These latter observations suggest that activating ligands on leukemia cells could counteract KIR-mediated inhibition and permit target-cell susceptibility in the absence of KIR incompatibility.
We chose to investigate the antileukemic potential of all CD56+-cell subsets in the context of CML, because previous studies showed that donor NK- and LAK-cell cytotoxicity can be readily generated against recipient CML in HLA-matched pairs, and NK-cell–mediated cytotoxicity against CML cells can be measured both by bulk cytotoxicity and by the CFU-GM inhibition assay.4 Furthermore, we previously described a correlation between NK/LAK activity after transplantation, and the probability of sustained molecular remission in patients with CML.21 Thus, CML represents an informative model for studying mechanisms of NK cytotoxicity in autologous, HLA-matched, and -mismatched combinations of NK effectors with leukemia targets. In preliminary experiments, HLA-identical NK cells from healthy donors were found to inhibit CFU-GM growth in a dose-dependent fashion (Figure 1). Somewhat surprisingly, in a family study, the NK-cell–mediated cytotoxicity against CML from the HLA-matched donor was superior to that of the other HLA-mismatched, KIR ligand–mismatched family members. Furthermore, there was no significant difference between NK-cell–mediated colony inhibition by the HLA-identical donor and by other partially matched or completely HLA-mismatched NK-cell donors (Table 2), suggesting that the KIR inhibitory pathway was either inoperative or counteracted by procytotoxic effector-target interactions. Nevertheless, after removal of HLA class I antigen by acid stripping, we exposed a KIR ligand inhibitory effect, because both matched and mismatched CML progenitors were rendered more susceptible to NK-cell cytotoxicity after acid stripping (Figure 2; Table 2). Although it is possible that acid treatment may have rendered the targets more susceptible through mechanisms other than MHC class I removal, these findings appear to confirm the importance of KIR interactions in modulating antileukemic cytotoxicity but indicate that KIR inhibition in the HLA-matched setting is nevertheless overcome by procytotoxic NK-CML interactions.
NKG2D expression by CD56+ cells of patients with CML and healthy donors. Cryopreserved PBLs of CML patient 10 or healthy donors were thawed and cultured in IL-2 and complete medium and analyzed directly or after 3 days (representative results from 3 patients with CML and 2 healthy donors). Numbers shown in each quadrant indicate the percentage of positive cells.
NKG2D expression by CD56+ cells of patients with CML and healthy donors. Cryopreserved PBLs of CML patient 10 or healthy donors were thawed and cultured in IL-2 and complete medium and analyzed directly or after 3 days (representative results from 3 patients with CML and 2 healthy donors). Numbers shown in each quadrant indicate the percentage of positive cells.
Detection of soluble MICA in CML sera. Soluble MICA was evaluated in the sera of 5 patients with CML and 6 healthy individuals, as indicated. MICA was detected by ELISA as described in “Patients, materials, and methods.” Leukocyte counts of the indicated patients are shown in Table 1. Error bars indicate plus or minus SD.
Detection of soluble MICA in CML sera. Soluble MICA was evaluated in the sera of 5 patients with CML and 6 healthy individuals, as indicated. MICA was detected by ELISA as described in “Patients, materials, and methods.” Leukocyte counts of the indicated patients are shown in Table 1. Error bars indicate plus or minus SD.
In further experiments, we showed that both CD56+ NK and NK-T subsets had comparable cytotoxicity to HLA-matched targets, confirming observations by Mackinnon et al suggesting that LAK cells have antileukemia activity.4 However, cytotoxicity and inhibition of proliferation of CD34+ cells by HLA-identical NK cells was restricted to CML and was not seen against autologous normal CD34 cells, suggesting that CML cells abnormally express NK-cell–activating ligands. Since MICA/B is the natural ligand for the NK-cell–activating molecule NKG2D, we looked for MICA/B expression by CML CD34 cells. We found MICA/B expression on CML but not on normal CD34 cells, which correlated with the ability to bind soluble NKG2D, implicating an NKG2D-MICA/B interaction in CML target susceptibility. In one patient with a high level of MICA/B expression and high affinity for NKG2D chimeric molecules, we showed by blocking studies with anti-NKG2D that NKG2D was the dominant mediator of cytotoxicity. However, killing of CML cells by HLA-matched CD56+ cells was also observed in 2 patients with low or absent expression of MICA/B, suggesting cytotoxicity through FAS/FASL interaction28 or through other NK-cell–activating ligands such as ULBP.17
Role of NKG2D in antileukemia activity of HLA-matched CD56+ cells. (A) CFU-GM colony inhibition assay of CD34+ CML cells from patient 2 by IL-2+IL-15–stimulated HLA-matched CD56+ cells in the presence or absence of anti-NKG2D mAb. IL-2+IL-15–stimulated HLA-matched PBLs served as a control. (B) CFU-GM colony inhibition assay of CD34+ CML cells from patient 14 by IL-2–stimulated HLA-matched CD56+ cells in the presence or absence of anti-NKG2D mAb. IL-2–stimulated HLA-matched CD56- cells and HLA-mismatched CD56+ cells were used as negative or positive controls, respectively. Error bars indicate plus or minus SD.
Role of NKG2D in antileukemia activity of HLA-matched CD56+ cells. (A) CFU-GM colony inhibition assay of CD34+ CML cells from patient 2 by IL-2+IL-15–stimulated HLA-matched CD56+ cells in the presence or absence of anti-NKG2D mAb. IL-2+IL-15–stimulated HLA-matched PBLs served as a control. (B) CFU-GM colony inhibition assay of CD34+ CML cells from patient 14 by IL-2–stimulated HLA-matched CD56+ cells in the presence or absence of anti-NKG2D mAb. IL-2–stimulated HLA-matched CD56- cells and HLA-mismatched CD56+ cells were used as negative or positive controls, respectively. Error bars indicate plus or minus SD.
These findings raised the paradox that while CML progenitor cells are apparently highly susceptible to NK-cell regulation in vitro, autologous NK cells are self-evidently unable to prevent the leukemic process in the patient. Since NK cells in CML have been extensively studied and found to be functional,27,29 we considered the possibility that leukemia cells avoid autologous NK cells through immune escape. Consistent with this was the finding that patients with CML have detectable and potentially high levels of MICA/B in their serum, as has been described in other malignancies.30,31 MICA/B shed in the serum could block NKG2D on CD56+ cells, thus preventing NK-cell activation by the target. In support of this possibility was the finding of reduced NKG2D expression by NK and NK-T cells from some patients with CML, which increased after incubation in serum-free medium and IL-2 (in the absence of soluble MICA/B).
The clinical evidence for an antileukemic potential of NK cells is compelling, but the context of achieving these effects through mismatched stem-cell transplants is problematic because of significant treatment-related risks. Since our results indicate that NK cells kill CD34+ CML cells in the presence of intact KIR–MHC class I inhibitory pathways, it might be possible to enhance autologous or HLA-matched NK–malignant-cell interactions to achieve the same degree of efficacy as KIR ligand–mismatched interactions, but with greater clinical safety. To achieve this goal, methods to overcome immune escape through MICA/B shedding and NKG2D blocking would have to be developed. Alternatively, agents that up-regulate MICA/B on the leukemia cell might be helpful. MICA/B has a limited distribution in normal tissues. MICA/B is preferentially expressed in the gastrointestinal tract and thymocortical epithelia,15 but its expression is also induced by cell stress, viral infection, and malignant transformation.32,33 One strategy would therefore be to provoke stress in CML cells by hyperthermia or with interferons, retinoic acid, or chemotherapeutic agents. Earlier studies demonstrated that pretreatment of leukemia cells with actinomycin D or metabolic inhibitors increases leukemia-cell sensitivity to NK cell–mediated cytotoxicity,34 but whether this effect is mediated by the up-regulation of NK cell–activating ligands on leukemia cells is not known.
Prepublished online as Blood First Edition Paper, July 26, 2005; DOI 10.1182/blood-2005-02-0479.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal