Abstract
Mutations in the perforin gene have been found in patients with hemophagocytic lymphohistiocytosis (HLH), a rare autosomal recessive disease. We describe a patient expressing perforin with amino acid changes A91V and W374X. The ability of cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells to lyse target cells is greatly reduced. Here we demonstrate that perforin from this patient is not recognized using an antibody raised against native perforin (δG9), but is readily detected using an antibody raised against a peptide epitope (2d4), suggesting that the epitope recognized by δG9 is destroyed by the change at A91V. Immunoblotting reveals no protein corresponding to the truncated transcript encoded by W374X, revealing that only perforin with the A91V change is expressed in CTLs from the patient. Patient CTLs show bands corresponding to the immature and intermediate forms of perforin, but the mature active form of perforin is absent or barely detectable. The conformational changes and impaired cleavage of A91V perforin are likely to explain the reduced cytotoxicity in CTLs and NK cells from this patient and are likely to contribute to the pathogenesis of HLH.
Introduction
Perforin plays a key role in the cytotoxicity of natural killer (NK) cells and cytotoxic T lymphocytes (CTLs).1 Perforin is stored as an active protein in specialized secretory lysosomes, known as lytic granules, of NK cells and CTLs.2 Upon target cell recognition, lytic granules polarize and release their contents at the immunologic synapse.3 Secreted perforin inserts into the lipid bilayer, polymerizing to form pores in the membranes of target cells and allowing entry of a series of granzymes that activate apoptotic pathways in the target.1 Perforin binds the target cells via a C2 domain that is able to bind phospholipid head groups in the presence of calcium. The C2 domain is hidden during biosynthesis by a carboxy-terminal propiece containing a large N-linked glycan, rendering perforin inactive.4 Removal of the propiece activates perforin by revealing the C2 domain. Processing occurs in a post-Golgi compartment by an as-yet-unidentified processing enzyme. Perforin is then stored as an active protein in the lytic granules, where it is complexed with proteoglycans that render the protein inactive at the low pH of the lysosomes.5
Genetic mutations in perforin give rise to approximately 30% of cases of the autosomal recessive disease familial hemophagocytic lymphohistiocytosis (HLH),6,7 while other mutations have been identified in proteins that are required for secretion of perforin.8 To date, the mutations identified in perforin result either in loss of a functional mRNA and complete loss of perforin protein, or production of a nonfunctional protein. Mutations giving rise to the production of nonfunctional perforin protein can map to the C2 lipid-binding domain or the C9 homology domain, thought to be responsible for pore formation. Additional mutations have been identified in the amino terminal domain of perforin, the function of which is not understood. A single nucleotide change of 272C>Tin perforin has been reported in several patients with HLH.6,9-11 Here we describe a patient expressing perforin A91V and demonstrate that this point mutation results in both loss of recognition by the antiperforin antibody, δG9, and severely impaired cleavage of perforin to its active processed form resulting in drastically reduced cytotoxicity against targets.
Patients, materials, and methods
Clinical diagnosis
The patient was a female and first child from unrelated healthy parents originating from southern Italy. This patient has been described as patient 6 in a previous study.12 At the age of 4.9 years, the patient developed an acute “flu-like” episode characterized by spiking fever, hepatosplenomegaly, and lymphadenomegaly. On admission, the patient had overt jaundice, anemia (70 g/L [7 g/dL]), neutropenia (0.8 × 109/L), thrombocytopenia (73 × 109/L), hypertriglyceridemia (8.31 μM [736 mg/dL]), and hypofibrinogenemia (0.53 g/L). The biochemical lab values were as follows: aspartate aminotransferase (AST), 260 IU/L; alanine aminotransferase (ALT), 103 IU/L; lactate dehydrogenase (LDH), 652 mg/dL; bilirubin, 237.7 μM (13.9 mg/dL); albumin, 28 g/L (2.8 g/dL); and activated partial thromboplastin time (APTT), 37 s. Cerebrospinal fluid (CSF) examination was normal. Infectious disease screening revealed the presence of Epstein-Barr virus (EBV)–specific immunoglobulin M (IgM) antibodies. Chest x-ray and the US study showed moderate enlargement of mediastinal and abdominal lymph nodes. Bone marrow aspiration showed normal cells and increased number of monocytes, with active erythrophagocytosis. The clinical diagnosis of HLH was also supported by evidence of complete loss of NK-cell activity against K562 targets in vitro using, as effector cells, peripheral blood lymphocytes (PBLs, containing 12% NK cells).12 The child was treated with chemoimmunotherapy (dexamethasone and etoposide) according to current standard recommendations of the Histiocyte society.13 The patient had a rapid and complete response within the first 8 weeks. At that point, based on the absence of familial disease and association with acute EBV infection, treatment was electively suspended. In the following months, immunologic and genetic work-up showed that the PBLs of the child showed a severely impaired perforin expression by CD16+ and CD56+ lymphocytes at flow cytometry. Mutation analysis of the perforin gene confirmed the presence of 2 mutations, 272C>T in exon 2, producing the Ala→Val substitution, and 1122G>A in exon 3, resulting in W374 and stop in the protein. These were the only changes found when the perforin gene was sequenced from 43 nucleotides upstream and 105 nucleotides downstream of exon 2, and 25 nucleotides upstream and 21 nucleotides downstream of exon 3. These mutations were documented for confirmation, one in each of the 2 parents. Approval was obtained from the Ospedale dei Bambini “G. Di Cristina” institutional review board for these studies. Informed consent was obtained from the patients or legal guardian, according to the Declaration of Helsinki.
The child remained in a good clinical condition under regular follow-up evaluations. Then 2 years later, she suddenly developed another overt episode of HLH, which was complicated by central nervous system (CNS) involvement (refractory seizures and nystagmus); treatment was started again with etoposide, dexamethasone, cyclosporine, and intrathecal methotrexate + prednisolone (weekly for 4 weeks). She rapidly improved, and within 2 months she underwent hematopoietic stem cell transplantation from a matched unrelated donor. At the time of writing, she is persistently asymptomatic at the age of 9 years.
The perforin-deficient patient shown in Figure 1 has been described as patient 1 in a previous study.12 Sequencing of the perforin gene from this patient shows a homozygous T deletion at nucleotide 50, producing a frameshift and a premature stop codon in the mRNA encoding perforin at nucleotide 150. No perforin protein is detected by Western blotting or immunolabeling in CD8+ CTLs derived from this patient.
Antibodies
The murine antibody against perforin, 2d4, was generated as previously described.4,14 Mouse monoclonal antibody against lysosomal-associated membrane protein-2 (Lamp2, H4B4) was obtained from the Developmental Studies Hybridoma Bank, Iowa City, IA. Murine antiperforin (δG9) and rabbit anti–cathepsin D were purchased from BDPharMingen (Oxford, United Kingdom) and Upstate Biotechnology (Charlottesville, VA), respectively. The fluorescein isothiocyanate (FITC)–conjugated antibody anti-CD8 and mouse antiactin antibodies were purchased from Sigma-Aldrich (Poole, United Kingdom). All secondary antibodies conjugated to cyanin 3 (Cy3), Cy5, FITC, and horseradish peroxidase (HRP), absorbed against cross-reactive species, were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
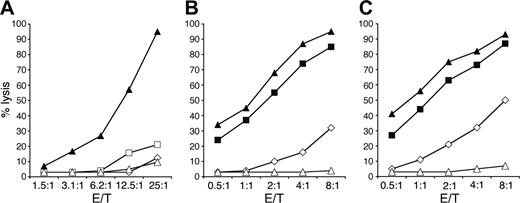
Defective lytic activity in CTLs and NK cells. (A) A CD8+ CTL clone derived from a healthy donor (▴) is compared with 2 different clones, derived from patient 612 expressing perforin A91V and W374X (□ and ⋄) and 1 clone derived from patient 112 lacking perforin expression (▵), for their cytolytic activity against P815 in the presence of anti-CD3 monoclonal antibody (mAb). (B-C) Polyclonal activated NK-cell populations derived from 2 healthy donors (▪ and ▴), from patient 6 (⋄), and patient 1 (▵) were tested in a cytolytic assay against the 51Cr-labeled FO-1 (B) and 721.221 (C) target cell lines. The percentages of target cell lysis at different effector-target (E/T) ratios are shown.
Defective lytic activity in CTLs and NK cells. (A) A CD8+ CTL clone derived from a healthy donor (▴) is compared with 2 different clones, derived from patient 612 expressing perforin A91V and W374X (□ and ⋄) and 1 clone derived from patient 112 lacking perforin expression (▵), for their cytolytic activity against P815 in the presence of anti-CD3 monoclonal antibody (mAb). (B-C) Polyclonal activated NK-cell populations derived from 2 healthy donors (▪ and ▴), from patient 6 (⋄), and patient 1 (▵) were tested in a cytolytic assay against the 51Cr-labeled FO-1 (B) and 721.221 (C) target cell lines. The percentages of target cell lysis at different effector-target (E/T) ratios are shown.
Cell culture
Peripheral blood mononuclear cells (PBMCs) from patients and controls were purified from blood samples by density gradient with Ficoll-Paque (Sigma-Aldrich); cells were washed 3 times and maintained in human T-cell medium (RPMI-1640, 5% AB serum, 100 U/mL recombinant interleukin-2 [rIL-2], and 50 μmol β-mercaptoethanol [Gibco, Paisley, United Kingdom]).
CTLs were isolated by limiting dilution: 0.3 cells/well were plated in 96 U-bottom–well plates, stimulated with 106 cells/mL irradiated (30 Gy) allogeneic lymphocytes and 1 μg/mL phytohemagglutinin (PHA; Sigma-Aldrich), and selected by CD8 staining. CTL clones were maintained at 37°C in 5% CO2 in human T-cell medium and stimulated with irradiated human peripheral blood cells and 1 μg/mL PHA approximately every 3 to 4 weeks. P815 mouse target cells were maintained in RPMI-1640 (Gibco), 10% fetal calf serum (FCS).
NK cells were purified from peripheral blood using the RosetteSep method (StemCell technologies, Vancouver, BC) following the manufacturer's instructions. NK cells were cultured on irradiated feeder cells (allogeneic PBMCs and 721.221 lymphoblastoid cell line) in the presence of 100 units/mL rIL2 (Proleukin; Chiron Italia, Milan, Italy) and 2 μg/mL PHA to obtain polyclonal NK-cell populations. Polyclonal activated NK-cell populations were more than 95% CD3–CD56+ by fluorescence-activated cell sorter (FACS) analysis when used for the cytotoxicity assays.
FACS analysis
Cells (2 × 105-1 × 106) were washed twice in phosphate-buffered saline (PBS) + 1% FCS and resuspended in cytofix/cytoperm (BDPharMingen) for 20 minutes on ice as described by the manufacturer; cells were then washed twice in perm/wash before antibody labeling with either δG9(IgG2b) antiperforin phycoerythrin (PE, 1:100; BDPharMingen) or a PE-conjugated isotype-matched control for 30 minutes on ice. Cells were washed 3 times in perm/wash and resuspended in PBS + 1% FCS prior to analysis on a FACScalibur (Becton Dickinson, Mountain View, CA).
Immunoblotting
CTL clones or NK cells were washed once in PBS, resuspended at 107 cell/mL in lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane] HCl [pH 8], 150 mM NaCl, 1 mM MgCl2, 1% Triton X-100) with complete protease inhibitor 25X (Amersham, Little Chalfont, United Kingdom), and incubated on ice for 30 minutes. Lysates were spun for 20 minutes at 16 000 g (8500 rpm) (Heraeus-Biofuge rotor no. 3043; Heraeus, Hanau, Germany) at 4°C. Where indicated, lysates were incubated with endoglycosidase H (Endo H; Boehringer Mannheim, Mannheim, Germany) at 37°C for 12 to 16 hours in 9 mM CaCl2, 50 mM sodium acetate [pH 5.5], 1 mM Pefabloc (Boehringer Mannheim). Lysates (10-μL aliquots) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on gels of 7.5% acrylamide for perforin detection under nonreducing conditions. Proteins were transferred to nitrocellulose (HybondC extra; Amersham) using a MiniTrans-Blot Apparatus (BioRad, Welwyn Garden City, United Kingdom) in 25 mM Tris (pH 8.3), 192 mM glycine, and 20% methanol. Filters were then blocked for one hour at room temperature (RT) in blocking buffer (PBS, 5% milk powder, and 0.2% Tween20) on a rocking platform and then incubated for 16 hours at 4°C with the antiperforin 2d4 murine antibody. Membranes were washed 3 times in PBS/0.1% Tween20 for 10 minutes each and incubated for one hour with HRP-labeled anti–mouse Ig secondary antibody (Jackson ImmunoResearch Laboratories), diluted in blocking buffer. Filters were washed 3 times in PBS/0.1% Tween20 for 10 minutes each and developed for 1 minute in SuperSignal detection reagents (Pierce, Rockford, IL) and exposed between 30 seconds to 10 minutes using Biomax film (Kodak, Rochester, NY). Rainbow Marker high range (1 mg/mL of each protein; Amersham) were loaded on each gel as molecular weight standards.
Cytotoxicity assays
Cytotoxicity assays were performed using chromium release for NK cells as previously described,15 or using a Cytotox96 nonradioactive kit (Promega, Madison, WI) for CTLs, according to the manufacturer's instructions. Briefly, CTL clones were counted and washed in RPMI-1640 without phenol red and plated in 96-well plates at E/T ratios from 25:1 to 0.75:1. P815 targets were added at 104 cells/well in a final volume of 100 μL using RPMI-1640 without phenol red in the presence of 1 μg/mL anti-CD3 (UCHT1; BD Pharmingen). LDH release was measured after 4 hours of incubation at 37°C. Supernatant (50 μL) was removed 45 minutes prior to the reading from each well and incubated with the reaction substrate, and the absorbance was read at 490 nm using a thermomax plate reader (Molecular Devices, Sunnyvale, CA). Percentage target cell lysis was calculated. All assays were performed in triplicate.
Immunofluorescence microscopy
CTL clones from patients and controls were washed in serum-free medium (RPMI-1640) at days 12 to 14 after stimulation, and each cell pellet was resuspended to a final concentration of 5 × 106 cell/mL in RPMI-1640 with no added serum. Samples were fixed for 5 minutes on ice with ice-cold methanol and washed 3 times with PBS and once in PBS + 1% bovine serum albumin (BSA; Sigma-Aldrich), and then blocked in PBS + 1% BSA for 15 to 30 minutes at RT.
Primary antibodies were diluted in PBS + 1% BSA and incubated at RT for 40 minutes, and then washed 3 times in PBS. The samples were blocked in PBS + 1% BSA for 5 minutes, and then the secondary antibodies were added after dilution in PBS + 1% BSA for 40 minutes. Samples were washed 3 times in PBS + 1% BSA and 4 to 6 times in PBS and then mounted in PBS, 90% glycerol, and 2.5% DABCO (1,4-diazabicyclooctane; Sigma-Aldrich). Samples were analyzed using an MRC-2000 BioRad laser scanning confocal microscope and images were processed using Metamorph software (Molecular Devices, Downington, PA).
Results
The cytotoxicity of both CD8+ CTL clones and NK cells derived from patient 612 with mutations at 272C>T and 1122G>A in the perforin gene, which give rise to a single amino acid change of A91V and a stop codon at W374, was tested (Figure 1). Figure 1A shows 2 separate clones from the patient compared with clones derived from a healthy donor and patient 1,12 who lacks perforin expression. One patient 6–derived clone shows no cytotoxicity, and the curve is superimposed above that of the CTL clone lacking perforin expression. The other clone gives up to 20% cytotoxicity at an effector-target (E/T) ratio of 25:1. Up to 30% cytotoxicity at an E/T ratio of 8:1 is seen with the polyclonal NK-cell population derived from patient 6 against target FO-1 (Figure 1B), and up to 50% against 721.221 (Figure 1C), while perforin-deficient NK cells from patient 112 are completely unable to kill either of these targets. These results show that CTL- and NK cell–expressing alleles of perforin encoding A91V and W374X show greatly impaired, although not completely ablated, cytotoxicity against targets.
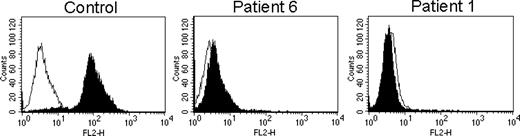
Cytofluorimetric analysis of perforin using δG9 mAb. FACS profiles of staining of control and patient CTLs with δG9 antiperforin. Nonspecific staining with an isotype-matched control antibody is shown by the open histograms. FL2-H indicates fluorescence channel 2.
Cytofluorimetric analysis of perforin using δG9 mAb. FACS profiles of staining of control and patient CTLs with δG9 antiperforin. Nonspecific staining with an isotype-matched control antibody is shown by the open histograms. FL2-H indicates fluorescence channel 2.
We examined the expression of perforin by FACS analysis using δG9, raised against native perforin.16 δG9 recognizes perforin in control CD8+ cells but not those derived from patient 6 or the perforin-null control, patient 1 (Figure 2). Similar profiles were obtained staining NK cells derived from the same donors (not shown). This indicates that the epitope recognized by δG9 is lost inA91V perforin. These findings were confirmed using immunofluorescent localization of perforin with δG9, which stains perforin in the granules of control CD8+ cells colocalizing the lysosomal protein cathepsin D. In patient 6–derived CTLs, only cathepsin D staining is visible, and perforin staining cannot be detected using δG9. There was no staining with δG9 in CTLs from the perforin-null patient 1 (Figure 3).
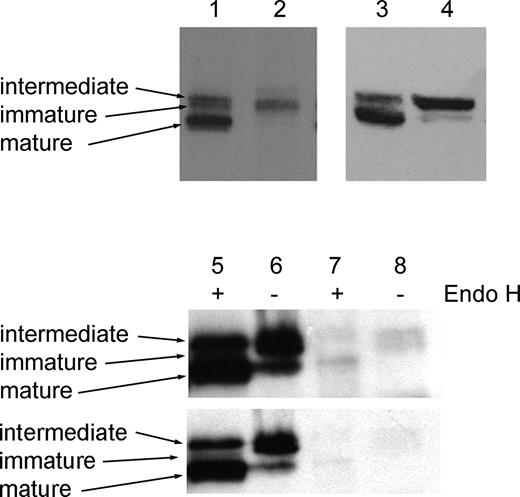
Although the antibody 2d4 does not detect perforin well by confocal microscopy, it detects all 3 biosynthetic forms of perforin by Western blotting. We therefore used 2d4 to examine cell lysates from control and patient CTLs (Figure 4). In lysates from control CTLs, 3 separate biosynthetic forms of perforin are seen (Figure 4 lanes 1 and 3). The 65-kDa immature form of perforin acquires N-linked glycans during biosynthesis to give rise to the 70-kDa intermediate form, which is subsequently cleaved to the 60-kDa active mature form of the protein, which represents the most abundant form as it accumulates in the lytic granule during activation of CTLs. In CTLs derived from patient 6, the immature and intermediate forms of perforin are found, but the mature form is absent (Figure 4 lane 2) or barely detectable (Figure 4 lane 4).
In addition to the A91V single amino acid change encoded on one perforin allele, patient 6 also possesses a mutation encoding W374X on the other allele. We do not detect any protein corresponding to this form (predicted molecular weight, 37-47 kDa) in spite of the fact that the epitope recognized by 2d4 would be expressed. This suggests either that the mRNA encoding W374X is unstable or that any protein is rapidly degraded. In this patient, only the A91V form of perforin appears to be expressed, allowing its functional defect to be studied.
The initial biosynthetic processing of the A91V perforin expressed is identical to that of control perforin (Figure 4 lanes 5-8). In lysates from control NK cells (which produce high levels of perforin), Endo H digestion selectively removes high mannose glycans found in the immature form, yielding a band of the same molecular weight as mature perforin, while the intermediate form is Endo H resistant4 (Figure 4 lanes 5 and 6, shorter exposure). Endo H digestion of A91V perforin (Figure 4 lanes 7 and 8, longer exposure) also shows the immature band yielding a lower molecular weight band (lane 7) as in the control (lane 5), with the intermediate band remaining Endo H resistant (lane 7). However the levels of A91V perforin after incubation at 37°C for 12 to 16 hours are lower than those seen in control lysates under the same conditions, with bands of less than half the level expected in a patient expressing protein from only one allele (Figure 4 lanes 5-8).
Patient CTLs do not stain with δG9 antiperforin antibody. Confocal microscopy of CD8+ CTL clones from control and patients 6 and 112 stained with δG9 antiperforin and cathepsin D (includes brightfield and all images merged [right]). Scale bar: 10 micrometers. See “Immunofluorescence microscopy” for details. Images were visualized with a Nikon TE300 inverted microscope equipped with a Plan Apo 100 ×/1.4 oil-immersion objective lens (Nikon UK, Kingston Upon Thames, United Kingdom), and with a Radiance 2000 confocal laser scanning microscope (Carl Zeiss, Welwyn Garden City, United Kingdom).
Patient CTLs do not stain with δG9 antiperforin antibody. Confocal microscopy of CD8+ CTL clones from control and patients 6 and 112 stained with δG9 antiperforin and cathepsin D (includes brightfield and all images merged [right]). Scale bar: 10 micrometers. See “Immunofluorescence microscopy” for details. Images were visualized with a Nikon TE300 inverted microscope equipped with a Plan Apo 100 ×/1.4 oil-immersion objective lens (Nikon UK, Kingston Upon Thames, United Kingdom), and with a Radiance 2000 confocal laser scanning microscope (Carl Zeiss, Welwyn Garden City, United Kingdom).
Processing of perforin to the mature form is impaired in patient CTLs. Western blots of CD8+ CTL lysates from control (lanes 1,3) and patient (lanes 2,4), showing immature, intermediate, and mature forms of perforin in control CTLs, and only the immature and intermediate forms in patient 612 . Longer (top) and shorter (bottom) exposures of lysates from NK cells from control (lanes 5-6) or patient 6 (lanes 7-8) were incubated in the presence (lanes 5,7) or absence (lanes 6,8) of Endo H and probed with 2d4-antiperforin. All samples were run on 7.5% acrylamide gels under nonreducing conditions.
Processing of perforin to the mature form is impaired in patient CTLs. Western blots of CD8+ CTL lysates from control (lanes 1,3) and patient (lanes 2,4), showing immature, intermediate, and mature forms of perforin in control CTLs, and only the immature and intermediate forms in patient 612 . Longer (top) and shorter (bottom) exposures of lysates from NK cells from control (lanes 5-6) or patient 6 (lanes 7-8) were incubated in the presence (lanes 5,7) or absence (lanes 6,8) of Endo H and probed with 2d4-antiperforin. All samples were run on 7.5% acrylamide gels under nonreducing conditions.
These results demonstrate that glycosylation of A91V perforin is normal, and A91V perforin passes from the endoplasmic reticulum through the Golgi stacks normally. However A91V perforin appears to be more susceptible to degradation than perforin with the alanine at residue 91.
In summary, our results show that A91V causes a conformational change that results in loss of recognition by δG9 antiperforin and reduces the production of the active, cleaved form of perforin even though glycosylation occurs normally and the protein passes through the Golgi apparatus.A91V perforin appears to be more sensitive to degradation. These observations are likely to explain the reduced lytic ability of CTLs and NK cells derived from this patient.
Discussion
We have previously shown that production of the active form of perforin requires cleavage of a heavily glycosylated propiece of approximately 20 amino acids from the carboxy-terminus of the protein.4 This exposes the phospholipid-binding C2 domain and initiates formation of the pore formed by polymerization of perforin monomers. In this report, we describe a patient with heterozygous point mutations in perforin. 1122G>A introduces a stop codon at amino acid 374, resulting in loss of stable protein production from this allele. A single nucleotide change, 272C>T, in the other allele produces a single amino acid change of A91V, which we show impairs the cleavage of perforin to the active form, resulting in loss of CTL and NK-cell cytotoxicity against targets.
Mutations resulting in HLH have been identified in several different domains of perforin.6,7,11,17 These include mutations in the leader sequence, the N-terminal, the membrane insertion (C9 homology), a cysteine-rich epidermal growth factor-like (EGF-like), and the C2 domain. To date, only one patient has been reported to possess mutations impairing the maturation of perforin to the active form.18 The mutations in perforin each encode single amino acid changes: F193L in the predicted membrane-insertion region and R410P in the C2 domain. These have been proposed to result in conformational changes that prevent cleavage to the active form of perforin. The F193L mutation reduced the ability of perforin to be immunoprecipitated with the δG9 antiperforin antibody, indicating that the epitope recognized by δG9 is disrupted by this change.18 Here we show that A91V results in loss of recognition by the antiperforin antibody, δG9, demonstrating that at least part of the epitope recognized by δG9 maps to the N-terminal region of perforin. Our results demonstrate a conformational change in A91V perforin, reflected by loss of δG9 antibody staining as well as an increased susceptibility to degradation. Intriguingly, A91V and F193L mutations, which both disrupt δG9 recognition, also impair the cleavage of perforin to its active, mature form. One possible explanation for this could be that the N-terminal region is involved in the targeting of perforin to the lytic granule and that the sorting motif is disrupted by both of these mutations. Alternatively, perforin may reach the granules but fail to undergo cleavage possibly because the protease binding site on perforin is disrupted. It is clear that A91V reaches the Golgi stacks, as glycosylation occurs normally. However it is not possible to determine whether perforin reaches the granules, as the epitope for δG9, the only antibody to work well by immunofluorescence, is lost (Figure 3). An alternative explanation is that the conformational changes that result in loss of recognition by δG9 also result in loss of recognition by the protease that cleaves perforin to its active form. The protease required to cleave perforin has not yet been identified, and it is possible that different proteases may fulfill this role in different cell types and/or different individuals. The considerable heterogeneity in the disease phenotype reported forA91V supports the idea that other protein or pathogen interactions may be involved.
The pathogenic role of the A91V change in perforin is currently debated. A91V has been identified in patients developing late onset HLH.9 The 2 siblings carrying the same compound heterozygous mutations in perforin reported here (A91V and W374X) developed HLH with adult onset and atypical presentation.9 We have also documented the same compound heterozygosity in one additional patient, with the same geographic origin, who developed HLH at the age of 3 months (M.A. and A.S., unpublished data, May 2001). The W374X mutation is located within the EGF-like domain, encoded by exon 3 of the perforin gene, and was found independently in different families of Turkish origin.19 The second mutation, A91V, appeared to be nonrandomly observed in Italian patients. In 2004, Busiello et al described atypical features of HLH in a family presenting 2 different mutations: R231H and A91V.10 In this family of 2 twins carrying A91V in a homozygous state and R231H in a heterozygous state, only 1 developed fatal HLH at 11 years, while the second twin, with the same pattern of perforin mutations, had no sign of HLH and had normal NK-cell activity. Both parents and 2 sisters were asymptomatic and heterozygous for A91V. The contribution of R231H is not known. Since late onset of HLH has been widely documented, the authors suggest that the asymptomatic twin remains at risk for developing the disease later on.10 Our study reveals that although cleavage of perforin to its active, mature form is greatly impaired, a small amount of the mature form can be detected biochemically (Figure 4). It is therefore possible that a low level of cytotoxicity may exist in the pool of CTLs and NK cells that may provide sufficient cytotoxic capability to combat viral infections for some period, with the disease only emerging in response to infections such as EBV, which elicits a strong CD8 response to control the virus.
Is A91V a frequent polymorphism? A homozygous A91V mutation has been reported in one asymptomatic subject.17 Zur Stadt et al studied 86 control DNA from healthy unrelated white persons and found a high prevalence of this mutation (n = 15, 17.5%).19 Furthermore, in their analysis of the North American population with HLH, Molleran Lee et al identified the A91V mutation in the heterozygous state in 7 individuals (3%).11 Of the patients, 3 had other biallelic disease-causing perforin (PRF1) mutations and 2 had HLH but no other observed PRF1 mutations; 1 asymptomatic sibling of a third patient with HLH (genotype unknown) demonstrated decreased levels of perforin protein and absent NK-cell function. Furthermore, one healthy control demonstrated slightly decreased levels of perforin and normal NK-cell function.11 The authors suggested that the finding of normal NK-cell activity supports the concept that this is a polymorphism and not a pathogenic mutation, despite reduced perforin protein expression, together with the finding that no patient with HLH and homozygous A91V mutation had been described so far. In a recent survey, some of us have identified the heterozygous A91V mutation in 3.9% of 127 healthy controls from southern Italy,20 in keeping with the findings by Molleran Lee et al11 in the North American population.
The biochemical and functional data presented here show that A91V causes a conformational change that results in loss of recognition by δG9 antiperforin, reduces the production of the active, cleaved form of perforin, and renders the protein more labile. These results suggest that A91V may represent a mutation that, being more frequent in some geographic spots than in others, may contribute significantly to the pathogenesis of HLH.
Prepublished online as Blood First Edition Paper, March 1, 2005; DOI 10.1182/blood-2004-09-3713.
Supported by grants from the Wellcome Trust (G.M.G.), Ministero Istruzione Università Ricerca–Fondo per gli Investimenti della Ricerca di Base (MIUR-FIRB) (L.N.), Associazione Italiana Ricerca sul Cancro (AIRC) (M.A., D.P., S.M., and L.M.), Associazione per la Ricerca sulle Sindromi Emofagocitiche-Istiocitosi (ARSE) (M.A.), Progetto obbiettivo Regione Sicilia (M.A.) and Fondazione Compagnia di San Paolo (D.P., S.M., and L.M.). S.M is recipient of a fellowship awarded by Fondazione Italiana per la Ricerca sul Cancero (FIRC).
C.T. and F.G. contributed equally to the work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 3. Patient CTLs do not stain with δG9 antiperforin antibody. Confocal microscopy of CD8+ CTL clones from control and patients 6 and 112 stained with δG9 antiperforin and cathepsin D (includes brightfield and all images merged [right]). Scale bar: 10 micrometers. See “Immunofluorescence microscopy” for details. Images were visualized with a Nikon TE300 inverted microscope equipped with a Plan Apo 100 ×/1.4 oil-immersion objective lens (Nikon UK, Kingston Upon Thames, United Kingdom), and with a Radiance 2000 confocal laser scanning microscope (Carl Zeiss, Welwyn Garden City, United Kingdom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/3/10.1182_blood-2004-09-3713/6/m_zh80150581730003.jpeg?Expires=1769013935&Signature=is~WozOvrVe3N-Hkl-OqrCcSAkGQwdFIyfH90UIzN3Hb~o1T2zA~EzQwRLo3gEqOoBL~~Xh~8BT~gFHizKBO12ugQAoTZLqJ786V~wIZDddJwvgM9K7oB6SOttzV9M7kZ36WzFGhLnXa6OKfzXQN9-zCk5idk2bsV-eH-gfjWG9MmiXQHaIrKb03~SgZVxv2EdRMOjBku-GWaDJWmvvKtsYcnD2Zf22AXMx7TeqrXDkeqbgiTpgs2lMrXifgO6xknigYZvg9Dgs-BiGgr4M8s9O088MUtagYmtf6yAZzl3hL7bye4crwuAieCTIqZ4gNflUXPxEbHoKnw8~Vf~BH6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal