Abstract
The receptor for hyaluronic acid–mediated motility (RHAMM/CD168) has been described as a leukemia-associated antigen. To define T-cell epitopes of RHAMM/CD168 toward specific immunotherapies for acute myeloid leukemia (AML), 10 potential HLA-A2–binding RHAMM/CD168 peptides (R1 to R10) were synthesized based on computer algorithms and screened by enzyme-linked immunospot (ELISPOT) analysis using CD8+ T cells isolated from peripheral blood (PB) of patients with AML and healthy donors. We found that CD8+ cells from 7 of 13 (54%) patients with AML presensitized with peptides R3 (ILSLELMKL) or R5 (SLEENIVIL) specifically recognized T2 cells pulsed with R3 (39%) or R5 (15%) peptide. In contrast, only 4 of 21 (19%) healthy volunteers had CD8+ cells reactive with R3- or R5-pulsed T2 cells after presensitization. The presence of R3 peptide–specific effector T cells in the peripheral blood of patients with AML could be confirmed by staining as HLA-A2/R3 peptide tetramer+ CCR7-CD45RA+ cells. In chromium-51 release assays, peptide-primed CD8+ T cells from patients with AML were able to lyse RHAMM/CD168 peptide–pulsed T2 cells, AML blasts, and dendritic cells generated thereof (AML DCs). Transfection of COS7 cells with RHAMM/CD168 cDNA revealed that peptides R3 and R5 are naturally processed epitopes of RHAMM/CD168 that are presented in an HLA-A2–restricted manner. In summary, RHAMM/CD168 is a promising target for immunotherapies in patients with AML, and we have therefore initiated a clinical vaccination trial with R3 peptide. Because RHAMM/CD168 is also expressed in various other hematologic malignancies and solid tumors, vaccines targeting this antigen may have even wider application.
Introduction
Treatment of patients with acute myeloid leukemia (AML) became more effective through the development of risk-adapted polychemotherapy and hematopoietic stem-cell transplantation (HSCT).1-3 Today, most patients with AML who are younger than 60 years of age reach a complete hematologic remission (CR). However, often the CR is not durable, and finally a high percentage of these patients relapse.1,3,4 Allogeneic HSCT is a therapeutic option for these patients1-3 based on the effects of total body irradiation and high-dose chemotherapy but also on the induction of a graft versus leukemia (GVL) effect.5 A compelling evidence for the power of the GVL effect is the fact that patients at relapse after allogeneic HSCT for chronic myeloid leukemia (CML) or AML can reach again a CR after donor lymphocyte infusion.6-10 This therapeutic benefit suggests the existence of leukemia-associated antigens (LAAs). To identify such LAAs, we employed earlier the method of serologic screening of a cDNA expression library (SEREX).11-14 The receptor for hyaluronic acid–mediated motility (RHAMM/CD168) was revealed to be the most promising LAA defined by this SEREX analysis: Messenger RNA for RHAMM/CD168 was demonstrated by reverse transcriptase–polymerase chain reaction (RT-PCR) and Western blot to be expressed in leukemic blasts of more than 80% of the patients with AML but not in peripheral blood mononuclear cells (PBMCs) or CD34+ stem cells of healthy volunteers. Among normal tissue, only testis, placenta, and thymus showed significant mRNA expression for the antigen RHAMM/CD168.12
To design T-cell–based immunotherapies for patients with AML expressing RHAMM/CD168 in leukemic blasts, it is a crucial step to characterize T-cell epitopes of the molecule.
We started the search for appropriate T-cell epitopes departing from different computer algorithms and evaluated these peptides in mixed lymphocyte peptide cultures (MLPCs) with PBMCs from patients with AML and healthy volunteers. Two peptides could be defined that were specifically recognized by T cells from more than 50% of patients with AML but only recognized by T cells from 19% of healthy volunteers. To prove that the peptides were naturally processed, RHAMM-negative COS7 cells were transfected by a plasmid vector containing the cDNA for the full-length molecule RHAMM/CD168. The results of T-cell responses against RHAMM/CD168 observed in enzyme-linked immunospot (ELISPOT) assays were confirmed in chromium-51 release assays and by tetramer staining to prove the specificity of the T-cell responses in patients with AML.
Patients, materials and methods
Samples from healthy volunteers and patients with AML
All samples were taken from patients with AML treated in the framework of clinical studies approved by the local ethics committee of the University of Ulm and from healthy blood donors. Both patients with AML and healthy blood donors gave their informed consent in accordance with the Declaration of Helsinki. PBMCs from healthy volunteers and patients with AML were prepared by Ficoll separation and stored for RNA preparation at –80°C. For cellular assays, Ficoll-separated PBMCs were tested freshly or cryopreserved in AB serum containing 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen. Peripheral blood samples anticoagulated by EDTA (ethylenediaminetetraacetic acid) and sera from patients with AML were collected at the time of diagnosis (blasts more than 90%) and in CR (blasts less than 5% in bone marrow and CR) after polychemotherapy with or without allogeneic stem-cell transplantation. Patients with AML, the therapeutic regimens they received, and the stage of disease at the time of sample collection are listed in Table 1.
Clinical characteristics of patients with AML
Patient no. . | Sex . | Age, y . | FAB classification . | Cytogenetics . | Treatment . | Allogeneic SCT . | Remission . |
|---|---|---|---|---|---|---|---|
| 1 | F | 44 | M2 | Normal | 2 × IVA, MIDAC | + | CR (5 y) |
| 2 | F | 31 | M3 | t(15;17) | ATRA, 4 × DA | - | CR (8 y) |
| 3 | M | 42 | M1 | 47,XY,+21 | Autologous PBSCT | - | CR (10 mo) |
| 4 | M | 56 | M2 | 47,XY,+8 | 2 × DA, 3 × high-dose AraC | - | CR (1 mo) |
| 5 | F | 56 | M3 | t(15;17) | 1 × AIDA, 2 × HAM | - | CR (1 y) |
| 6 | M | 72 | M5b | t(9;11) | 2 × ICE, 1 × HAM | + | CR (2 y) |
| 7 | F | 37 | RA | Normal | No treatment | - | MDS (RA) |
| 8 | M | 55 | M2 | del 9 | 2 × ICE, 1 × HD AraC | - | CR (1 mo) |
| 9 | F | 20 | M3 | t(15;17) | 4 × ABVD + rad; 1 × AIDA, 1 × IC, 2 × HAM | - | CR (1 mo) |
| 10 | M | 52 | M4 | t(4;11;9) | 2 × ICE, 1 × HAM | - | CR (1 y) |
| 11 | M | 48 | M0 | t(3;14) | 2 × ICE, 1 × HAM | - | Relapse |
| 12 | M | 49 | M1 | t(10;11) | 2 × ICE, 2 × HAM | - | CR (1 y) |
| 13 | M | 69 | M1 | Normal | 2 × ICE, 1 × HAM | - | Relapse (only BM) |
Patient no. . | Sex . | Age, y . | FAB classification . | Cytogenetics . | Treatment . | Allogeneic SCT . | Remission . |
|---|---|---|---|---|---|---|---|
| 1 | F | 44 | M2 | Normal | 2 × IVA, MIDAC | + | CR (5 y) |
| 2 | F | 31 | M3 | t(15;17) | ATRA, 4 × DA | - | CR (8 y) |
| 3 | M | 42 | M1 | 47,XY,+21 | Autologous PBSCT | - | CR (10 mo) |
| 4 | M | 56 | M2 | 47,XY,+8 | 2 × DA, 3 × high-dose AraC | - | CR (1 mo) |
| 5 | F | 56 | M3 | t(15;17) | 1 × AIDA, 2 × HAM | - | CR (1 y) |
| 6 | M | 72 | M5b | t(9;11) | 2 × ICE, 1 × HAM | + | CR (2 y) |
| 7 | F | 37 | RA | Normal | No treatment | - | MDS (RA) |
| 8 | M | 55 | M2 | del 9 | 2 × ICE, 1 × HD AraC | - | CR (1 mo) |
| 9 | F | 20 | M3 | t(15;17) | 4 × ABVD + rad; 1 × AIDA, 1 × IC, 2 × HAM | - | CR (1 mo) |
| 10 | M | 52 | M4 | t(4;11;9) | 2 × ICE, 1 × HAM | - | CR (1 y) |
| 11 | M | 48 | M0 | t(3;14) | 2 × ICE, 1 × HAM | - | Relapse |
| 12 | M | 49 | M1 | t(10;11) | 2 × ICE, 2 × HAM | - | CR (1 y) |
| 13 | M | 69 | M1 | Normal | 2 × ICE, 1 × HAM | - | Relapse (only BM) |
FAB indicates French-American-British; F, female; M, male; RA, refractory anemia; IVA, idarubicin, etoposide, and cytarabine (AraC); DA, daunorubicin and cytarabine; ATRA, all-trans retinoic acid; ICE, idarubicin, cytarabine, and etoposide; HAM, high-dose cytarabine and mitoxantrone; MIDAC, mitroxantrone and intermediate-dose cytarabine; ABVD, doxorubicin bleomycin, vinblastine, and dacarbazine (for Hodgkin disease); PBSCT, peripheral blood stem cell transplantation; AIDA, all-trans retinoic acid and idarubicin; and AraC,.
Immunoscreening
For the immunoscreening, phages containing the full-length cDNA for the LAA gene RHAMM/CD168 were used.12 Serologic screening of sera of patients with AML was performed as described.11-13 Serologic results were compared with the mRNA expression of RHAMM/CD168 in AML blasts of the same patients at the time of diagnosis.
Conventional RT-PCR
Messenger RNA was prepared from PBMCs or tumor samples by using mRNA QuickPrep Micro purification kits (Amersham Pharmacia Biotech, Little Chalfont, England). A total of 2.0 μg of each mRNA sample was subjected to cDNA synthesis (Superscript II; Gibco, Frederick, MD). PCR for RHAMM/CD168 was performed as described12,14 using the indicated conditions and ingredients.
Real-time RT-PCR
The mRNA expression of RHAMM/CD168 was quantified by real-time RT-PCR using the light cycler SYBR Green I technology according to the manufacturer's instructions and the primers described elsewhere.14 The amount of mRNA of RHAMM/CD168 was normalized to the housekeeping gene TATA-box binding protein (TBP)15 and copy numbers calculated.14
Definition of RHAMM/CD168-derived peptides
RHAMM/CD168-derived peptide sequences with HLA-A*0201–binding motifs were predicted using 2 different computer algorithms based on known binding affinities.16,17 Candidate peptides were also checked for their cleavage pattern by a prediction algorithm for proteasomal cleavage designated “PAProC.”18 Peptides were excluded that showed no appropriate sequence after proteasomal cleavage. Ten peptides representing major histocompatibility complex (MHC) class-I–restricted RHAMM epitopes with favorable predicted binding to the HLA-A*0201 motif are listed in Table 2. The influenza matrix protein (IMP)–derived peptide GILGFVFTL served as a positive control.19 Peptides were synthesized by Thermo Electron (Ulm, Germany) to a minimum of 95% purity as measured by high-performance liquid chromatography.
RHAMM/CD168-derived T-cell epitope peptides predicted by computer algorithms
No. . | Peptide sequence . | Peptide position . | SYFPEITHI ranking . | BIMAS ranking . | PAProC proteasome type . |
|---|---|---|---|---|---|
| R1 | KLLEYIEEI | 232-240 | 1 | 2 | PII, PIII |
| R2 | KLQEELNKV | 671-679 | 2 | 1 | PI-PIII |
| R3 | ILSLELMKL | 165-173 | 4 | 4 | PI-PIII |
| R4 | KLQVTQRSL | 194-202 | 15 | > 20 | PIII |
| R5 | SLEENIVIL | 274-282 | 3 | > 20 | PII |
| R6 | NLNAEMQNL | 313-321 | > 20 | 8 | PI-PIII |
| R7 | SLLQQEKEL | 344-352 | 10 | 5 | PI-PIII |
| R8 | KLKGKEAEL | 419-427 | 6 | > 20 | PII |
| R9 | QLQLDAFEV | 594-602 | > 20 | 3 | PI |
| R10 | QLKSEVSKL | 650-658 | 11 | > 20 | PII, PIII |
No. . | Peptide sequence . | Peptide position . | SYFPEITHI ranking . | BIMAS ranking . | PAProC proteasome type . |
|---|---|---|---|---|---|
| R1 | KLLEYIEEI | 232-240 | 1 | 2 | PII, PIII |
| R2 | KLQEELNKV | 671-679 | 2 | 1 | PI-PIII |
| R3 | ILSLELMKL | 165-173 | 4 | 4 | PI-PIII |
| R4 | KLQVTQRSL | 194-202 | 15 | > 20 | PIII |
| R5 | SLEENIVIL | 274-282 | 3 | > 20 | PII |
| R6 | NLNAEMQNL | 313-321 | > 20 | 8 | PI-PIII |
| R7 | SLLQQEKEL | 344-352 | 10 | 5 | PI-PIII |
| R8 | KLKGKEAEL | 419-427 | 6 | > 20 | PII |
| R9 | QLQLDAFEV | 594-602 | > 20 | 3 | PI |
| R10 | QLKSEVSKL | 650-658 | 11 | > 20 | PII, PIII |
The designation, the position of the peptides (according to accession no. U29343), and the ranking numbers based on HLA- binding prediction algorithms (http://www.syfpeithi.de, 16 http://bimas.dcrt.nih.gov/molbio/hla_bind/17 ) are indicated. The digestion of RHAMM/CD 168 by the proteasome types I to III (PI-PIII) was analyzed by using the PAProC algorithm (http://www.paproc.de18 ). The respective proteasome types producing these potential peptide sequences are listed.
Culture of cell lines
The human cell lines were cultured in a standard medium consisting of RPMI 1640 (Biochrom, Berlin, Germany) supplemented with 10% (vol/vol) AB serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin. T2 cells and the COS7 cell line were obtained from Deutsche Sammlung von Zellen und Mikroorganismen (Braunschweig, Germany).
Generation of dendritic cells (DCs)
DCs were generated from AML blasts as described earlier.20 Briefly, peripheral blood from patients with AML containing more than 90% leukemic blasts was submitted to Ficoll separation, a subsequent plastic adhesion step, and thereafter to 6 days of cell culture in a standard medium supplemented with 10% AB serum, 100 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) (Immunex, Seattle, WA), and 1000 U/mL interleukin-4 (IL-4) (Strathmann, Hannover, Germany). After 6 days of culture, the medium was completely changed and additionally supplemented with 50 ng/mL tumor necrosis factor-α (TNF-α) (Strathmann). DCs were harvested on day 8 and revealed homogenously a typical phenotype with dendritic veils in light microscopy and with expression of CD86 and HLA class-I and -II molecules of more than 97% as determined by flow cytometry. CD80 and CD83 were expressed at more than 70% (data not shown).
Mixed lymphocyte peptide culture (MLPC)
PBMCs from healthy volunteers or patients with AML were separated by Ficoll and subsequently selected by magnetic beads through a magnetic-activated cell separation (MACS) column. More than 95% purity was reached in the CD8+ fraction. CD8– antigen-presenting cells (APCs) were irradiated with 30 Gy and pulsed with a RHAMM/CD168–derived peptide (Table 2) or the IMP control at a concentration of 20 μg/mL for 2 hours. After coincubation with CD8+ T lymphocytes overnight, the MLPC was supplemented with 10 U/mL IL-2 and 20 ng/mL IL-7 on day +1. Every 3 days, half of the medium was renewed with medium containing IL-2 and IL-7. On day +7, the medium was changed and new irradiated and peptide-pulsed CD8– APCs were added as 1 week before. Again, the MLPC was supplemented with IL-2 and IL-7 on the following day. After a total of 14 days of culture, T cells were harvested and evaluated for their specific cytotoxicity in a standard chromium-51 release assay against T2 cells pulsed with the RHAMM/CD168 peptides, with the positive control IMP peptide or without peptide. The cell line K562 was used as a control for T-cell–mediated cytotoxicity, because K562 expresses RHAMM, but not HLA class-I molecules, thus being a target to natural killer (NK) cells but not to specific T lymphocytes.
IFN-γ ELISPOT
Interferon-γ (IFN-γ) ELISPOT assays were performed as described earlier.21 Briefly, 96-well nitrocellulose plates (Millipore, Schwalbach, Germany) were coated with IFN-γ monoclonal antibodies (mAbs) (Mabtech, Hamburg, Germany) and incubated overnight at 4°C. After washing with phosphate-buffered saline (PBS), plates were blocked with 10% human AB serum (German Red Cross, Ulm, Germany) for 2 hours at 37°C. Presensitized CD8+ T lymphocytes (1 × 104) and 4 × 104 target cells (peptide-pulsed PBMCs) were added to each well. After incubation in RMPI medium overnight, plates were washed with PBS supplemented with 0.05% Tween 20. IFN-γ mAbs (0.2 μg/mL) were added to each well, and plates were incubated for 2 hours at room temperature. Cells were washed with streptavidin-alkaline phosphatase (1 μg/mL; Mabtech) for 2 hours. Plates were washed, and BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) (Sigma-Aldrich, Munich, Germany) was used for colorization according to the manufacturer's instructions. Plates were dried up at room temperature and thereafter evaluated by the use of an ELISPOT reader (CTL, Reutlingen, Germany).
Granzyme B ELISPOT assay
The granzyme B ELISPOT assay was performed as previously described21 to determine specific lysis of RHAMM/CD168 (peptide)–positive target cells according to the manufacturer's instructions (BD, San Diego, CA).
Tetramer staining
HLA-A2/R3 peptide phycoerythrin (PE) tetramer specifically was synthesized at the Lausanne Branch of the Ludwig Institute for Cancer Research. CD8+ T cells (1 × 106), stimulated with irradiated CD8– APCs in the presence of the R3 peptide or an irrelevant (MAGE3 [melanoma-associated antigen 3])–derived peptide, or in the absence of any peptide (as described above), were stained with HLA-A2/R3 peptide PE tetramer at a concentration of 2 μg/mL in the dark and incubated for 40 minutes at room temperature. Thereafter, for 2-color staining, 10 μL anti-CD8 fluorescein isothiocyanate (FITC) (BD, Heidelberg, Germany) was added at 4°C for 20 minutes in the dark. For 4-color staining, additionally to tetramer-PE, pure anti-CCR7 rat and secondary goat anti–rat allophycocyanin, anti-CD8 peridinin chlorophyll protein (PerCP), and anti-CD45RA FITC were used. After washing twice, stained cells were fixed with 0.5% paraformaldehyde (Sigma, Munich, Germany) for 10 minutes and then analyzed by flow cytometry.
Cloning of RHAMM/CD168 and/or HLA-A*0201 into the plasmid vector pBudCE4.1
The clone of RHAMM/CD168 was generated by cloning the full-length cDNA of the gene into the eukaryotic pBudCE4.1 expression vector (Invitrogen, Carlsbad, CA). Briefly, the full-length cDNA of RHAMM/CD168 (2130 base pair [bp]) was amplified by PCR (template: cDNA of AML blasts) with the forward primer 5′-cac cat gtc ctt tcc taa ggc g-3′ and the reverse primer 5′-tta ctt cca tga ttc ttg aca ctc-3′ and cloned using the pcDNA3.1 Directional TOPO expression kit (Invitrogen). The correct sequence of the insert was ascertained by sequence analysis using an ABI PRISM 310 capillary genetic analyzer (BD). The 2.13 kb KpnI-XhoI fragment obtained by the PCR was then ligated to the KpnI and XhoI sites of pBudCE4.1 expression vector, which was designated pBudCE4.1/(RHAMM/CD168). Plasmid pcDNA3.1/HLA-A*0201 was a generous gift from Thomas Wölfel (Third Department of Internal Medicine, University of Mainz, Germany). To generate the plasmids pBudCE4.1/HLA-A*0201 and pBudCE4.1/HLA-A*0201&RHAMM/CD168, the 1.2 kb HindIII-XbaI fragment from pcDNA3.1/HLA-A*0201 was subcloned into the HindIII and XbaI sites of the pBudCE4.1 and the pBudCE4.1/(RHAMM/CD168) expression vector.
Transfection of COS7 cells by lipofection
Transient transfection of these plasmids (4 μg) was carried out using Lipofectamine 2000 (Invitrogen Life Technologies) into COS7 cells in 6-well plates according to the manufacturer's instructions. After 36 hours, cells were harvested to perform a chromium-51 cytotoxicity assay.
Chromium-51 release cytotoxicity assay
AML blasts, DCs generated thereof, as well as RHAMM/CD168-, RHAMM/CD168&HLA-A2–, HLA-A2–, and mock-transfected COS7 cells, or T2 cells pulsed with 20 μM RHAMM-derived peptide R3 were labeled with 51CrO4 (chromium-51) for 2 hours as described earlier.22 After washing, labeled cells were incubated with CD8+ T cells at effector-target (E/T) ratios of 40:1 to 2.5:1 for 16 hours at 37 °C. Radioactivity in the supernatant from all wells of round-bottomed 96-well plates (Becton Dickinson) was measured by a gamma counter (PerkinElmer, Boston, MA). The percentage of specific lysis was calculated as follows: (chromium-51 release in the test well–spontaneous chromium-51 release)/(maximum chromium-51 release–spontaneous chromium-51 release).
Immunocytology for the detection of RHAMM/CD168 protein expression
The method has been described elsewhere.23 Briefly, cytospins of mock-, single-, and double-transfected COS7 cells, as well as K562 cells as a positive control, were incubated with a monoclonal anti-CD168 antibody (clone 2D6; Novocastra, Newcastle upon Tyne, United Kingdom) at a 1:100 dilution. A polyclonal biotinylated Fab antibody to mouse immunoglobulin G (IgG) reactive with all mouse isotypes and a streptavidin-biotinylated peroxidase complex (all obtained from Amersham, High Wycombe, United Kingdom) served as a detection system for the primary antibody. 3-Amino-9-ethylcarbazole (Sigma, St Louis, MO) was used as substrate for the enzyme. The cytospin preparations were faintly counterstained with Harris hematoxylin. Negative controls were performed without primary antibody.
Results
Patient characteristics
All patients included in the present study tested positive for both RHAMM/CD168 in (quantitative) RT-PCR and HLA-A2 in flow cytometry as described in the “Materials and methods.” Most of the patients showed a CR of their disease; 2 patients with AML were examined after allogeneic stem-cell transplantation. One patient showed a relapse, and 1 has a myelodysplastic syndrome (MDS) (refractory anemia, myeloblasts in bone marrow less than 5%). The cytogenetic findings and clinical characteristics are listed in Table 1.
To identify patients for further characterization of the T-cell immune response, we screened patients with AML for RHAMM/CD168 by RT-PCR and serology as described in “Materials and methods.”
Specific cellular immune responses to RHAMM/CD168-derived peptides
Ten HLA-A2–binding RHAMM/CD168-derived peptides (Table 2) were identified using 3 different computer algorithms as outlined in “Materials and methods.” These peptides (R1-R10) were synthesized and tested for their potential to elicit specific T-cell responses in 21 healthy volunteers and 13 patients with AML (with the clinical characteristics indicated in Table 1). ELISPOT measuring release of IFN-γ and granzyme B was used to assay reactivity of peptide-sensitized CD8+ T cells with test peptides presented by T2 cells.
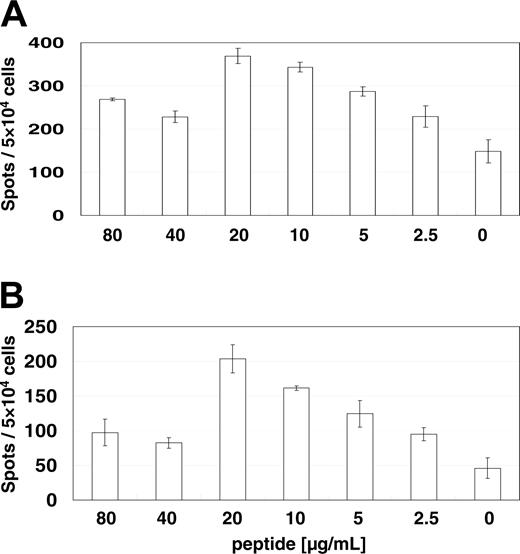
The T-cell reaction to the RHAMM/CD168-derived peptide R3 was demonstrated to be dependent on the peptide concentration as analyzed by ELISPOTs for IFN-γ (Figure 1A) and granzyme B (Figure 1B) indicating peptide-specific T-cell responses. The optimal dose of 20 μg peptide per milliliter was used in consecutive assays.
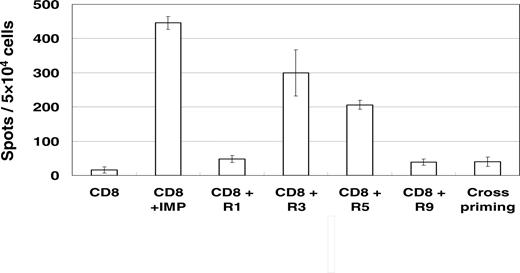
Figure 2 shows representative results for a granzyme B release by CD8+ T cells from an AML patient in a long-term complete remission over 8 years. A high number of reactive CD8+ T cells were counted after stimulation with the RHAMM/CD168-derived peptides R3 and R5. Also, a high granzyme B release was detected for the IMP peptide as a positive control. No T-cell reaction was found for the negative control (T2 cells without peptide) and for the RHAMM/CD168-derived peptides R1 and R9. Cross-reactivity was excluded by presensitizing CD8+ T cells with autologous CD8– APCs pulsed with the R3 peptide and thereafter evaluating their granzyme B release upon presentation of an unrelated MAGE3-derived peptide by T2 cells.
Determination of the optimal peptide concentration for MLPC. The reaction is specific because it is dependent on peptide concentration analyzed by ELISPOT assays for IFN-γ (A) and granzyme B (B). Specific CD8+ T-cell activation evaluated by IFN-γ and granzyme B release was dependent on the R3 peptide concentration indicating peptide-specific T-cell answers. The optimal dose of 20 μg peptide per milliliter was used in consecutive assays. Error bars indicate standard deviation.
Determination of the optimal peptide concentration for MLPC. The reaction is specific because it is dependent on peptide concentration analyzed by ELISPOT assays for IFN-γ (A) and granzyme B (B). Specific CD8+ T-cell activation evaluated by IFN-γ and granzyme B release was dependent on the R3 peptide concentration indicating peptide-specific T-cell answers. The optimal dose of 20 μg peptide per milliliter was used in consecutive assays. Error bars indicate standard deviation.
In healthy volunteers, specific immune responses were found in ELISPOT assays at a low frequency for the RHAMM/CD168-derived peptide R3 (5%), R5 (14%), and R9 (5%). In contrast, in patients with AML CD8+-specific immune responses were found especially for the RHAMM/CD168-derived peptide R3 (39%) but also at lower frequency for the peptide R5 (15%). No T-cell responses against the other RHAMM/CD168-derived peptides could be detected in patients with AML.
Tetramer staining of CD8+ T lymphocytes recognizing peptide R3 in the context of HLA-A2
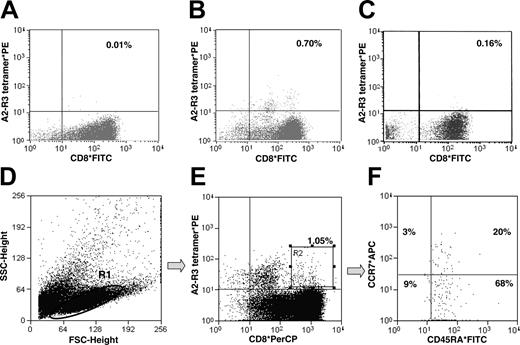
CD8+ lymphocytes from 5 patients with AML were subjected to one round of stimulation with autologous CD8 APCs in the presence or absence of the R3 peptide or in the presence of the irrelevant MAGE3-derived peptide (FLWGPRALV). Fluorescence-activated cell sorter (FACS) analysis with double staining by anti-CD8 antibodies and HLA-A2/R3 tetrameric complexes revealed a frequency of CD8+ T cells specifically recognizing the R3 peptide ranging from 0.60% to 0.70% in the presence of the R3 peptide (Figure 3B), in contrast with 0.01% to 0.04% in the absence of the peptide (Figure 3A), or up to 0.16% in the presence of the irrelevant peptide MAGE3 (Figure 3C). T lymphocytes (Figure 3D; gate R1) elicited by stimulation through the R3 peptide were gated for CD8+ HLA-A2/R3 tetramer+ cells (Figure 3E; gate R2) and further characterized as predominantly (68%) CCR7-CD45RA+ effector T cells (Figure 3F).
Specific immune responses (assessed by ELISPOT assays for granzyme B) against RHAMM/CD168 R3- and R5-pulsed T2 cells in an AML patient (patient 2 inTable 1) with a long-term complete remission over 8 years. No T-cell reaction was found for the negative control (T2 cells without peptide) and for the RHAMM/CD168-derived peptides R1 and R9. Cross-reactivity against an irrelevant peptide was excluded. All assays were performed in triplicate. The figure shows the results of a representative experiment. Error bars indicate standard deviation.
Specific immune responses (assessed by ELISPOT assays for granzyme B) against RHAMM/CD168 R3- and R5-pulsed T2 cells in an AML patient (patient 2 inTable 1) with a long-term complete remission over 8 years. No T-cell reaction was found for the negative control (T2 cells without peptide) and for the RHAMM/CD168-derived peptides R1 and R9. Cross-reactivity against an irrelevant peptide was excluded. All assays were performed in triplicate. The figure shows the results of a representative experiment. Error bars indicate standard deviation.
Multicolor staining of R3-specific T lymphocytes. CD8+ lymphocytes from an AML patient were subjected to one round of stimulation with autologous CD8 APCs in the presence (B) or absence (A) of the R3 peptide or an irrelevant MAGE3-derived peptide (C) as described in “Materials and methods.” A difference in frequency of R3-specific T cells could be noted. To further characterize R3-specific CD8+ T cells, lymphocytes were gated (D; gate R1) and stained with HLA-A2/R3 tetramers and anti-CD8*PerCP (E; gate R2). Lymphocytes from gate R2 were analyzed for their counterstaining of CCR7 and CD45RA (F). Most (68%) were revealed to be CD8+ HLA-A2/R3 tetramer+ CCR7-CD45RA+ effector T cells. The figure displays representative results. SSC indicates side scatter; FSC, forward scatter; APC, allophycocyanin (panel F only).
Multicolor staining of R3-specific T lymphocytes. CD8+ lymphocytes from an AML patient were subjected to one round of stimulation with autologous CD8 APCs in the presence (B) or absence (A) of the R3 peptide or an irrelevant MAGE3-derived peptide (C) as described in “Materials and methods.” A difference in frequency of R3-specific T cells could be noted. To further characterize R3-specific CD8+ T cells, lymphocytes were gated (D; gate R1) and stained with HLA-A2/R3 tetramers and anti-CD8*PerCP (E; gate R2). Lymphocytes from gate R2 were analyzed for their counterstaining of CCR7 and CD45RA (F). Most (68%) were revealed to be CD8+ HLA-A2/R3 tetramer+ CCR7-CD45RA+ effector T cells. The figure displays representative results. SSC indicates side scatter; FSC, forward scatter; APC, allophycocyanin (panel F only).
Peptide R3-primed CD8+ T cells lyse RHAMM/CD168+ AML blasts and DCs generated thereof
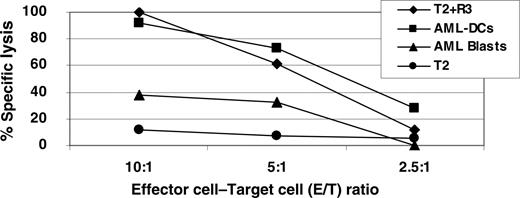
chromium-51 release assays showed a strong specific lysis of R3-pulsed T2 cells and AML DCs in contrast to unmanipulated AML blasts (Figure 4). Stimulated CD8+ T cells generated with the RHAMM/CD168 peptide R3 were able to recognize 40% of AML blasts at an E/T ratio of 10:1. In contrast, 100% of the AML DCs were killed at this E/T ratio showing similar lysis by specific T lymphocytes like R3-pulsed T2 cells. T2 cells pulsed with the unrelated MAGE3 peptide were not recognized (data not shown).
Simultaneous humoral and cellular immune responses to RHAMM/CD168
Simultaneous humoral and cellular immune responses to RHAMM/CD168 occur in patients with AML. Data from one exemplary patient are displayed in Figure 5: serologic results (Figure 5A) to RHAMM/CD168, mRNA expression of the gene (Figure 5B-C), and specific cellular immune responses against RHAMM/CD168 in a AML patient (Figure 5D). The specific lysis of T2 cells pulsed with the RHAMM/CD168 R3 peptide is demonstrated in Figure 5D: Specific CD8+ T cells generated of MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse R3-pulsed T2 cells but not T2 cells alone.
High specific lysis of RHAMM/CD168 R3-pulsed T2 cells and AML DCs in contrast to unmanipulated AML blasts as evaluated by chromium-51 release assay. Stimulated R3-primed CD8+ T cells were able to recognize AML blasts at a certain level, but AML DCs killed to 100% at an E/T ratio of 10:1. Primed T lymphocytes were also able to lyse R3-pulsed T2 cells. T2 cells were used as a negative control.
High specific lysis of RHAMM/CD168 R3-pulsed T2 cells and AML DCs in contrast to unmanipulated AML blasts as evaluated by chromium-51 release assay. Stimulated R3-primed CD8+ T cells were able to recognize AML blasts at a certain level, but AML DCs killed to 100% at an E/T ratio of 10:1. Primed T lymphocytes were also able to lyse R3-pulsed T2 cells. T2 cells were used as a negative control.
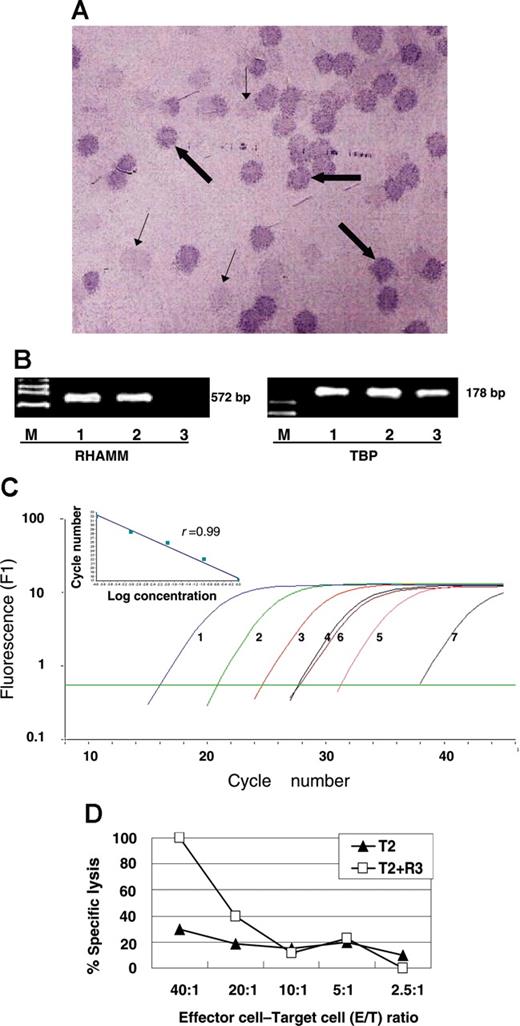
Positive serologic results to RHAMM/CD168, mRNA expression, and specific cellular immune responses against RHAMM/CD168 in a 69-year-old AML patient with a normal karyotype. Positive serologic immune responses against RHAMM/CD168 were detected using the phage plaque assay (A): bold arrows point to positive plaque-forming units (PFUs); thin arrows, to negative PFUs from the background of the cDNA library. Pictures of immunocytological stainings were taken with an Axiophot microscope with 25 × objective lens (Zeiss, Oberkochen, Germany) and a digital camera (JVC, Friedberg, Germany) coupled to a commercially avalable imaging system (Diskus, Königswinter, Germany). Positive mRNA expression was assessed by conventional RT-PCR (B). Lane M represents marker VI; lane 1, AML patient; lane 2, K562; and lane 3, healthy volunteer. In real-time RT-PCR, high expression of RHAMM/CD168 could be detected (C). Curve 1 represents 6.19 × 106 copies; curve 2, 6.19 × 105 copies; curve 3, 6.19 × 104 copies; curve 4, 6.19 × 103 copies; curve 5, 6.19 × 102 copies; curve 6, AML blasts; and curve 7, PBMCs of healthy volunteers (HV). The inset demonstrates the regression analysis with a regression coefficient of 0.99. Specific lysis of T2 cells pulsed with the RHAMM/CD168 R3 peptide occurred as demonstrated in panel D: specific CD8+ T cells generated of MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse R3-pulsed T2 cells but not T2 cells alone.
Positive serologic results to RHAMM/CD168, mRNA expression, and specific cellular immune responses against RHAMM/CD168 in a 69-year-old AML patient with a normal karyotype. Positive serologic immune responses against RHAMM/CD168 were detected using the phage plaque assay (A): bold arrows point to positive plaque-forming units (PFUs); thin arrows, to negative PFUs from the background of the cDNA library. Pictures of immunocytological stainings were taken with an Axiophot microscope with 25 × objective lens (Zeiss, Oberkochen, Germany) and a digital camera (JVC, Friedberg, Germany) coupled to a commercially avalable imaging system (Diskus, Königswinter, Germany). Positive mRNA expression was assessed by conventional RT-PCR (B). Lane M represents marker VI; lane 1, AML patient; lane 2, K562; and lane 3, healthy volunteer. In real-time RT-PCR, high expression of RHAMM/CD168 could be detected (C). Curve 1 represents 6.19 × 106 copies; curve 2, 6.19 × 105 copies; curve 3, 6.19 × 104 copies; curve 4, 6.19 × 103 copies; curve 5, 6.19 × 102 copies; curve 6, AML blasts; and curve 7, PBMCs of healthy volunteers (HV). The inset demonstrates the regression analysis with a regression coefficient of 0.99. Specific lysis of T2 cells pulsed with the RHAMM/CD168 R3 peptide occurred as demonstrated in panel D: specific CD8+ T cells generated of MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse R3-pulsed T2 cells but not T2 cells alone.
The RHAMM/CD168-derived peptide R3 is a naturally processed T-cell epitope
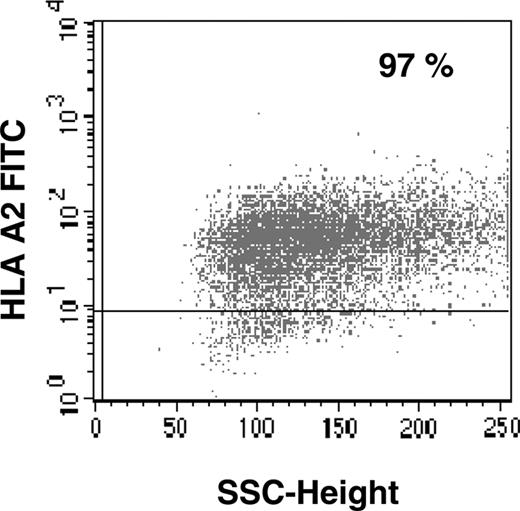
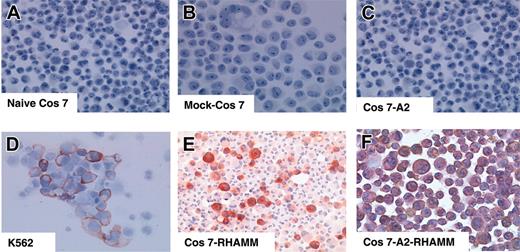
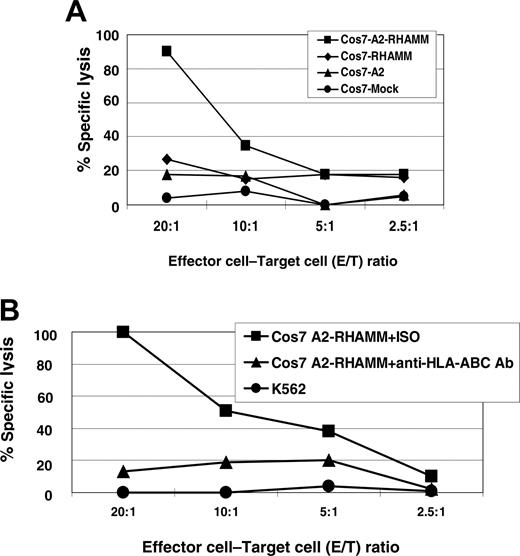
To demonstrate that the RHAMM/CD168 peptide R3 is naturally processed, COS7 cells were stably transfected by a plasmid vector containing HLA-A2&RHAMM/CD168 showing a transfection efficiency of more than 97% (Figure 6). The RHAMM/CD168 protein is expressed in most cells of leukemia patients and leukemia cell lines (eg, K562). Immunocytologic staining (ICS) for RHAMM/CD168 in RHAMM/CD168&HLA-A2 double-transfected or RHAMM/CD168 single-transfected COS7 cells showed a strong expression of the gene at the protein level, whereas RHAMM/CD168 protein was revealed not to be expressed in COS7 cells mock transfected with the vector or transfected with an HLA-A2–encoding vector alone (Figure 7). T lymphocytes generated in an MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse COS7 cells double transfected with HLA-A2 and RHAMM/CD168 but not mock-transfected or solely HLA-A2– or RHAMM/CD168-transfected cells (Figure 8A). Addition of an anti–HLA-ABC antibody but not an IgG isotype antibody abolished the lysis of RHAMM/CD168&HLA-A2 double transfectants by R3-primed T lymphocytes (Figure 8B). K562 cells lacking HLA-ABC molecules but expressing RHAMM/CD168 could not be lysed by R3-primed T cells (Figure 8B).
FACS analysis of HLA-A2/RHAMM/CD168 double-transfected COS7 cells. Flow cytometry for HLA-A2 was performed to detect HLA-A2&RHAMM/CD168 double-transfected COS7 cells. A stable transfection up to more than 97% was achieved after positive selection with antibiotics. For flow cytometry, no appropriate mAb against RHAMM/CD168 was available.
FACS analysis of HLA-A2/RHAMM/CD168 double-transfected COS7 cells. Flow cytometry for HLA-A2 was performed to detect HLA-A2&RHAMM/CD168 double-transfected COS7 cells. A stable transfection up to more than 97% was achieved after positive selection with antibiotics. For flow cytometry, no appropriate mAb against RHAMM/CD168 was available.
Discussion
Targeted immunotherapies require the definition and characterization of appropriate antigens. Two such antigens are under current clinical investigation in peptide vaccination trials for patients with AML24,25 (ie, proteinase 3 and the Wilms tumor gene-1 [WT1]). Proteinase 3 is expressed in AML and CML patients, and a cytotoxic T-lymphocyte (CTL) epitope designated PR1 has been characterized.26,27 However, proteinase 3 is expressed in various normal tissues and is a synonym for the autoantigen of the Wegener granulomatosis (C-ANCA).14,28 Therefore, immunotherapies targeting proteinase 3 might elicit autoimmune reactions. However, such reactions have not been observed in ongoing clinical trials. The second candidate, WT1, is expressed in AML blasts but also in CD34+ stem cells of healthy donors.28-32 Immunotherapies using WT1-derived peptides might therefore induce CTLs against these progenitor cells of the normal early hematopoiesis as observed in patients with MDS after vaccination with a WT1 peptide.25
Immunocytologic staining for RHAMM/CD168 in naive versus transfected COS7 cells. The cell line K562 served as a positive control (D). ICS for RHAMM/CD168 in RHAMM/CD168 single-transfected COS7 cells (E) and RHAMM/CD168&HLA-A2 permanently double-transfected COS7 cells (F) showed a strong expression of RHAMM/CD168 in all transfected cells, whereas RHAMM/CD168 protein was not expressed in naive COS7 cells (A), COS7 cells mock transfected with the vector pBudCE4.1 (B), or COS7 cells solely transfected with HLA-A*0201 but not with RHAMM/CD168 (C). All photomicrographs were taken at a magnification × 100.
Immunocytologic staining for RHAMM/CD168 in naive versus transfected COS7 cells. The cell line K562 served as a positive control (D). ICS for RHAMM/CD168 in RHAMM/CD168 single-transfected COS7 cells (E) and RHAMM/CD168&HLA-A2 permanently double-transfected COS7 cells (F) showed a strong expression of RHAMM/CD168 in all transfected cells, whereas RHAMM/CD168 protein was not expressed in naive COS7 cells (A), COS7 cells mock transfected with the vector pBudCE4.1 (B), or COS7 cells solely transfected with HLA-A*0201 but not with RHAMM/CD168 (C). All photomicrographs were taken at a magnification × 100.
The RHAMM/CD168-derived peptide R3 is naturally processed. R3-primed effector cells showed specific lysis of RHAMM/CD168&HLA-A2–transfected COS7 cells. COS7 cells were transfected with the plasmid vector pBudCE4.1 containing RHAMM/CD168 and/or HLA-A2. T lymphocytes generated in an MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse COS7 cells double transfected with HLA-A2 and RHAMM/CD168 but not mock-transfected or solely HLA-A2– or RHAMM/CD168-transfected cells (A). Addition of an anti–HLA-ABC antibody but not an IgG isotype antibody (ISO) abolished the lysis of T2 pulsed with the R3 peptide by R3-primed T lymphocytes (B). K562 cells lacking HLA-ABC molecules but expressing RHAMM/CD168 could not be lysed by R3-pulsed T cells (B).
The RHAMM/CD168-derived peptide R3 is naturally processed. R3-primed effector cells showed specific lysis of RHAMM/CD168&HLA-A2–transfected COS7 cells. COS7 cells were transfected with the plasmid vector pBudCE4.1 containing RHAMM/CD168 and/or HLA-A2. T lymphocytes generated in an MLPC with the RHAMM/CD168-derived peptide R3 were able to lyse COS7 cells double transfected with HLA-A2 and RHAMM/CD168 but not mock-transfected or solely HLA-A2– or RHAMM/CD168-transfected cells (A). Addition of an anti–HLA-ABC antibody but not an IgG isotype antibody (ISO) abolished the lysis of T2 pulsed with the R3 peptide by R3-primed T lymphocytes (B). K562 cells lacking HLA-ABC molecules but expressing RHAMM/CD168 could not be lysed by R3-pulsed T cells (B).
Because the vaccination with the above-mentioned antigens might raise clinical problems, we investigated in the present study the novel leukemia antigen RHAMM/CD168 previously defined by us in a SEREX analysis.12 RHAMM/CD168 is a glycosaminoglycan and extracellular matrix molecule that plays a fundamental role in cell growth, differentiation, and motility.33 Overexpression of RHAMM/CD168 is essential for ras-mediated transformation,34 and it is associated with the development of metastases.35-37 Mitosis is directly linked to RHAMM/CD168 by suppressing cyclin-dependent kinase-2 (CDK2) and cyclin B1 expression. Moreover, RHAMM/CD168 might contribute to the differentiation and maturation of leukemic blasts like CD44, another receptor for hyaluronic acid.38 Therefore, immune therapies targeting RHAMM/CD168 might not only target an immunogenic antigen but also a gene critically involved in cell cycle, differentiation, and proliferation.
We defined RHAMM/CD168 as a new immunogenic antigen in myeloid leukemias and several solid tumors by means of the SEREX method.14 SEREX has been used to define numerous tumor-associated antigens (TAAs) in solid tumors.39-42 However, none of SEREX-defined CT/CG antigens hitherto tested in PCR assays was expressed in AML blasts.13,14,43
RHAMM/CD168 was revealed to be a promising antigen because of its highly exquisite mRNA expression pattern in normal tissues and its protein expression in about 80% of patients with AML and the incidence of positive humoral immune responses in approximately 40% of the patients with AML. A good correlation of mRNA expression and IgG antibody titers was noted. In this study, we could demonstrate by immunocytologic staining that RHAMM/CD168 is expressed both in the cytoplasma and on the surface of leukemic blasts, opening new strategies for potentially using RHAMM/CD168-targeting antibody therapies in analogy to the CD33-directed antibody therapy with gemtuzumab ozogamicin.44,45
The focus of the present study was to define T-cell epitopes derived from RHAMM/CD168. The analysis of 10 potential HLA-A2–binding peptides revealed 2 peptides designated R3 and R5 that were recognized by CD8+ cells in the context of this HLA class-I type. T-cell responses to either one of these peptides could be detected in 54% of the patients with AML, indicating that patients with AML in long-term remission are immunocompetent and that R3 and R5 are immunogenic. Taken together with the incidence of high-titer IgG antibodies against RHAMM/CD168 in 40% of patients with AML,12 these data characterize RHAMM/CD168 as another SEREX antigen eliciting both humoral and cellular immune responses like NY-ESO1 and SSX-2.46 R3 and R5 are naturally processed antigen epitopes, because RHAMM/CD168&HLA-A*0201 double-transfected but not RHAMM/CD168 or HLA-A*0201 single-transfected COS7 cells were recognized by R3/R5-primed T lymphocytes. The presentation of RHAMM/CD168-derived epitope peptides was clearly HLA class I restricted as demonstrated by antibody-blocking experiments and by the deficiency of the RHAMM/CD168-specific T cells to lyse RHAMM/CD168+ K562 cells lacking HLA class I molecules.
Our data for RHAMM/CD168-specific CD8+ T-cell reactivity in patients with AML obtained by ELISPOT assays for both IFN-γ and granzyme B production could be confirmed in chromium-51 release cytotoxicity assays and by tetramer staining. The T-cell responses were related to the concentration of the epitope peptide in the assays, supporting the specificity of the T-cell response. The specificity of the immune responses was also confirmed by ELISPOT and cytotoxicity assays of T cells primed by RHAMM/CD168, which were unable to recognize unrelated peptides (cross-priming).
The higher lytic efficiency of RHAMM/CD168-specific T cells against DCs generated from AML blasts when compared with the mere AML blasts indicates the potential of these professional antigen-presenting cells maintaining the expression of the leukemia antigen to improve the recognition of RHAMM/CD168 through up-regulation of costimulatory molecules and cytokine release.20 The production of AML DCs is cumbersome and costly. Augmentation of the T-cell response to RHAMM/CD168 might also be feasible by the increase of the concentration of RHAMM/CD168 epitope peptides for the presentation toward T cells. The present study provided evidence that CD8+ T cells from patients with AML are able to recognize RHAMM/CD168-derived epitope peptides in the context of HLA-A2. Such presensitized T lymphocytes were able to efficiently lyse RHAMM/CD168+ autologous AML blasts. We therefore initiated a clinical peptide vaccination trial for patients with AML, MDS, or multiple myeloma using the most immunogenic epitope peptide R3 as a cancer vaccine. RHAMM/CD168 is also expressed in patients with other hematologic malignancies12,37,47 and solid tumors,33-36 which suggests a potentially broader clinical application of cancer vaccines and mAbs targeting RHAMM/CD168.
Prepublished online as Blood First Edition Paper, April 12, 2005; DOI 10.1182/blood-2004-12-4787.
Supported by a generous grant from the German José Carreras Leukemia Foundation (J.G., M.W., H.D., M.S.) and grants from the German Academic Exchange Service (DAAD) and Catholic Academic Exchange Service (KAAD) (K.G.).
J.G. and L.L. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ms Götz, Ms Szmaragowska, and Ms Nerbas for their excellent technical support and highly appreciate Ms Götz's taking care of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal