Abstract
Scott syndrome (SS) is a bleeding disorder characterized by a failure to expose phosphatidylserine (PS) to the outer leaflet of the platelet plasma membrane. Because the adenosine triphosphate (ATP)–binding cassette transporter A1 (ABCA1) is implicated in the exofacial translocation of PS, we assessed its role in the pathophysiology of a patient with SS. Substantially reduced levels of ABCA1 mRNA were found in the patient's leukocytes, compared with controls. The SS patient was heterozygous for a novel missense mutation c.6064G>A (ABCA1 R1925Q), absent from unaffected family members and controls. Both mutant and wild-type alleles were reduced in mRNA expression, and no causative mutation for this phenomenon was identified in the ABCA1 gene or its proximal promoter, suggesting a putative second mutation in a trans-acting regulatory gene may also be involved in the disorder in this patient. In vitro expression studies showed impaired trafficking of ABCA1 R1925Q to the plasma membrane. Overexpression of wild-type ABCA1 in SS lymphocytes complemented the Ca2+-dependent PS exposure at the cell surface. These data identify a mutation in ABCA1 that contributes to the defective PS translocation phenotype in our patient with SS.
Introduction
Scott syndrome (SS) is a rare, moderately severe bleeding disorder (Online Mendelian Inheritance in Man [OMIM] database: 262890). Because only 3 patients (one American, one French, and one British) have been identified, the inheritance pattern is unknown,1,2 and the rarity of affected individuals precludes the use of conventional mapping approaches to identify the underlying genetic lesions. Hemostatic parameters of close relatives of patients with SS exhibit significant though clinically silent defects, indicating that any defective gene has substantial penetrance in the heterozygous state,1,2 and do not preclude the possibility that the SS phenotype reflects the interaction of 2 (or more) defective genes.
The defining characteristic of SS is the absence of Ca2+-stimulated exposure of phosphatidylserine (PS) from the inner leaflet of the plasma membrane bilayer to the cell surface, which in platelets normally provides an appropriate surface for the assembly of the tenase and prothrombinase complexes of the coagulation network. This defect is also observed in Epstein-Barr virus (EBV)–transformed lymphocytes of patients with SS. The mechanism by which PS is translocated to the cell surface following Ca2+ stimulation is controversial, and several proteins have been suggested as playing a role. However, both phospholipid scramblase 13-5 and P-glycoprotein4,6 appear to be normal in patients with SS.
The adenosine triphosphate (ATP)–binding cassette transporter A1 (ABCA1) has been implicated in PS translocation both by genetic disruption in mice7 and chemical inhibition.8 It is not yet clear whether ABCA1 translocates PS directly or acts as a regulator of another protein.9 Mutations in ABCA1 underlie Tangier disease (TD) and familial high-density lipoprotein (HDL) deficiency,10-13 conditions characterized by the absence of plasma HDL and greatly increased susceptibility to coronary heart disease. In addition, patients with TD have been shown to exhibit severely disrupted platelet function, consistent with the high platelet expression of this protein.14,15 Further, in abca1-deficient mice aberrant PS translocation is reflected in a hemorrhagic diathesis characteristic of SS. We therefore hypothesized that defects in ABCA1 underlie SS and assessed the expression and function of ABCA1 in a recently characterized patient with SS.16,17
Patients, materials, and methods
Patients
The SS proband is a 59-year-old white woman of British origin. Clinical features have been described previously.16,17 Family members were evaluated separately. Controls for analysis of ABCA1 mRNA expression were healthy volunteers between 23 and 71 years old. For mRNA analysis, 10 mLwhole blood was taken after an overnight fast. Approval was obtained from the Hammersmith Research Ethics Committee. Informed consent was provided in accordance with the Declaration of Helsinki.
Lipid profile
Lipid parameters of the proband were assessed at the lipid clinic of the Hammersmith Hospital. Blood for lipid parameters was obtained after 12-hour overnight fast. Serum total cholesterol, triglyceride, and HDL cholesterol levels were determined by automated methods using commercial kits and interassay controls. Levels of serum apolipoprotein A-I, apolipoprotein B, and lipoprotein (a) were measured using an automated immunoturbidimetric assay (Beckman Coulter, Galway, Ireland). Low-density lipoprotein (LDL) cholesterol was calculated from the standard formula as follows: LDL cholesterol (mM) = Total cholesterol – (HDL cholesterol + triglyceride/2.2).18 Apolipoprotein E phenotype was performed by immunoblotting.19
Nucleotide sequence analysis
DNA was extracted from whole blood using Nucleon DNA isolation kit (Tepnel Life Sciences, Manchester, United Kingdom) according to the manufacturer's instructions. The coding region and proximal promoter (395 base pair (bp) upstream from the ATG) of ABCA1 were amplified from gDNA by polymerase chain reaction (PCR) with primers located 50 to 70 bp from each splice junction and from cDNA amplified by reverse transcription (RT) PCR from leukocyte RNA with primers that produced 9 overlapping fragments (primer and amplification details available on request). Amplification products were sequenced on both strands and the sequences compared with ABCA1 cDNA sequence (GenBank accession no. AF285167). The nucleotide substitution is designated as recommended by a working group on nomenclature.20
Analysis of polymorphisms in DNA by WAVE analysis
PCR fragments of ABCA1 exons 6 and 42 were amplified with the following primers: exon 6, 5′-GGACCCAGCTTCCAATCTTCATAATCC-3′ and 5′-GCCTCACATTCCGAAAGCATTAGTGC-3′; exon 42, 5′-GTGGTTTATAGTCCTGCCTTCCAC-3′ and 5′-ACGAGCATCGTTGCTTGATTGGGT-3′.
PCR products were denatured at 94°C, followed by cooling to 25°C over 25 minutes to enhance heteroduplex formation, and analyzed on a WAVE Nucleic Acid Fragment Analysis System (Transgenomic, Omaha, NE).
ABCA1 gene expression in leukocytes
cDNA preparation. Leukocytes were isolated from 10 mL whole blood and RNA was extracted and reverse transcribed as described previously.21 For TaqMan analysis, the cDNA was diluted 4-fold with nuclease-free water.
Real-time quantitative RT-PCR (TaqMan). Single-tube TaqMan analysis was performed on an ABI prism 7700 sequence detection system (Applied Biosystems, Warrington, United Kingdom) with 300 nM forward and reverse primers in the presence of 200 nM 5′FAM-3′TAMRA–tagged probe located in exon 3 of the ABCA1 gene as described recently.21 The internal standard was glyceraldehyde-3-phosphate dehydrogenase (G3PDH), assayed with commercially supplied reagents (Applied Biosystems). Reactions were carried out in triplicate. The amount of ABCA1 mRNA in leukocytes of the patient and diverse control populations was calculated according to the standard curve method described in Applied Biosystems User bulletin no. 2. ABCA1 mRNA of all control subjects was expressed relative to that of G3PDH and calculated as relative (x-fold) expression as compared to the patient with SS.
Quantitative mRNA measurements of exon skipping (LightCycler). Previously, an alternative transcript of the ABCA1 gene has been reported with an out-of-frame deletion of exon 3, which results in a truncated protein of 74 amino acids presumably devoid of function.22 To assess the ratio of full-length to alternative transcript, a quantitative PCR method was developed using SYBR Green I and LightCycler technology (Roche, Penzberg, Germany). A reverse primer spanning exon boundaries 4 and 5 (5′-GCTTCAAGTTTGAGCTGGAT-3′) was combined either with a forward primer spanning exon junctions 2 and 3′, 5′-CTATGAACATGAATGCCATT-3′) to assess the 278-nucleotide (nt) full-length ABCA1 cDNA fragment including exon 3, or with a forward primer spanning the junction of exon 2 and 4 to measure only the 138-nt aberrant transcript. 18S RNA primers (forward 5′-AAGTCTTTGGGTTCCGGG-3′; reverse 5′-GGACATCTAAAGGGCATCACA-3′) amplifying a 365-bp fragment were used for normalization. PCRs contained 3 mM MgCl2, 0.4 μM forward and reverse primer, and 1 μL LightCycler DNA Master SYBR Green I (10×, Roche). Before amplification, a preincubation step (60 seconds at 95°C) was performed to activate FastStart DNA polymer and to ensure complete denaturation of the cDNA. LightCycler PCR was performed with 40 cycles using following amplification conditions: denaturation 15 seconds at 95°C, annealing 10 seconds at 62°C, and elongation 25 seconds at 72°C. To each amplification cycle a fourth segment with an elevated temperature fluorescence acquisition point was added to remove unspecific signals before SYBR Green I quantification (3 seconds at 80°C, 84°C, and 85°C for the full-length product, the aberrant product and 18S, respectively). Amplified products underwent melting curve analysis after the last cycle to specify the integrity of amplification.
Both transcripts of the SS proband were analyzed using the Fit Points and 2nd Derivative Maximum calculation described in the LightCycler Relative Quantification Software and compared to a leukocyte control population. ABCA1 mRNA of all control subjects was expressed relative to that of 18S RNA and calculated as x-fold expression as compared to the SS patient. The relative distribution between the 2 transcripts is expressed as the full-length–aberrant transcript ratio.
Plasmids/DNA constructs
Full-length human ABCA1 cDNA was generated by RT-PCR of mRNA obtained from leukocytes of a healthy individual and the patient as described previously.13 Wild-type cDNA was cloned into pGEM-11Zf vector (Promega, Madison, WI) and replaced with the corresponding cDNA fragment (Asp718-HpaI) containing the patient's mutation (c.6064G>A). No other single nucleotide polymorphisms were introduced as verified by sequence analysis.
Enhanced green fluorescent protein (EGFP) was fused in frame to the C-terminus of ABCA1 by an overlapping PCR strategy as described previously.13,23 The PCR product was cloned into pCR 4Blunt-TOPO vector (Invitrogen, Paisley, United Kingdom). BamHI-Not I ABCA1 cDNA fragments in the pGEM-11Zf constructs were replaced with the ABCA1-EGFP hybrid. The resulting wild-type and mutant ABCA1-EGFP cDNAs were cloned with Sal I-Not I into pCI-neo mammalian expression vector (Promega) and all constructs verified by complete sequencing.
Expression and localization of ABCA1-EGFP fusion proteins in HEK 293 cells
Transfection, fixation, immunocytochemistry, and confocal imaging of HEK 293 cells was performed as previously described.13 In brief, HEK 293 cells were plated on poly-L-lysine–coated (Sigma, St Louis, MO) glass cover slips 12 hours before transfection. Cells were transfected in 6-well plates according to the calcium phosphate transfection protocol (Clontech, Palo Alto, CA) with 2 μg DNA per well. Cells were washed twice with phosphate buffer saline solution (PBS) after six hours to remove plasmid DNA and fixed in 4% formaldehyde–4% sucrose 24 hours after transfection for localization studies. Fixed cells were permeabilized with 0.1% Triton X-100 (Sigma, Saint Louis, MO) for 4 mintues and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 15 mg/mL; 1:10 000 dilution; Molecular Probes, Eugene, OR) for nuclear DNA. In order to identify the intracellular localization of the wild-type and mutant ABCA1 proteins, cells were also stained with the mouse monoclonal antibody against sarco/endoplasmic reticulum calcium adenosine trophosphatase (SERCA 2, clone IID8; Affinity Bioreagents, Golden, CO) for 2 hours at room temperature after blocking with 0.2% fish-skin gelatin (Sigma) in PBS. The antibody against SERCA 2 was detected by goat anti–mouse immunoglobulin G (IgG) secondary antibody conjugated to Alexa 568 (1:400 dilution for 30 minutes at room temperature; Molecular Probes). Cells were imaged with a Leica SP confocal microscope equipped with a 63×/1.32 PlanApoChromat oil-immersion objective lens (Leica, Wetzlar, Germany). EGFP was excited with the 488-nm line of an argon laser; Alexa 568, with the 568-nm line of a krypton laser. To avoid bleedthrough, the fluorophores were excited sequentially. The emitted fluorescence was collected separately through a triple-dichroic mirror (488/568/663). The emission filter bands for EGFP and Alexa-568 fluorescence were restricted to 500-552 nm and 594-620 nm, respectively. DAPI staining of nuclear DNA was excited with the 351-nm line of an ultraviolet (UV) laser, and emission fluorescence collected with a 396-508–nm bandpass filter. Stacks of confocal sections separated by 1-μm increments were taken and images analyzed with Metamorph 5.0v1 software (Universal Imaging, Downington, PA). Figures were assembled for publication with Adobe Photoshop 6.0 software (Adobe Systems, San Jose, CA).
Analysis of allelic frequency
PCR amplification of leukocyte cDNA with primers (position 5702 and 6457) spanning the mutation was performed using standard PCR conditions and cloned using the TOPO-TA cloning kit (Invitrogen) according to the manufacturer's instructions. Direct DNA sequence analysis was performed on an ABI 3700 Prism automated sequencer.
Preparation of wild-type ABCA1 expressing SS EBV-transformed B cells
The full-length wild-type human ABCA1 cDNA9 was cloned into a bicistronic retroviral vector SPsLdS,24 modified to contain the neomycin-resistance cassette (kindly provided by Attila Ilias, Institute of Enzymology, Budapest, Hungary). The generation of retroviral particles and establishment of stable cell lines expressing the ABCA1 protein in SS EBV-transformed B cells (SS-ABCA1+) were achieved using the method of Ujhelly et al25 with minor modifications. Briefly, Phoenix-eco packaging cell line26 was transfected by calcium phosphate coprecipitation (Gibco, Karlsruhe, Germany). The cell-free viral supernatant was collected and used immediately to transduce PG13 packaging cells.24,25 The retroviruses produced by the G418-selected PG13 cell lines were used for SS EBV-transformed B-cell transduction. To prepare stable, ABCA1-expressing cell population, we used a sequential selection method as described by Kiffmeyer et al.27
RNA preparation and RT-PCR of cell lines
Total RNA was isolated by a single-step method using TriZol (Invitrogen). For each reaction 1 μg total RNA was reverse-transcribed into cDNA according to the manufacturer's protocol (Promega) using random hexamer oligonucleotides. The resulting cDNA was then subjected to PCR by using specific primers to detect human ABCA1 cDNA, but not the genomic ABCA1 sequence. RT-PCR analysis for the housekeeping Abelson (ABL) gene transcript served as control for total RNA preparation and the RT reaction.28 Oligonucleotides designed for PCR analysis of ABCA1 or ABL cDNAs were: 5′-ACAAGATGCTGAGGGCTGAT-3′ and 5′-CCCAAGACTATGCAGCAATG-3′ or 5′-GGGCTCATCACCACGCTCCA-3′ and 5′-CTGCCGGTTGCACTCCCTCA-3′, respectively.
Analysis of PS exposure by annexin V–binding assay
SS and control EBV-transformed B cells were harvested and resuspended in annexin-binding buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) prior to addition of propidium iodide (PI; 19 μg/mL) and Alexa Fluor 488-conjugated annexin V (1:80 vol/vol; Molecular Probes, Eugene, OR). Fluorescence was measured on a FACScalibur (Becton Dickinson, Heidelberg, Germany) flow cytometer; the simultaneous presence of PI in the samples monitored possible cell death. After the addition of either A23187 (1 μM; Boehringer Mannheim, Mannheim, Germany) or dimethyl sulfoxide (DMSO; as control), changes in annexin V–Alexa Fluor 488 binding over time were recorded at 1- or 5-minute intervals for 15 or 50 minutes.
In addition, effects of glyburide, cyclosporin A, verapamil, KO143, PSC833, and DMSO (as control) on the A23187-induced changes in the binding of annexin V–Alexa Fluor 488 were measured. Before addition of A23187 the cells were preincubated with annexin V–binding buffer for 10 minutes either with DMSO or with these agents at the concentrations indicated; changes in annexin V–Alexa Fluor 488 induced by A23187 binding over time were then recorded. Data from live cells, manually gated on the basis of excluding PI-stained cells, were analyzed with CellQuest software (Becton Dickinson). Ca2+-dependent annexin V binding was calculated by subtracting the values measured in the presence of DMSO from the values detected in the presence of A23187. Each data point represents the mean value of at least 3 independent experiments. KO143 was a generous gift from Drs. J. Allen and G. Koomen (Division of Experimental Therapy, The Netherlands Cancer Institute, and Laboratory of Organic Chemistry, University of Amsterdam, Amsterdam, The Netherlands). PSC 833 was a gift from Novartis Pharma (Basel, Switzerland). Other chemicals were from Sigma. Inhibitors and A23187 were dissolved in DMSO.
Results
ABCA1 gene expression
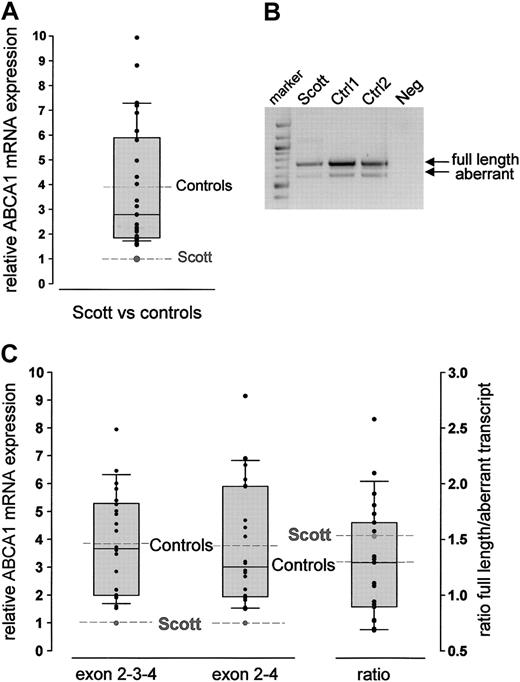
Total RNA was prepared from leukocytes isolated from a recently reported SS proband17 and a series of healthy controls (n = 30; aged 30-59). ABCA1 mRNA levels were quantified by real-time RT-PCR relative to expression of G3PDH using a TaqMan probe located in exon 3 of the ABCA1 gene. ABCA1 mRNA was readily detectable in total leukocyte RNA from controls and the patient. The relative amount of ABCA1 transcript detected including exon 3 varied within the control group (mean 3.90; Figure 1A). However, the relative ABCA1 mRNA expression level in the SS patient was very low and lay outside the 10th percentile limits (1.76) of the controls.
An alternative ABCA1 transcript with an out-of-frame deletion of exon 3, introducing a premature stop codon after 74 amino acids, has been reported in a number of cell lines.22 We therefore investigated the expression of the 2 alternative transcripts in leukocytes of the SS patient and controls (n = 22, aged 23-71 years). Both transcripts were found in controls and in the proband (Figure 1B). Quantitative RT-PCR analysis using amplicons specific for the wild-type (exon 2-3-4) and aberrant (exon 2-4) transcripts revealed extremely low expression of the wild-type cDNA in the SS patient compared with controls (mean of controls, 3.85; Figure 1C). Analysis of the aberrant transcript demonstrated that the patient had similarly reduced levels of the alternative, nonfunctional transcript (mean of controls, 3.77; Figure 1C). Because the relative abundance of wild-type to aberrant transcript was similar for the controls (mean, 1.29) and the SS patient (mean, 1.53; Figure 1C), low ABCA1 mRNA levels in the patient are not due to preferential alternative splicing.
Sequence analysis
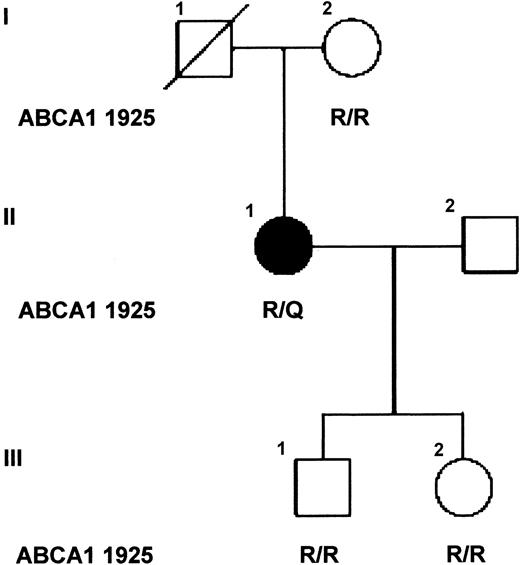
Sequence analysis of the promoter region, exons, and flanking intronic sequence of the patient's ABCA1 gene revealed no sequence changes that might explain the aberrant levels of ABCA1 mRNA observed in the leukocytes of the SS patient. However, several known polymorphisms,29-31 and heterozygosity for a novel single base pair substitution c.6064G>A in exon 42, predicted to substitute R1925 with glutamine, were identified (Table 1). The only available first-degree relatives of the patient, her mother and 2 children, are unaffected and homozygous wild-type for R1925 (Figure 2). The R1925Q variant was not found in 164 alleles of a control population of British origin,32 evidenced by WAVE analysis. For comparison, the allele frequency of the polymorphism in exon 6 (R219K) in this population was 0.22, similar to that observed in other European populations (0.254).33
Nucleotide substitutions in ABCA1 in the SS proband
Exon . | Nucleotide* . | Codon† . | Base substitution . | Amino acid substitution . | Comments . | Reference (variant db SNP rsID) . |
|---|---|---|---|---|---|---|
| 1 | 273 | 5′ UTR | hmz G | None | Polymorphism | Pullinger et al29 |
| 2 | 368-70 | Leu 26 | hmz CTG | None | CTG in general sequence | Pullinger et al29 |
| 5 | 764 | Leu 158 | htz A/G | None | Polymorphism | Clee et al30 (rs 2230805) |
| 6 | 947 | Arg 219 | htz G/A | R→K | Polymorphism | Wang et al31 (rs 2230806) |
| 8 | 1226 | Pro 312 | htz C/T | None | Polymorphism | (rs 2274873) |
| 14 | 2330 | Ile 680 | hmz C | None | Polymorphism | Wang et al31 (rs 7031748) |
| 16 | 2763 | Ile 825 | hmz G | I→V | Polymorphism | Wang et al31 (rs 4149312) |
| 17 | 2930 | Met 883 | hmz A | M→I | Polymorphism | Wang et al31 (rs 4149313) |
| 42 | 6064 | Arg 1925 | htz G/A | R→Q | Novel, rare | Present paper |
Exon . | Nucleotide* . | Codon† . | Base substitution . | Amino acid substitution . | Comments . | Reference (variant db SNP rsID) . |
|---|---|---|---|---|---|---|
| 1 | 273 | 5′ UTR | hmz G | None | Polymorphism | Pullinger et al29 |
| 2 | 368-70 | Leu 26 | hmz CTG | None | CTG in general sequence | Pullinger et al29 |
| 5 | 764 | Leu 158 | htz A/G | None | Polymorphism | Clee et al30 (rs 2230805) |
| 6 | 947 | Arg 219 | htz G/A | R→K | Polymorphism | Wang et al31 (rs 2230806) |
| 8 | 1226 | Pro 312 | htz C/T | None | Polymorphism | (rs 2274873) |
| 14 | 2330 | Ile 680 | hmz C | None | Polymorphism | Wang et al31 (rs 7031748) |
| 16 | 2763 | Ile 825 | hmz G | I→V | Polymorphism | Wang et al31 (rs 4149312) |
| 17 | 2930 | Met 883 | hmz A | M→I | Polymorphism | Wang et al31 (rs 4149313) |
| 42 | 6064 | Arg 1925 | htz G/A | R→Q | Novel, rare | Present paper |
Variant db SNP rsID: rsID is the dbSNP-assigned reference SNP ID.
UTR indicates untranslated region; hmz, homozygous; htz, heterozygous.
Based on published sequence for cDNA (GenBank accession no. AF285167).
Based on ATG start codon = bp 291-3 in cDNA sequence above.
To investigate whether there was differential expression of the 2 alleles, a PCR product spanning the c.6064G>A mutation was generated by RT-PCR from total RNA isolated from leukocytes of the SS patient. The PCR products were cloned and sequenced. Of 33 independent clones sequenced, 17 clones had the wild-type sequence, whereas 16 had the c.6064G>A mutation, indicating that both alleles were equally expressed in the ABCA1 leukocyte mRNA population of the SS patient.
Relative expression of ABCA1 mRNA in leukocytes. (A) Quantitative analysis of ABCA1 mRNA using TaqMan technology and a fluorescent probe located in exon 3. Analysis was performed using the standard curve method. The control population is visualized by the box plots indicating the 10th and 90th percentile (error bars), the median (solid horizontal line), and the mean (dashed line). The relative amount of ABCA1 mRNA in each control is represented as x-fold expression of the SS patient (set as 1, dashed line). Measurements were performed in triplicate. Because ABCA1 mRNA expression did not differ between men and women, a mixed control population is shown. (B) Agarose gel electrophoresis (1.5% agarose gel) of RT-PCR products amplified from leukocyte RNA from the SS patient and 2 controls using primers spanning exon 1-6. The 2 bands correspond to the wild-type (746-bp) and the alternatively spliced (606-bp) transcript. Lane 1, 100-bp marker; lane 4: negative control. (C) Quantitative analysis of the 2 transcripts described in panel B using LightCycler technology. The SS proband showed reduced ABCA1 mRNA levels for both the appropriately spliced (exon 2-3-4) and the alternatively spliced (exon 2-4) transcript. The ratio of the 2 transcripts did not differ between the patient and the controls (column 3). Analyses were performed using the FitPoints calculation method as described in “Patients, materials, and methods.” The control population is visualized by the box plots (details are described in panel A).
Relative expression of ABCA1 mRNA in leukocytes. (A) Quantitative analysis of ABCA1 mRNA using TaqMan technology and a fluorescent probe located in exon 3. Analysis was performed using the standard curve method. The control population is visualized by the box plots indicating the 10th and 90th percentile (error bars), the median (solid horizontal line), and the mean (dashed line). The relative amount of ABCA1 mRNA in each control is represented as x-fold expression of the SS patient (set as 1, dashed line). Measurements were performed in triplicate. Because ABCA1 mRNA expression did not differ between men and women, a mixed control population is shown. (B) Agarose gel electrophoresis (1.5% agarose gel) of RT-PCR products amplified from leukocyte RNA from the SS patient and 2 controls using primers spanning exon 1-6. The 2 bands correspond to the wild-type (746-bp) and the alternatively spliced (606-bp) transcript. Lane 1, 100-bp marker; lane 4: negative control. (C) Quantitative analysis of the 2 transcripts described in panel B using LightCycler technology. The SS proband showed reduced ABCA1 mRNA levels for both the appropriately spliced (exon 2-3-4) and the alternatively spliced (exon 2-4) transcript. The ratio of the 2 transcripts did not differ between the patient and the controls (column 3). Analyses were performed using the FitPoints calculation method as described in “Patients, materials, and methods.” The control population is visualized by the box plots (details are described in panel A).
Family pedigree. The affected individual is indicated by the solid symbol and unaffected relatives by open symbols. The amino acids at codon 1925 of ABCA1 are shown for each individual tested (R/R, homozygote; R/Q heterozygote). Individual I.1 died following a myocardial infarct and previously had undergone amputation of his right arm following an accident without excessive bleeding, and thus is likely to be unaffected.
Family pedigree. The affected individual is indicated by the solid symbol and unaffected relatives by open symbols. The amino acids at codon 1925 of ABCA1 are shown for each individual tested (R/R, homozygote; R/Q heterozygote). Individual I.1 died following a myocardial infarct and previously had undergone amputation of his right arm following an accident without excessive bleeding, and thus is likely to be unaffected.
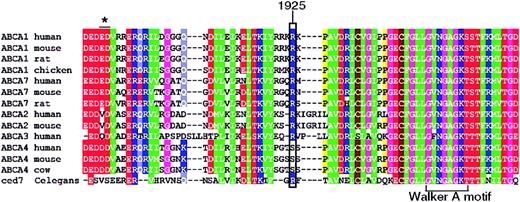
R1925 is predicted to lie 20 amino acids N-terminal of the Walker A motif in the second nucleotide-binding domain and is conserved in all known mammalian ABCA1 transporters (Figure 3). Substitution of R1925 with glutamine disrupts a stretch of 5 highly charged amino acids (RRKRK), which is strongly conserved between all members of the A family of ABC transporters. Interestingly, ABCA7, which is also believed to efflux cellular phospholipids but not cholesterol,34 is identical to ABCA1 at position 1925 and the homologous region of the Caenorhabditis elegans protein ced735 shares significant sequence similarity with ABCA1 and ABCA3 (Figure 3), pointing to the functional importance of this residue.
Localization studies
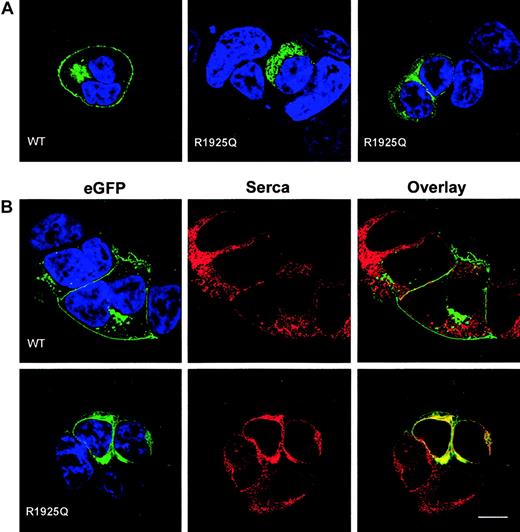
To test the impact of the R1925Q mutation on intracellular trafficking of ABCA1, we introduced the c.6064G>A mutation into a full-length cDNA encoding ABCA1 fused at its carboxyterminus to EGFP, and expressed the fusion protein in HEK 293 cells. The wild-type ABCA1-EGFP fusion protein is known to be expressed and to function normally in transfected cells.7,36 As expected,7,36 in cells transfected with wild-type ABCA1-EGFP, fluorescence was present mainly at the cell surface, with punctate intracellular vesicles visible in some cells and possible location in the Golgi in others (Figure 4A-B left panel). In contrast, little or no fluorescence was detected on the surface of cells transfected with the ABCA1 R1925Q-EGFP construct (Figure 4A middle and right panels; Figure 4B left panel). Instead fluorescence was observed largely in the endoplasmic reticulum (ER), evidenced by colocalization studies with sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA2), a marker of the ER37 (Figure 4B right panels).
Amino acid sequence. Amino acid sequence alignment of mammalian sequences encoding ABCA1, ABCA2, ABCA3, ABCA4, ABCA7 and C elegans ced7, spanning the region mutated in the SS patient. The mutated residue (ABCA1 R1925Q) is indicated by a rectangle. The Walker A motif of the ATP-binding domain is marked. The asterisk depicts a 2-amino acid deletion found in a patient with HDL deficiency.
Amino acid sequence. Amino acid sequence alignment of mammalian sequences encoding ABCA1, ABCA2, ABCA3, ABCA4, ABCA7 and C elegans ced7, spanning the region mutated in the SS patient. The mutated residue (ABCA1 R1925Q) is indicated by a rectangle. The Walker A motif of the ATP-binding domain is marked. The asterisk depicts a 2-amino acid deletion found in a patient with HDL deficiency.
Complementation of Ca2+-stimulated PS exposure in SS lymphocytes by ABCA1
To test the role of ABCA1 in Ca2+-stimulated PS translocation, we expressed wild-type ABCA1 in an EBV-transformed B-cell line obtained from the SS patient using a retroviral expression system. RT-PCR analysis showed that the stable, G418-selected SS-ABCA1+ cell line has elevated levels of ABCA1 mRNA expression as compared with the parental SS EBV-transformed B cells (Figure 5A). Expression of ABCA1 protein in EBV-transformed B cells (wild-type, SS-parental, and SS-ABCA1+) was below the level of detection by 2 anti-ABCA1 antibodies (Novus Biologicals, Little-ton, CO; and as described elsewhere9 ) in immunoblots or by immunofluorescent staining, though ABCA1 was apparent in transfected HEK cells (data not shown). The low-level expression of ABCA1 protein in transduced B cells despite high mRNA levels suggests posttranslational control; evidence that ABCA1 is posttranslationally regulated has been presented.38-41 Nevertheless, the low level of ABCA1 expression was sufficient to restore A23187-stimulated PS exposure similar to wild-type levels (Figure 5B). Furthermore, PS translocation in SS-ABCA1+ cells was completely blocked by the ABCA1 inhibitor glyburide8,42 (Figure 6). Although basal binding of annexin V to SS-parental and SS-ABCA1+ cells was somewhat higher than that to wild-type cells, this is likely to reflect biologic variation between transformed cells unrelated to disease because the phenotype was not apparent in fresh lymphocytes.17 These results show that expression of wild-type ABCA1 complements the defective Ca2+-activated PS translocation in the parental SS cells.
Expression of ABCA1-EGFP fusion proteins in HEK 293 cells. (A) Confocal sections of transiently transfected HEK 293 cells expressing wild-type ABCA1-EGFP (left) and ABCA1 R1925Q-EGFP (middle and right). EGFP fluorescence is shown in green and DAPI staining of nuclear DNA in blue. The mutation caused severe alteration in ABCA1 trafficking to the plasma membrane as compared to the wild type (WT). (B) HEK 293 cells expressing wild-type ABCA1-EGFP and ABCA1 R1925Q-EGFP were stained for SERCA 2 (red) as marker for the ER. The same confocal sections are shown for EGFP (green, left column), SERCA 2 (red, middle column) and colocalization of EGFP and SERCA 2 in the overlay (yellow, right column). Colocalization of ABCA1-SERCA2 was observed in cells transfected with mutant ABCA1, implying retention of R1925Q in the ER. In contrast, wild-type ABCA1-EGFP localized predominantly to the plasma membrane and no colocalization was detected. Scale bar indicates 10 μm.
Expression of ABCA1-EGFP fusion proteins in HEK 293 cells. (A) Confocal sections of transiently transfected HEK 293 cells expressing wild-type ABCA1-EGFP (left) and ABCA1 R1925Q-EGFP (middle and right). EGFP fluorescence is shown in green and DAPI staining of nuclear DNA in blue. The mutation caused severe alteration in ABCA1 trafficking to the plasma membrane as compared to the wild type (WT). (B) HEK 293 cells expressing wild-type ABCA1-EGFP and ABCA1 R1925Q-EGFP were stained for SERCA 2 (red) as marker for the ER. The same confocal sections are shown for EGFP (green, left column), SERCA 2 (red, middle column) and colocalization of EGFP and SERCA 2 in the overlay (yellow, right column). Colocalization of ABCA1-SERCA2 was observed in cells transfected with mutant ABCA1, implying retention of R1925Q in the ER. In contrast, wild-type ABCA1-EGFP localized predominantly to the plasma membrane and no colocalization was detected. Scale bar indicates 10 μm.
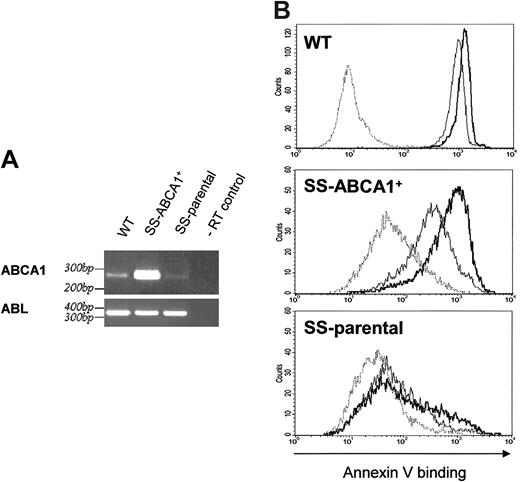
Expression of ABCA1 in SS-parental lymphocytes increased Ca2+-stimulated PS exposure on the cell surface. (A) Verification of the elevated level of ABCA1 mRNA in the transduced and G418-selected cell line by RT-PCR. Agarose gel electrophoresis of the PCR products using primers for ABCA1 or ABL and following templates: cDNAs obtained by reverse transcription of total RNA prepared from control EBV-transformed B cells (WT), from the stable SS-ABCA1+ cell line, or from the parental EBV transformed SS B cells (SS-parental). The last lane shows a negative control using total RNA prepared from the SS-ABCA1+ cells (-RT control) as template for the PCR. (B) Flow cytometry analysis of A23187-induced annexin V*488 binding for control wild-type (WT; top), ABCA1-expressing SS (SS-ABCA1+; middle) or SS-parental cells (bottom). Representative experiments show annexin V–Alexa Fluor 488 binding in the absence of A23187 at t0 (dotted lines), or annexin V–Alexa Fluor 488 binding induced by A23187 for 3 minutes (thin solid lines) or 10 minutes (bold solid lines). Ca2+-dependent PS exposure of SS-ABCA1+ cells was significantly increased compared to the SS-parental cell line.
Expression of ABCA1 in SS-parental lymphocytes increased Ca2+-stimulated PS exposure on the cell surface. (A) Verification of the elevated level of ABCA1 mRNA in the transduced and G418-selected cell line by RT-PCR. Agarose gel electrophoresis of the PCR products using primers for ABCA1 or ABL and following templates: cDNAs obtained by reverse transcription of total RNA prepared from control EBV-transformed B cells (WT), from the stable SS-ABCA1+ cell line, or from the parental EBV transformed SS B cells (SS-parental). The last lane shows a negative control using total RNA prepared from the SS-ABCA1+ cells (-RT control) as template for the PCR. (B) Flow cytometry analysis of A23187-induced annexin V*488 binding for control wild-type (WT; top), ABCA1-expressing SS (SS-ABCA1+; middle) or SS-parental cells (bottom). Representative experiments show annexin V–Alexa Fluor 488 binding in the absence of A23187 at t0 (dotted lines), or annexin V–Alexa Fluor 488 binding induced by A23187 for 3 minutes (thin solid lines) or 10 minutes (bold solid lines). Ca2+-dependent PS exposure of SS-ABCA1+ cells was significantly increased compared to the SS-parental cell line.
To confirm that the restoration of the ability of SS-ABCA1+ cells to expose PS in response to stimulation with Ca2+ ionophore was due to increased (but low level) expression of ABCA1, we assessed its sensitivity to inhibitors. In preliminary experiments we found that glyburide, an inhibitor of ABCA1,8,42 suppressed Ca2+-stimulated PS exposure by control cells (Figure 6A). In contrast, inhibitors of other ABC transporters at concentrations that fully blocked the respective transporters (1-10 μM cyclosporine A (CsA), 30 μM verapamil (VRP), 1 μM PSC833, and 1 μM KO14343-47 ) had no effect on PS translocation. These results are consistent with other data indicating that ABCA1,7,8 but not transporters such as MDR1,4 regulates loss of lipid asymmetry. The data do not support, however, a role for ABCG2 as has been suggested elsewhere.48 Glyburide completely blocked the remaining Ca2+ ionophore-stimulated PS exposure in SS-parental cells, consistent with residual expression of ABCA1. Further, because glyburide completely blocked Ca2+-ionophore–stimulated PS exposure by SS-ABCA1+ cells (Figure 6B), the data confirm that expression of wild-type ABCA1 reverses the defect in PS translocation of SS cells.
Discussion
SS is a mild bleeding disorder characterized by a defect in Ca2+-stimulated exposure of PS at the outer leaflet of hematopoietic cell membranes. ABCA1 has been proposed as a PS translocase7,8 or translocase regulator9 and its potential role in the pathophysiology of SS was investigated. We identified a novel missense mutation in ABCA1 (R1925Q) in the SS patient, which results in severely impaired trafficking and reduced expression of functional ABCA1 protein at the cell surface. We also demonstrated severely reduced steady-state levels of ABCA1 mRNA in leukocytes of an SS patient. This could not be explained by a change in the ratio of an alternatively spliced aberrant transcript. Sequence analysis of the ABCA1 gene of the SS patient revealed no difference in the proximal promoter region, in intron-exon junctions, or within the coding region that might explain the reduced ABCA1 mRNA levels. Hence, an additional unidentified mutation, potentially in a trans-acting regulatory gene, is hypothesized to account for reduced ABCA1 mRNA from both alleles. This putative second mutation and the incomplete loss of ABCA1 activity may account for the phenotypic differences observed between SS and TD.
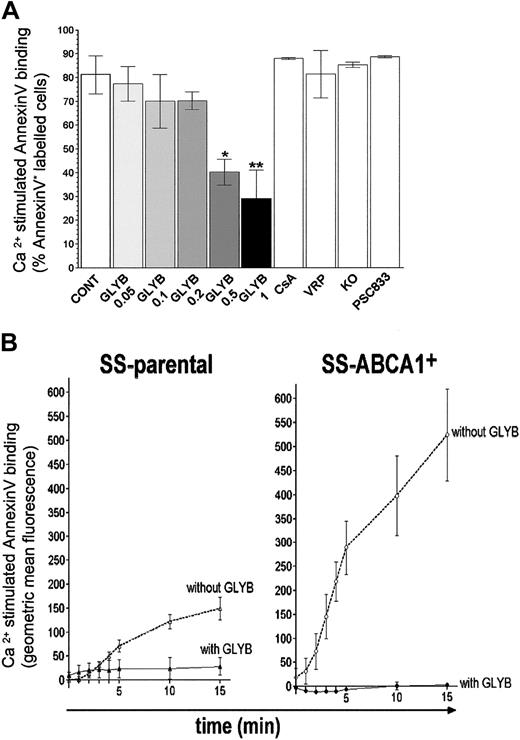
Glyburide inhibits Ca2+-stimulated PS exposure in B lymphocytes. (A) Control wild-type EBV-transformed B cells were preincubated with the vehicle DMSO (CONT), or DMSO containing 0.05, 0.1, 0.2, 0.5, 1 mM glyburide (GLYB), 10 μM cyclosporine A (CsA), 30 μM verapamil (VRP), 1 μM KO143, or 1 μM PSC833 for 10 minutes. Ca2+-activated PS exposure was measured after 10 minutes stimulation by A23187. Each data point represents the mean value of at least 3 independent experiments. **P < .005; *P < .01. (B) Glyburide inhibits Ca2+-activated PS exposure by SS cells transduced with ABCA1. Ca2+-stimulated PS translocation was analyzed by an annexin V binding assay. Cells were preincubated either with 1 mM glyburide or with DMSO only before addition of A23 187 at t0, and changes in annexin V binding over time were recorded. Ca2+-stimulated annexin V binding was calculated by subtracting the values measured in the presence of DMSO from the values detected in the presence of ionophore A23187 and is expressed as an increase in fluorescence (geometric mean of annexin V binding) by cells in the PI– cell population. Each data point represents the mean value of at least 3 independent experiments; error bars represent SD. SS-parental indicates EBV-transformed B lymphocytes from the SS patient; SS-ABCA1+ indicates SS-parental cells transduced with ABCA1.
Glyburide inhibits Ca2+-stimulated PS exposure in B lymphocytes. (A) Control wild-type EBV-transformed B cells were preincubated with the vehicle DMSO (CONT), or DMSO containing 0.05, 0.1, 0.2, 0.5, 1 mM glyburide (GLYB), 10 μM cyclosporine A (CsA), 30 μM verapamil (VRP), 1 μM KO143, or 1 μM PSC833 for 10 minutes. Ca2+-activated PS exposure was measured after 10 minutes stimulation by A23187. Each data point represents the mean value of at least 3 independent experiments. **P < .005; *P < .01. (B) Glyburide inhibits Ca2+-activated PS exposure by SS cells transduced with ABCA1. Ca2+-stimulated PS translocation was analyzed by an annexin V binding assay. Cells were preincubated either with 1 mM glyburide or with DMSO only before addition of A23 187 at t0, and changes in annexin V binding over time were recorded. Ca2+-stimulated annexin V binding was calculated by subtracting the values measured in the presence of DMSO from the values detected in the presence of ionophore A23187 and is expressed as an increase in fluorescence (geometric mean of annexin V binding) by cells in the PI– cell population. Each data point represents the mean value of at least 3 independent experiments; error bars represent SD. SS-parental indicates EBV-transformed B lymphocytes from the SS patient; SS-ABCA1+ indicates SS-parental cells transduced with ABCA1.
The role of ABCA1 in this SS patient was confirmed by complementation with wild-type ABCA1 in transformed lymphocytes from the SS patient and further supported by pharmacologic data. Increased expression of wild-type ABCA1 in SS-parental cells restored the wild-type phenotype of Ca2+-induced PS translocation. Although ABCA1 protein levels were low in EBV-transformed B-cell lines (wild-type, SS-parental, and SS-ABCA1+), the restored ability of SS-ABCA1+ cells to expose PS was blocked by glyburide, an inhibitor of ABCA1. These expression and pharmacologic studies strongly support the hypothesis that the SS phenotype observed in our proband is at least in part dependent on reduced ABCA1 activity.
Despite the severely reduced ABCA1 mRNA levels in lymphocytes, the 59-year-old proband's lipid profile (Table 2) revealed only mild mixed (type IIb) hyperlipidemia characterized by elevations of both total cholesterol and triglyceride serum levels, and she did not show clinical features of TD or HDL deficiency, which are phenotypes of other characterized mutations in ABCA1.10-12,33 Interestingly, a novel case of TD without symptoms of atherosclerotic disease has been reported,49 despite harboring a null mutation that results in a nonfunctional ABCA1 protein. In addition, some patients heterozygous for ABCA1 mutations show greater than 50% HDL cholesterol levels, and single nucleotide polymorphisms (SNPs) in this gene associated with increased severity of atherosclerosis are not always associated with changes in lipid levels.33 Bone marrow transplants in ABCA1–/– mice also suggest that blood-derived cellular expression of ABCA1 does not influence plasma lipid levels.50,51 Taken together these data indicate that changes in ABCA1 activity can occur without alterations in the steady-state plasma lipid levels33 as observed for this SS patient.
Lipid profile of the SS proband
Parameter . | Baseline level . | Desirable level or range . |
|---|---|---|
| Total cholesterol, mM | 6.5 | <5.0 |
| Triglyceride, mM | 2.3 | <1.7 |
| HDL-cholesterol, mM | 1.3 | >1.3 |
| LDL-cholesterol, mM | 4.1 | <3.0 |
| Total cholesterol/HDL cholesterol | 4.9 | <5.0 |
| Apolipoprotein B, mg/dL | 122 | <120 |
| Apolipoprotein A-1, mg/dL | 141 | 112-201 |
| Lipoprotein (a), mg/dL | 102 | 0-30 |
| Apolipoprotein E phenotype | E2/E3 | NA |
Parameter . | Baseline level . | Desirable level or range . |
|---|---|---|
| Total cholesterol, mM | 6.5 | <5.0 |
| Triglyceride, mM | 2.3 | <1.7 |
| HDL-cholesterol, mM | 1.3 | >1.3 |
| LDL-cholesterol, mM | 4.1 | <3.0 |
| Total cholesterol/HDL cholesterol | 4.9 | <5.0 |
| Apolipoprotein B, mg/dL | 122 | <120 |
| Apolipoprotein A-1, mg/dL | 141 | 112-201 |
| Lipoprotein (a), mg/dL | 102 | 0-30 |
| Apolipoprotein E phenotype | E2/E3 | NA |
NA indicates not applicable.
ABCA1 has been implicated in several cellular functions, namely, transport of lipids from the Golgi to the plasma membrane,52 externalization of PS,7 uptake of apoptotic cells,8 secretion of interleukin 1β,53 and formation of plasma HDL.10-12,31,33 How ABCA1 is associated with these seemingly disparate processes and how its deficiency results in a variety of disease phenotypes is unclear. It is possible that the common feature linking these functions with both SS and TD relates to the formation of outward membrane protrusions such as microvilli, filopodia, lamellipodia, blebs, and spikes (echinocytes), consistent with our observation that defective microvesicle formation is the most pronounced phenotype in fresh SS lymphocytes.17
Alterations in hemostatic parameters and disrupted platelet function have been observed in TD patients,15 and data on a patient with a prolonged bleeding time have been reported,54 highlighting a potential role of ABCA1 in bleeding disorders. It has been clearly shown that ABCA1 is expressed on platelets and that lack of ABCA1 expression either in TD patients or ABCA1-deficient mice severely disrupts platelet function.7,15,52 Stimulation of platelets from TD patients deficient in ABCA1 with either collagen or A23187 did not induce detectable differences in PS exposure as assayed by annexin V binding.15 In contrast, we have demonstrated that our SS patient with severely reduced levels of functional ABCA1 has a defect in PS exposure in lymphocytes in response to A23187.17 These differences may be due to methodologic differences because changes in PS translocation are more obvious at early stages after stimulation with A23187.17 Alternatively, they may reflect differences due to the second unidentified mutation in our SS patient. It is also possible that ABCA1 might have a distinct role in blood cells compared with tissue-derived cells.
Prepublished online as Blood First Edition Paper, March 24, 2005; DOI 10.1182/blood-2004-05-2056.
Supported by the Swiss National Science Foundation (fellowship 823A-056551 [C.A.]), the Hungarian Academy of Sciences (Bolyai fellowship [K.S.]), Orszagos Tudomanyos Kutatasi Alapprogramok (OTKA) (research grant T038337), and the Medical Research Council, United Kingdom.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Anne Soutar for help with the nucleotide sequence analysis and helpful critical discussions. We are grateful to Balazs Sarkadi who supported this work by his expert advice and by critical reading of the manuscript. Furthermore, the authors are indebted to Mandy Bennett for the WAVE analysis, to Tamara Stelzl and Clare Neuwirth for technical assistance, as well as to Selina Raguz and Soni Soumian for advice and help with laboratory techniques. We would also like to thank the patient and her family for their kind cooperation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal