Abstract
Glycoprotein (GP) Ibα, a member of the leucine-rich repeat (LRR) protein family, mediates platelet adhesion to immobilized von Willebrand factor (VWF). We investigated the role in VWF binding of charged residues in the LRR region of GP Ibα that are conserved in human, canine, and murine proteins. Substitution of His86 with either Ala or Glu resulted in a gain of VWF-binding function as judged by increased VWF binding in the presence of the modulators ristocetin and botrocetin and by enhanced adhesion of Chinese hamster ovary (CHO) cells expressing the mutant GP Ibα to immobilized VWF under conditions of flow. This is the first report of a gain-of-function phenotype resulting from mutations in the LRR region of GP Ibα. Because His86 is 2 nm away from the region of GP Ibα with the largest surface of contact with VWF, the data suggest that the LRRs regulate GP Ibα affinity for VWF allosterically.
Introduction
Platelet adhesion at high shear stress to sites of blood vessel injury is initiated by the binding of the platelet glycoprotein (GP) Ib-IX-V complex to its ligand on the exposed subendothelium, von Willebrand factor (VWF). The VWF-binding site is contained within the amino terminus of GP Ibα, the largest polypeptide of the GP Ib-IX-V complex. This region of GP Ibα contains 8 tandem leucine-rich repeats (LRRs) and binds VWF largely through electrostatic interactions.1,2 Single, relatively conserved amino acid changes in the LRR region have been reported to result either in the absence (Asn158Lys3 ) or the dysfunction (Leu57Phe,4 Cys65Arg,5 Ala156Val6,7 ) of GPIbα, leading to Bernard-Soulier syndrome, but all the reported patient and experimental mutations that produce gain-of-function have been reported to lie in a region C-terminal to the LRRs. Here, we investigate the role in VWF binding of charged residues within the GP Ibα LRR region conserved in humans, dogs, and mice.
Materials and methods
Site-directed mutagenesis
Mutagenesis was performed directly on the GP Ibα cDNA cloned into the mammalian expression vector pDX8 using a commercial polymerase chain reaction (PCR)–based mutagenesis kit (QuikChange; Stratagene, La Jolla, CA). Mutated GP Ibα cDNAs were sequenced to verify the mutations.
Cell lines and transfections
Chinese hamster ovary (CHO) cells expressing GP Ibα, GP Ibβ, and GP IX (CHOαβIX)8 were grown in α–minimal essential medium (α-MEM; Life Technologies, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 400 μg/mL G418, 80 μg/mL methotrexate, 400 μg/mL hygromycin, 100 U/mL penicillin, and 100 U/mL streptomycin. To develop stable cell lines expressing GP Ibα mutants, we transfected the mutant GP Ibα cDNAs and a hygromycin-resistance marker (LipofectAMINE2000; Life Technologies) into CHO βIX cells, which stably express GP Ibβ and GP IX.9 Transfected cells were grown in progressively increasing hygromycin concentrations, to a final concentration of 400 μg/mL. Cells were then repeatedly sorted for GP Ib-IX expression with antibody WM23–coupled magnetic beads according to the procedure described by the manufacturer (Dynabeads; Dynal, Lake Success, NY).
Flow cytometry
The surface expression and conformation of mutant GP Ibα polypeptides were examined by flow cytometry after labeling individual aliquots of cells with one of the following GP Ibα monoclonal antibodies: AN51 (DAKO, Carpinteria, CA), MB45 (Pelicluster, Amsterdam, The Netherlands), Hip1 (BD PharMingen, San Diego, CA), 6D1 (Dr Barry Coller, Rockefeller University, New York, NY), AP1 (Dr Dermot Kenny, The Royal College of Surgeons, Dublin, Ireland), AK2 (Dr Michael Berndt, Monash University, Melbourne, Australia), SZ2 (Research Diagnostics, Flanders, NJ), and WM23 (Dr Michael Berndt). Surface expression of the mutant polypeptides was determined using WM23 because its epitope is not affected by mutations in the GP Ibα N-terminus.10,11 In brief, adherent cells were detached with versene (Life Technologies), then washed with and resuspended in Tris-buffered saline (TBS) containing 0.5% bovine serum albumin (BSA). The cells (5 × 105) were then incubated with 2 μg/mL antibody for 60 minutes at room temperature, washed twice, and incubated with a fluorescein isothiocyanate (FITC)–conjugated rabbit antimouse secondary antibody (Zymed Laboratories, South San Francisco, CA) for 30 minutes. Cells were then rewashed twice and analyzed on a flow cytometer (Epics XL; Beckman Coulter, Fullerton, CA) stimulating with laser light at 488 nm and collecting light emitted at more than 520 nm.
Enzyme-linked immunosorbent assay for ristocetin- and botrocetin-mediated VWF binding
Ristocetin- and botrocetin-mediated VWF binding to cells expressing wild-type or mutant GP Ib-IX complex was measured as described previously.12 Cells (2 × 105) were plated on wells of a 24-well plate coated with 0.01% poly-L-lysine. After 36 hours, the cells were washed twice with phosphate-buffered saline (PBS)/0.5% BSA and were incubated for 20 minutes at room temperature with either GP Ibα antibodies (1 μg/mL) or increasing concentrations of VWF (0-4 μg/mL) in the presence of 1.5 mg/mL ristocetin or 2.0 μg/mL botrocetin. For cells expressing mutants that displayed gain-of-function, VWF binding was examined at various modulator concentrations. For ristocetin, the cells were incubated at ristocetin concentrations of 0, 0.1, 0.8, and 1.6 mg/mL, and for botrocetin, they were incubated at concentrations of 0, 0.25, and 2.0 μg/mL; all incubations occurred in the presence of 4 μg/mL VWF. Nonspecific binding was determined in the absence of modulators or in the presence of saturating concentrations of GP Ibα antibodies, includingAK2,AP1, and SZ2.AK210,13 and AP114,15 inhibit ristocetin-induced binding of VWF to GP Ib and adhesion of GP Ib-IX complex–expressing cells to VWF under flow. SZ2 preferentially inhibits botrocetin-dependent VWF binding.13,16 Unbound VWF was removed by washing 4 times with 0.1 M sodium acetate/0.5× PBS. The cells were then incubated in α-MEM/5% fetal bovine serum (FBS) with either horseradish peroxidase (HRP)–conjugated rabbit anti–human VWF (DAKO) or HRP-conjugated goat antimouse antibody (Pierce, Rockford, IL) diluted for 10 minutes at 4°C. The cells were then washed twice with TBS/0.5% BSA. To normalize for the slight differences in GP Ibα expression between the cell lines, the data are expressed as a ratio of VWF binding to WM23 binding.
Cell adhesion under flow
The system includes a parallel-plate flow chamber and an inverted-stage microscope (Eclipse TE300; Nikon, Garden City, NY) equipped with a high-speed digital camera (Quantix; Photometrics, Tucson, AZ). The parallel-plate flow chamber was composed of a polycarbonate slab, a silicon gasket, and a glass coverslip held together by vacuum such that the coverslip forms the bottom of the chamber. Coverslips were coated with 20 μg/mL human VWF. Cell suspensions in TBS were drawn through the chamber with a Harvard syringe pump; the wall shear stress was proportional to the flow rate.17 The binding of gain-of-function mutants was examined on coverslips coated at VWF concentrations of either 4, 10, or 20 μg/mL.
In studies of GP Ib–mediated cell adhesion and rolling on VWF, suspensions of mutant CHO αβIX, wild-type CHO αβIX, or control CHO βIX cells (1 × 106 cells/mL) were injected into the chamber, and the cells were allowed to settle for 1 minute onto the immobilized VWF. The chamber was then perfused with TBS containing 0.5% BSA, generating wall shear stresses of 2.5, 10, or 20 dyne/cm2. The rolling of cells was recorded for 2 minutes on videotape. Acquired images were analyzed off-line using MetaMorph software (Universal Images, West Chester, PA) to quantify the number of adherent cells and their rolling velocities.18 Cells considered to be rolling were those that translocated over the matrix while maintaining constant contact. Rolling velocity was defined as the distance a cell travels during a defined period (μm/s).
Statistical analysis
Groups were compared using the nonpaired t test. All data are presented as the mean plus or minus SEM.
Results
Conserved charged amino acids in the LRR maintain the conformation of GP Ibα
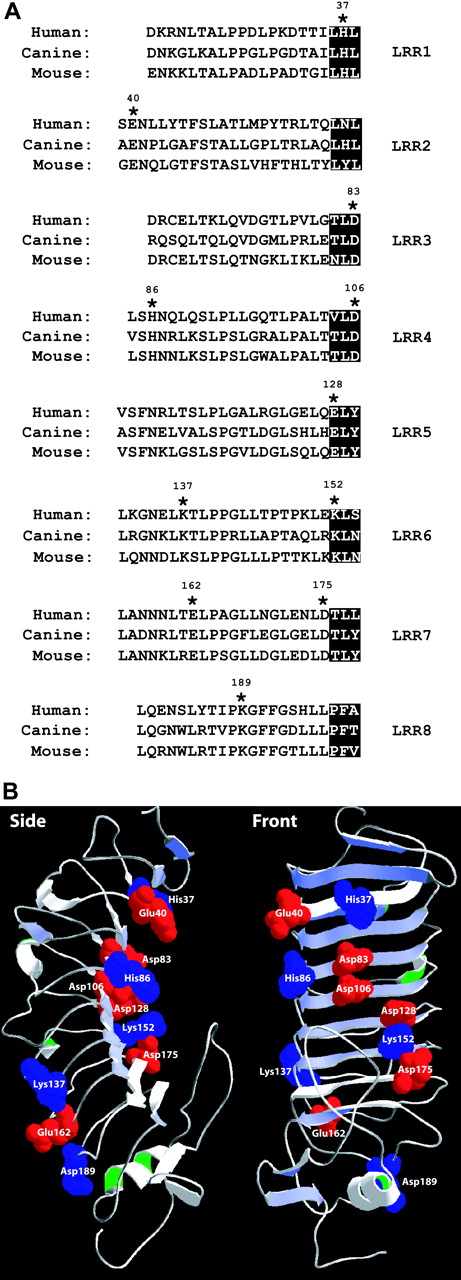
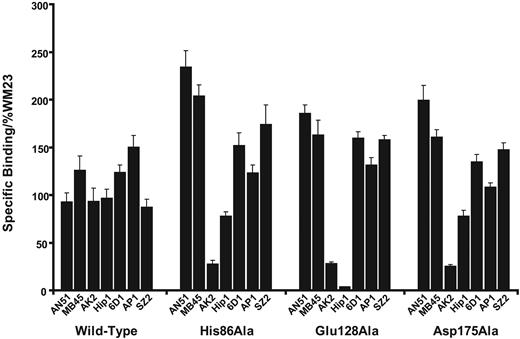
The following 11 charged residues identical in human, canine, and murine GP Ibα were mutated to Ala: His37, Glu40, Asp83, His86, Asp106, Glu128, Lys137, Lys152, Glu162, Asp175, and Lys189 (Figure 1A). These residues are distributed throughout the GP Ibα LRRs (Figure 1B). Mutant cDNAs were transfected into CHOβIX cells and selected for stable expression through continued culture in hygromycin-containing medium. All mutants were expressed on the cell surface and, when they were tested for function, at comparable surface levels (Figure 2A). We evaluated the possibility that the mutations produced long-range changes in GP Ibα conformation using a panel of monoclonal antibodies with defined epitopes, most of which are conformation sensitive. Epitopes were located in the LRRs (AN51, MB45, AK2, Hip1, and 6D1), the C-terminal flanking region (AP1), or the anionic sulfated region (SZ2) (Figure 2B). To adjust for any differences in surface expression, binding was normalized to the binding of WM23, the epitope of which is unaffected by the mutations. As shown in Table 1 and Figure 3, the monoclonal antibody–binding patterns of these mutants were vastly different. Of particular interest, the binding of AN51 and SZ2 was significantly enhanced when Asp83, His86, Glu128, or Asp175 was converted to Ala (Figure 3). Because AN51 recognizes a conformation-sensitive epitope within the GP Ibα N-terminus (His1-Ile35),14 the increased binding of this antibody likely reflects a change in the overall conformation of the GP Ibα N-terminal flank (the sequence N-terminal to the LRRs). Although SZ2 recognizes sodium dodecyl sulfate (SDS)–denatured GP Ibα,19 its increased binding to the mutants may reflect increased epitope exposure in the folded polypeptide. Together our data indicate that charged conserved amino acids in the LRR region are important for maintaining the conformation of GP Ibα.
Binding of anti-GP Ib monoclonal antibodies to GP Ibα mutants
. | AN51 . | MB45 . | AK2 . | Hip1 . | 6D1 . | AP1 . | SZ2 . |
|---|---|---|---|---|---|---|---|
| Wild-type | 93 ± 9 | 126 ± 15 | 93 ± 14 | 96 ± 10 | 124 ± 7 | 150 ± 12 | 88 ± 8 |
| His37Ala | 108 ± 14 | 147 ± 23 | 36 ± 2 | 13 ± 1 | 101 ± 16 | 103 ± 12 | 81 ± 6 |
| Glu40Ala | 125 ± 10 | 132 ± 12 | 34 ± 2 | 8 ± 0 | 114 ± 7 | 106 ± 5 | 101 ± 10 |
| Asp83Ala | 173 ± 11 | 134 ± 12 | 63 ± 2 | 79 ± 0 | 110 ± 10 | 106 ± 3 | 137 ± 8 |
| His86Ala | 234 ± 17 | 204 ± 12 | 28 ± 4 | 78 ± 5 | 152 ± 13 | 123 ± 9 | 174 ± 21 |
| Asp106Ala | 68 ± 19 | 87 ± 10 | 71 ± 1 | 71 ± 7 | 84 ± 6 | 88 ± 4 | 56 ± 5 |
| Glu128Ala | 186 ± 9 | 163 ± 16 | 28 ± 1 | 4 ± 0 | 160 ± 7 | 131 ± 8 | 158 ± 4 |
| Lys137Ala | 96 ± 19 | 106 ± 3 | 74 ± 3 | 81 ± 0 | 100 ± 10 | 122 ± 8 | 71 ± 9 |
| Lys152Ala | 128 ± 13 | 129 ± 15 | 58 ± 6 | 93 ± 12 | 117 ± 5 | 113 ± 11 | 113 ± 2 |
| Glu162Ala | 86 ± 19 | 111 ± 17 | 74 ± 2 | 86 ± 11 | 97 ± 9 | 110 ± 8 | 64 ± 5 |
| Asp175Ala | 199 ± 15 | 161 ± 8 | 25 ± 1 | 78 ± 6 | 135 ± 8 | 109 ± 4 | 147 ± 8 |
| Lys189Ala | 79 ± 24 | 96 ± 14 | 75 ± 1 | 72 ± 3 | 90 ± 7 | 95 ± 3 | 56 ± 7 |
. | AN51 . | MB45 . | AK2 . | Hip1 . | 6D1 . | AP1 . | SZ2 . |
|---|---|---|---|---|---|---|---|
| Wild-type | 93 ± 9 | 126 ± 15 | 93 ± 14 | 96 ± 10 | 124 ± 7 | 150 ± 12 | 88 ± 8 |
| His37Ala | 108 ± 14 | 147 ± 23 | 36 ± 2 | 13 ± 1 | 101 ± 16 | 103 ± 12 | 81 ± 6 |
| Glu40Ala | 125 ± 10 | 132 ± 12 | 34 ± 2 | 8 ± 0 | 114 ± 7 | 106 ± 5 | 101 ± 10 |
| Asp83Ala | 173 ± 11 | 134 ± 12 | 63 ± 2 | 79 ± 0 | 110 ± 10 | 106 ± 3 | 137 ± 8 |
| His86Ala | 234 ± 17 | 204 ± 12 | 28 ± 4 | 78 ± 5 | 152 ± 13 | 123 ± 9 | 174 ± 21 |
| Asp106Ala | 68 ± 19 | 87 ± 10 | 71 ± 1 | 71 ± 7 | 84 ± 6 | 88 ± 4 | 56 ± 5 |
| Glu128Ala | 186 ± 9 | 163 ± 16 | 28 ± 1 | 4 ± 0 | 160 ± 7 | 131 ± 8 | 158 ± 4 |
| Lys137Ala | 96 ± 19 | 106 ± 3 | 74 ± 3 | 81 ± 0 | 100 ± 10 | 122 ± 8 | 71 ± 9 |
| Lys152Ala | 128 ± 13 | 129 ± 15 | 58 ± 6 | 93 ± 12 | 117 ± 5 | 113 ± 11 | 113 ± 2 |
| Glu162Ala | 86 ± 19 | 111 ± 17 | 74 ± 2 | 86 ± 11 | 97 ± 9 | 110 ± 8 | 64 ± 5 |
| Asp175Ala | 199 ± 15 | 161 ± 8 | 25 ± 1 | 78 ± 6 | 135 ± 8 | 109 ± 4 | 147 ± 8 |
| Lys189Ala | 79 ± 24 | 96 ± 14 | 75 ± 1 | 72 ± 3 | 90 ± 7 | 95 ± 3 | 56 ± 7 |
Values are percentages of the values obtained with the WM23 antibody and represent the mean ± SEM.
Location of GP Ibx mutations. (A) Alignment of the leucine-rich repeats (LRRs) of human, canine, and murine GP Ibα. The 11 conserved charged amino acids are indicated by asterisks. The β strands in the LRRs of GP Ibα are highlighted. (B) Location of residues mutated in the current study within the structure of the GP Ibα N-terminus. Acidic residues appear in red; basic residues appear in blue.
Location of GP Ibx mutations. (A) Alignment of the leucine-rich repeats (LRRs) of human, canine, and murine GP Ibα. The 11 conserved charged amino acids are indicated by asterisks. The β strands in the LRRs of GP Ibα are highlighted. (B) Location of residues mutated in the current study within the structure of the GP Ibα N-terminus. Acidic residues appear in red; basic residues appear in blue.
Surface expression of mutant GP Ibα and location of antibody epitopes. (A) Surface expression of GP Ibα in the cell lines tested was evaluated at the time of the functional studies by flow cytometry after the cells were labeled with antibody WM23. Error bar indicates SEM. (B) Schema showing the locations of the epitopes of antibodies used to evaluate the conformational consequences of the mutations.
Surface expression of mutant GP Ibα and location of antibody epitopes. (A) Surface expression of GP Ibα in the cell lines tested was evaluated at the time of the functional studies by flow cytometry after the cells were labeled with antibody WM23. Error bar indicates SEM. (B) Schema showing the locations of the epitopes of antibodies used to evaluate the conformational consequences of the mutations.
The results with Hip1 were also interesting. This antibody inhibits VWF binding induced by ristocetin and botrocetin, and its epitope has been mapped to a region within the GP Ibα LRRs spanning residues 59 to 81.14,20 Of interest, 3 mutations— His37Ala, Glu40Ala, and Glu128Ala—outside the region containing the assigned epitope effectively abolished Hip1 binding. All the affected residues are close to each other in GP Ibα structure (Figure 1B), suggesting that they may contribute to the Hip1 epitope. Of the 4 mutations, only Glu128Ala disrupted ristocetin-induced VWF binding, indicating that this mutation also may have a long-range conformational effect.
Alteration of VWF-binding function by the LRR mutants
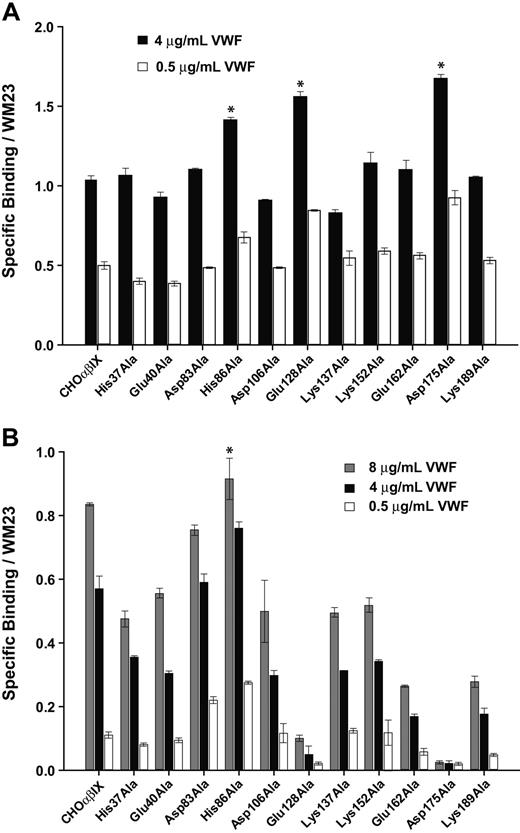
We then examined the interaction of cells expressing mutant GP Ibα with VWF in the presence of modulators and under flow.16 In addition to binding AN51 and SZ2 at higher levels, cells expressing mutants His86Ala, Glu128Ala, and Asp175Ala also exhibited a substantially altered interaction with VWF in the presence of modulators (Figure 4) and under flow (Table 2), exhibiting either gain-of-function with respect to VWF binding (His86Ala) or loss-of-function (Glu128Ala and Asp175). Mutants His86Ala, Glu128Ala, and Asp175Ala bound significantly more VWF than wild-type GP Ibα in the presence of botrocetin, further evidence that GP Ibα conformation was substantially altered by the mutations (Figure 4A). This enhanced binding is consistent with the increased binding of each of these mutants by SZ2, an antibody that binds the anionic sulfated region of GP Ibα and inhibits botrocetin-induced binding of VWF.14 Of interest, even subtle mutations of the anionic sulfated region inhibit botrocetin-induced VWF binding.21,22
Rolling velocity of CHO cells expressing GP Ibα mutant
. | 2.5 dyne/cm2 . | 10 dyne/cm2 . | 20 dyne/cm2 . |
|---|---|---|---|
| CHOαβ9 | 107 ± 34 | 180 ± 54 | 221 ± 67 |
| His37Ala | 153 ± 70 | 302 ± 94 | 517 ± 109 |
| Glu40Ala | 159 ± 72 | 223 ± 77 | 378 ± 98 |
| Asp83Ala | 110 ± 41 | 173 ± 46 | 214 ± 75 |
| His86Ala | 73 ± 22 | 136 ± 37 | 178 ± 30 |
| Asp106Ala | 133 ± 48 | 309 ± 92 | 373 ± 77 |
| Glu128Ala | No adhesion | No adhesion | No adhesion |
| Lys137Ala | 121 ± 38 | 260 ± 80 | 457 ± 93 |
| Lys152Ala | 142 ± 49 | 260 ± 93 | 314 ± 85 |
| Glu162Ala | 146 ± 76 | No adhesion | No adhesion |
| Asp175Ala | No adhesion | No adhesion | No adhesion |
| Lys189Ala | 142 ± 48 | No adhesion | No adhesion |
. | 2.5 dyne/cm2 . | 10 dyne/cm2 . | 20 dyne/cm2 . |
|---|---|---|---|
| CHOαβ9 | 107 ± 34 | 180 ± 54 | 221 ± 67 |
| His37Ala | 153 ± 70 | 302 ± 94 | 517 ± 109 |
| Glu40Ala | 159 ± 72 | 223 ± 77 | 378 ± 98 |
| Asp83Ala | 110 ± 41 | 173 ± 46 | 214 ± 75 |
| His86Ala | 73 ± 22 | 136 ± 37 | 178 ± 30 |
| Asp106Ala | 133 ± 48 | 309 ± 92 | 373 ± 77 |
| Glu128Ala | No adhesion | No adhesion | No adhesion |
| Lys137Ala | 121 ± 38 | 260 ± 80 | 457 ± 93 |
| Lys152Ala | 142 ± 49 | 260 ± 93 | 314 ± 85 |
| Glu162Ala | 146 ± 76 | No adhesion | No adhesion |
| Asp175Ala | No adhesion | No adhesion | No adhesion |
| Lys189Ala | 142 ± 48 | No adhesion | No adhesion |
Units for all values are μm/s.
Monoclonal antibody binding patterns of the GP Ibα mutants. The binding of monoclonal antibodies with epitopes within the GP Ibα ligand–binding domain was compared with the binding of WM23, which binds outside the region. Values represent the ratio of the mean fluorescence intensity of each monoclonal antibody to that of WM23. Error bars indicate SEM.
Monoclonal antibody binding patterns of the GP Ibα mutants. The binding of monoclonal antibodies with epitopes within the GP Ibα ligand–binding domain was compared with the binding of WM23, which binds outside the region. Values represent the ratio of the mean fluorescence intensity of each monoclonal antibody to that of WM23. Error bars indicate SEM.
In contrast to botrocetin-induced binding, each of the 11 mutants except His86Ala showed decreased ristocetin-induced VWF binding compared with wild-type GP Ibα (Figure 4B). In particular, the mutations Glu128Ala and Asp175Ala almost completely abolished ristocetin-induced VWF binding.
The hydrodynamic interaction of mutant-expressing cells with immobilized VWF closely resembled the pattern of VWF binding induced by ristocetin but not that induced by botrocetin, consistent with our earlier findings.12,15 As seen in Table 2, cells expressing wild-type GP Ibα adhered to the matrix and rolled at mean velocities of 107 μm/s, 161 μm/s, and 220 μm/s at 2.5, 10, and 20 dyne/cm,2 respectively. Changes to the species-conserved charged residues profoundly affected the interaction. The mutants Glu128Ala and Asp175Ala failed to adhere to the surface or roll at any shear stress. Mutants Glu162Ala and Lys189Ala attached to the VWF surface at 2.5 dyne/cm2 but were unable to attach at shear stresses of 10 dyne/cm2 or higher (20 dyne/cm2).
Mutation of His86 to Ala or Glu leads to gain of VWF binding function
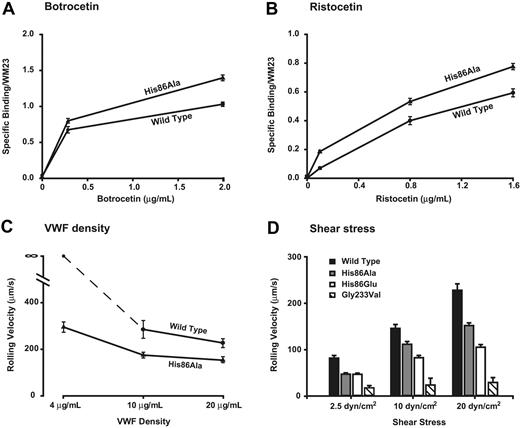
CHO cells expressing His86Ala GP Ibα exhibited gain-of-function in every assay. They bound more VWF in the presence of botrocetin (Figure 5A) or ristocetin (Figure 5B) at all concentrations and rolled on the VWF surface more slowly than cells expressing wild-type GP Ibα (Table 2). Moreover, His86Ala cells were able to attach to a VWF surface coated at a density below the density needed to support the attachment of cells expressing wild-type GP Ibα, an indication of increased receptor-ligand affinity (Figure 5C).
We also investigated the effect of converting His86 to Glu, a residue of opposite charge. Cells expressing the His86Glu mutant displayed even greater gain-of-function than the His86Ala-expressing cells, rolling more slowly over the VWF surface at the higher shear stresses of 10 and 20 dyne/cm2 (Figure 5D).
Discussion
To date, 4 gain-of-function mutations of GP Ibα have been identified. Gly233Val and Met239Val are natural mutations that associate with platelet-type VWD23 ; Asp235Val and Lys237Val are artificial mutants.1 All 4 mutations are found in a disulfide loop between Cys209 and Cys248, and all, with the exception of Asp235Val, have been postulated to interact directly with the VWF A1 domain based on the crystal structure of the GP Ibα–A1 complex.2 We compared one of these mutants, Gly233Val, with the 2 gain-of-function mutants described here. Gly233Val cells rolled more slowly on VWF than either of the His86 mutants, indicating that the Gly233Val mutation has a more profound effect on the affinity of the GP Ibα–VWF bond (Figure 5D). This is not altogether surprising given that the mutations in the C-terminal β switch of GP Ibα (residues 227-241), including Gly233Val, lead to a more extensive and stable interaction of the GP Ibα C-terminal region with VWF.24 On the other hand, changes in the LRRs were not seen by superimposition of several crystal GP Ibα structures.2,24,25 Therefore, the mechanism by which mutations within the LRR produce gain-of-function (this study) or loss-of-function remains unclear. Loss-of-function mutations included those associated with Bernard-Soulier Syndrome (Cys65Arg, Leu57Phe,4 Ala156Val6,7 ) and those described in this report (Glu162Ala, Lys189Ala). None of the affected amino acids is in direct contact with the VWF A1 domain.2,24,25 The discrepancy between the crystal structures and the gain- and loss-of-function associated with mutations of the LRR may have to do with the difference in the bond formed under the static conditions used for obtaining crystals and the dynamic bond formed in the process of platelet attachment from flow and rolling on the VWF surface. In the latter situation, the GP Ibα–VWF bond is likely to experience considerable tensile stress and torque before it is broken.
Modulator-induced VWF binding. Cells expressing either wild-type or mutant GP Ibα were incubated with increasing concentrations of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. Cells were then incubated with HRP-conjugated anti–human VWF antibody. Results were normalized based on GP Ibα expression, as determined by the binding of WM23. Error bars indicate SEM. *Significant differences (P < .05).
Modulator-induced VWF binding. Cells expressing either wild-type or mutant GP Ibα were incubated with increasing concentrations of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. Cells were then incubated with HRP-conjugated anti–human VWF antibody. Results were normalized based on GP Ibα expression, as determined by the binding of WM23. Error bars indicate SEM. *Significant differences (P < .05).
Mutation of His86 to either Ala or Glu leads to gain of VWF binding function. Mutants His86Ala and His86Glu were tested for their ability to bind VWF in the presence of ristocetin (A) or botrocetin (B) at different modulator concentrations and a constant VWF concentration (4.0 μg/mL). The protocol for measuring the modulator-induced VWF binding is similar to that depicted in the legend to Figure 4. (C-D) In the assay of cell adhesion under flow, the cells expressing wild-type or mutant GP Ibα were incubated on immobilized VWF (20 μg/mL) for 1 minute in a parallel-plate flow chamber, after which the chamber was perfused with PBS at flow rates that generated wall shear stresses of 2.5, 10, or 20 dyne/cm2. Rolling velocities of the mutant-expressing cells were measured and compared with those of cells expressing wild-type GP Ibα. (C) Effects of VWF coating density on cell rolling at 20 dyne/cm2. (D) Effects of shear stress on the rolling velocity of mutant-expressing cells at a constant VWF-coating density (20 μg/mL VWF). Error bars indicate SEM.
Mutation of His86 to either Ala or Glu leads to gain of VWF binding function. Mutants His86Ala and His86Glu were tested for their ability to bind VWF in the presence of ristocetin (A) or botrocetin (B) at different modulator concentrations and a constant VWF concentration (4.0 μg/mL). The protocol for measuring the modulator-induced VWF binding is similar to that depicted in the legend to Figure 4. (C-D) In the assay of cell adhesion under flow, the cells expressing wild-type or mutant GP Ibα were incubated on immobilized VWF (20 μg/mL) for 1 minute in a parallel-plate flow chamber, after which the chamber was perfused with PBS at flow rates that generated wall shear stresses of 2.5, 10, or 20 dyne/cm2. Rolling velocities of the mutant-expressing cells were measured and compared with those of cells expressing wild-type GP Ibα. (C) Effects of VWF coating density on cell rolling at 20 dyne/cm2. (D) Effects of shear stress on the rolling velocity of mutant-expressing cells at a constant VWF-coating density (20 μg/mL VWF). Error bars indicate SEM.
Alternatively, because the crystal structures are based on complexes between isolated domains and our studies involved the intact receptor and ligand, the interaction with other regions of either protein could influence the results. It is unlikely that all the mutations with altered function affect a VWF-binding site directly. Rather, the associated increase in the binding of 2 monoclonal antibodies suggests that the interaction of receptor and ligand is altered allosterically. That allosteric or topologic alterations of GP Ibα can enhance VWF binding is supported by the recent characterization of a naturally occurring gain-of-function GP Ibα mutant lacking residues 421 to 429, a deletion within the macroglycopeptide region far from the ligand-binding domain.26
A recent study using alanine-scanning mutagenesis to identify GP Ibα residues essential for VWF binding examined 5 residues also evaluated in our study (Asp83Ala, His86Ala, Glu128Ala, Glu162Ala, and Lys189Ala).27 The investigators in this study failed to detect increased ristocetin-induced VWF binding in the His86Ala mutant but did find an increase in botrocetin-induced binding. Several methodologic differences likely account for the differences between these 2 studies. The first, and most obvious, is that the study by Shimizu et al27 used a recombinant fragment consisting of the N-terminal 293 amino acids of GP Ibα fused to a C-terminal flag tag, whereas we expressed the mutations in the context of the intact GP Ibα polypeptide expressed on the cell surface as part of the GP Ib-IX complex. The presentation and orientation of the polypeptide undoubtedly influence its ability to bind VWF. This point is highlighted by the fact that although both studies found that the mutation altered the binding of the monoclonal antibody AN51, increased binding of SZ2 was only seen in our study, a likely consequence of the fact that the N-terminal fragment in the study of Shimizu et al27 was attached to and immobilized by a C-terminal flag tag. Further, we tested ristocetin-induced VWF binding at several concentrations of ristocetin and observed the largest difference between the mutant and wild-type GP Ibα at the lowest ristocetin concentration (0.1 mg/mL).
It is clear that, despite the availability of elegant crystal structures of the GP Ibα N-terminus in complex with the VWF A1 domain, much remains to be learned about how the receptor and the ligand interact physiologically, particularly considering the exposure to hydrodynamic shear stress, the presence of other domains and polypeptides in the intact molecules, their interactions with other proteins, and the constraints imposed by the restriction of the GP Ib complex to the surface of the platelet.
Prepublished online as Blood First Edition Paper, June 2, 2005; DOI 10.1182/blood-2005-02-0514.
Supported by National Institutes of Health gants HL64796 and P50 HL65967 (J.A.L.).
Y.P. and C.N.S. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal