Abstract
Multiple cooperating mutations that deregulate different signaling pathways are required to induce cancer. Identifying these cooperating mutations is a prerequisite for developing better combinatorial therapies for treating cancer. Here we show that cooperating cancer mutations can be identified through oncogenic-retrovirus-induced insertional mutagenesis. Among 13 myeloid leukemias induced by transplanting into mice bone marrow cells infected in vitro with a replication-defective retrovirus carrying the Sox4 oncogene, 9 contained insertional mutations at known or suspected cancer genes. This likely occurred because rare bone marrow cells, in which the oncogenic retrovirus happened to integrate and in which it mutated a cooperating cancer gene, were selected because the host harbored a cooperating cancer mutation. Cooperativity between Sox4 and another gene, Mef2c, was subsequently confirmed in transplantation studies, in which deregulated Mef2c expression was shown to accelerate the myeloid leukemia induced by Sox4. Insertional mutagenesis of cooperating cancer genes by a defective oncogenic retrovirus provides a new method for identifying cooperating cancer genes and could aid in the development of better therapies for treating cancer. (Blood. 2005;106:2498-2505)
Introduction
Cancer results from a multistep process that requires several cooperating mutations for its occurrence.1 Determining the nature of these cooperating mutations and the signaling pathways they affect represents a major challenge for future cancer research. Replication-competent murine leukemia viruses that themselves lack cancer genes have proven to be valuable tools for identifying cooperating cancer genes. These viruses induce tumors by integrating into the genome and insertionally mutating cellular proto-oncogenes, tumor-suppressor genes, or both. Tumor development requires multiple rounds of viral infection. Initial rounds mutate genes important for cancer initiation, and subsequent rounds mutate genes involved in tumor progression.2 Through cloning and analyzing the multiple retroviral integrations present in each tumor, it has become possible to identify genes that cooperate to induce cancer.3 This approach has a number of drawbacks, however, that limit its use for identifying cooperating cancer genes. Tumors generated by replication-competent retroviruses are oligoclonal. Even when 2 genes are insertionally mutated in the same tumor, it is often difficult to tell whether they are mutated in the same cancer cell. Retroviruses also often target many genes infrequently rather than a few genes frequently. Thus, it is sometimes difficult to identify multiple tumors with integrations in the same two genes, which is an important indicator of cooperating cancer genes.
Until recently, it had been assumed that insertional mutagenesis after replication-defective retroviral infection was rare because these viruses only integrate into the genome at the time of initial infection. However, an IL2RG-containing retrovirus was recently found to induce T-cell lymphomas in patients with the SCID-X1 mutation after retroviral gene therapy by insertionally mutating the LMO2 proto-oncogene.4,5 Subsequent studies suggest that IL2RG might act as an oncogene when expressed from the LTR of the retrovirus used for therapy, and it has been speculated that oncogenic IL2RG might cooperate with insertionally mutated LMO2 in leukemia induction, explaining why lymphomas occur so frequently in these gene therapy trials.6 In addition, a defective Moloney murine leukemia virus encoding the platelet-derived growth factor B-chain (PDGFB) was shown to induce malignant brain tumors after the intracranial inoculation of mice.7 Analysis of the retroviral integration sites in these tumors identified several loci that were targeted in more than one tumor, suggesting that these loci harbor cancer genes that cooperate with PDGFB in tumor induction, though this has not been formally proven.
Leukemia develops in mice that receive transplanted bone marrow cells infected with replication-defective retrovirus carrying the multidrug resistance 1 (Mdr1) or the fluorescence protein gene, but only when the cells are infected with high titers of the virus.8 Leukemias contain multiple defective retroviral integrations (typically more than 10 integrations per leukemic clone) and have insertional mutations in genes that are likely to represent cooperating cancer genes, though again this has not been confirmed. The suggestion is that replication-defective retroviruses that lack oncogenes can also induce leukemia provided the cells are infected with enough virus to ensure independent insertional mutations in cooperating cancer genes.
Here we have tested the notion that defective oncogenic retroviruses induce leukemia in part through the insertional mutagenesis of cooperating cancer genes, and we show for the first time that this is a viable means for identifying cooperating cancer genes. We also show that mice receiving Sox4 virus-infected marrow develop myeloid leukemia, confirming that Sox4 is a leukemia gene.
Materials and methods
Retrovirus construction
The murine Sox4 coding region was amplified by reverse transcription-polymerase chain reaction (RT-PCR), cloned into the HpaI site of MSCV-neo (Clontech, Palo Alto, CA), and verified by sequencing. Similarly, the murine Mef2c coding region was cloned into MSCV-puro vector (Clontech). To obtain stable clones producing high-titer helper-free virus, all constructs were transfected into ecotropic GP+E86 packaging cells using calcium-phosphate precipitation, and viral supernatant from resultant G418 or puromycin-resistant clones were titered on NIH-3T3 cells using standard procedures. Clones with titers of 1 × 106 colony-forming units (CFUs)/mL for all constructs were used in this study.
Splinkerette PCR
Genomic DNA prepared from spleens of animals with leukemia was digested with NlAIII (NEB) or MseI and ligated to the splinkerette linker overnight. Nested PCR was then performed, as described,9 on ligation reactions using splinkerette-specific primers and primers recognizing the long terminal repeat (LTR) of murine stem-cell virus (MSCV). Products were then separated on 2% agarose gels, purified using MiniElute columns (Qiagen, Valencia, CA), and sequenced directly using the PRISM Big Dye Cycle Sequencing kit (Perkin Elmer, Shelton, CT) and an ABI model 373A DNA Sequencer (Applied Biosystems, Foster City, CA). Primer sequences are available on request.
Retroviral transduction and bone marrow transplantation
Bone marrow cells were harvested from 8- to 12-week-old C57BL/6-Ly5.1 mice 4 days after intraperitoneal injection of 5-fluorouracil (5-FU; 150 mg/kg body weight) and were expanded in Dulbecco modified Eagle medium (DMEM) plus 15% fetal bovine serum in the presence of murine stem-cell factor (SCF; 100 ng/mL), interleukin-6 (IL-6; 10 ng/mL), and IL-3 (6 ng/mL) for 2 days. Retroviral infection was performed in 6-well culture dishes by coculturing expanded bone marrow cells (0.5-1 × 106 cells/well) on a layer of 3 × 106 packaging cells irradiated at 179 cGy/min for 2 days. When cotransductions were conducted, cells of different packaging lines were plated together at 1.5 × 106 cells/well. Transduced cells (3-10 × 105) resuspended in phosphate-buffered saline (PBS) were then injected into the tail veins of lethally irradiated C57BL/6-Ly5.2 mice (single dose, 179 cGy/min) along with 5 × 105 supporting bone marrow cells from unirradiated C57BL/6-Ly5.2 mice. For secondary transplantation, 1 × 106 bone marrow cells from primary recipients with leukemia were injected along with supporting bone marrow cells.
In vitro clonogenic progenitor assay
Bone marrow cells (7.5 × 103) harvested from cocultivation with viral producer cells were plated on 35-mm dishes in a 1.1-mL culture mixture containing 0.35% low-gelling temperature agarose in Iscove medium supplemented with 20% fetal bovine serum, 100 ng/mL SCF, 50 ng/mL IL-6, and 30 ng/mL IL-3 in the presence of 1.3 mg/mL G418, 1.3 μg/mL puromycin, or both. Colonies were counted after 7 days.
Northern and Southern hybridizations and real-time RT-PCR
Total RNA was prepared from spleens of healthy and leukemic animals using Trizol (Invitrogen, Carlsbad, CA). Fifteen micrograms of each RNA sample was then resolved, transferred, and hybridized using standard procedures. The full-length coding regions of Sox4 cDNA were used as probes for hybridization. The β-actin probe was amplified as described previously.10 Southern blotting was performed using standard procedures. Spleen DNA was digested and hybridized with probes, as indicated in Figures 1, 2, and 5. Neomycin- and puromycin-specific probes were excised from MSCV-neo by BamHI and EagI double digestion and MSCV-puro by HindIII and ClaI digestion. Locus-specific probes were amplified by PCR from genomic DNA; primer sequences are available on request. For real-time RT-PCR, oligo-dT-primed cDNA samples were prepared from total RNA using Superscript (Invitrogen), and quantitative-PCR (Q-PCR) analysis was performed using Syber Green Chemistry (Qiagen) according to the manufacture's instructions in 10-μL final volume in 384-well microtiter plates. Gene-specific primer sequences and thermocycling conditions are available on request.
Results
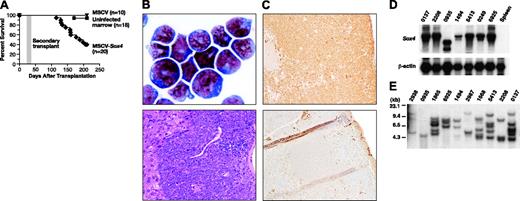
To determine whether defective retroviruses carrying activated oncogenes can induce tumors through the insertional mutagenesis of cooperating cancer genes, we transplanted C57BL/6-Ly5.1 bone marrow cells, infected in vitro for 2 days with MSCV carrying Sox4 (MSCV-Sox4) and a neomycin (neo) selection marker, into lethally irradiated C57BL/6-Ly5.2 hosts. Sox4 is an HMG-box transcription factor and a frequent insertional mutagenesis target of replication-competent retroviruses in murine B-cell lymphomas and myeloid leukemias.11-13 (http://RTCGD.ncifcrf.gov). Sox4 knockout mice display defects in B- and T-cell expansion, suggesting that Sox4 is a positive regulator of cell proliferation.14,15 Bone marrow cells used for infection were harvested from mice injected 4 days earlier with 5-FU and were cultured for 2 days in vitro in the presence of SCF, IL-3, and IL-6 to increase the number of cycling progenitor cells. The cells were then infected by coculture with packaging cells stably expressing 1 × 106 CFU/mL MSCV-Sox4 for 2 days. Transduction efficiencies, estimated by in vitro colony-forming assays in the presence of G418, were between 30% and 50%. Approximately 2 to 5 × 103 colony-forming cells (CFCs) were then transplanted into each lethally irradiated recipient. Combined results for 4 experiments are shown in Figure 1A. More than 60% of the mice that underwent transplantation fell ill in 4 to 7 months, whereas mice that received uninfected marrow or marrow infected with a similar titer of MSCV vector alone did not. The marrow from sick mice was filled with myeloid blasts with partial differentiation, and the numbers of other hematopoietic cells, including B cells and red blood cells, were severely reduced (Figure 1B). Myeloid blasts also infiltrated nonhematopoietic tissues, including the liver and kidney (Figure 1B; data not shown). Myeloid blasts were myeloperoxidase (MPO) positive, indicating that they were partially differentiated (Figure 1C). Leukemias are transplantable, as evidenced by the fact that myeloid leukemia developed in all secondary recipients that received 1 × 106 bone marrow cells harvested from primary leukemic recipients 29 to 37 days after transplantation (Figure 1A). Northern blot analysis showed that leukemic cells expressed high levels of retrovirally encoded Sox4 mRNA compared with normal spleen or bone marrow (Figure 1D; data not shown). This observation, and the fact that mice that received transplanted empty MSCV-infected marrow did not develop leukemia, illustrates the important role of Sox4 in disease induction.
If leukemias were induced in part by the insertional mutagenesis of cooperating cancer genes during MSCV-Sox4 integration, we would have expected them to be derived from one or a few infected cells because the probability of mutating a cooperating gene during integration should be low. To determine whether this was the case, we characterized the MSCV-Sox4 integrations in 10 leukemias by Southern blot analysis. Spleen DNA from leukemic mice was digested with HindIII, which cuts once in the provirus, and was hybridized with a neo-specific probe. Each band on the Southern blot, therefore, should represent an independent MSCV-Sox4 integration. As shown in Figure 1E, 9 of 10 leukemias harbored multiple MSCV-Sox4 integrations. One, leukemia 0935, contained only a single MSCV-Sox4 integration. In many types of leukemia, the relative hybridization intensity of the different integrations was the same, suggesting these leukemias were derived from a single infected cell. In others, integrations of 2 different intensities were observed, suggesting they were derived from 2 infected cells. These results are expected if insertional mutagenesis of Sox4 cooperation occurs and are rare events during MSCV-Sox4 integration.
Myeloid leukemia developed in mice that received transplanted MSCV-Sox4-infected bone marrow cells. (A) Survival curve of lethally irradiated mice that received transplanted MSCV-Sox4-infected marrow, uninfected marrow, or empty MSCV-infected marrow. The area shaded in gray represents the survival of secondary transplant recipient mice (n = 8) that received leukemia cells from a primary recipient. (B) (top) Wright-Giemsa staining of representative leukemic bone marrow. (bottom) Hematoxylin and eosin staining of a representative liver section showing leukemia-cell infiltration. Original magnifications, × 400 (left) and × 100 (bottom). (C) Normal (top) and leukemic (bottom) spleens stained with anti-MPO antibody. Original magnification, × 100. (D) Northern blot analysis of Sox4 expression in 7 myeloid leukemias induced by MSCV-Sox4 infection or normal spleen RNA. β-Actin was included to control for RNA loading. The Sox4 transcript expressed in leukemia 0935 was smaller than expected, suggesting a rearrangement in the MSCV-Sox4 virus present in this leukemia. (E) Southern blotting of leukemic spleen DNA using an MSCV-specific probe. Samples were digested with HindIII, which cuts once in the viral construct. Each band represents a separate Sox4 viral integration.
Myeloid leukemia developed in mice that received transplanted MSCV-Sox4-infected bone marrow cells. (A) Survival curve of lethally irradiated mice that received transplanted MSCV-Sox4-infected marrow, uninfected marrow, or empty MSCV-infected marrow. The area shaded in gray represents the survival of secondary transplant recipient mice (n = 8) that received leukemia cells from a primary recipient. (B) (top) Wright-Giemsa staining of representative leukemic bone marrow. (bottom) Hematoxylin and eosin staining of a representative liver section showing leukemia-cell infiltration. Original magnifications, × 400 (left) and × 100 (bottom). (C) Normal (top) and leukemic (bottom) spleens stained with anti-MPO antibody. Original magnification, × 100. (D) Northern blot analysis of Sox4 expression in 7 myeloid leukemias induced by MSCV-Sox4 infection or normal spleen RNA. β-Actin was included to control for RNA loading. The Sox4 transcript expressed in leukemia 0935 was smaller than expected, suggesting a rearrangement in the MSCV-Sox4 virus present in this leukemia. (E) Southern blotting of leukemic spleen DNA using an MSCV-specific probe. Samples were digested with HindIII, which cuts once in the viral construct. Each band represents a separate Sox4 viral integration.
To determine whether any MSCV-Sox4 integrations were located near cancer genes, we cloned and sequenced 50 integrations from 13 leukemias by splinkerette PCR (Table 1). We then searched these integration site sequences against the mouse genome sequence through BLAST17 and tried to determine whether any were located near cancer genes. Interestingly, 3 leukemias contained integrations that were located in slightly different positions in the first intron of Sfpi1 (Table 1; Figure 2A, upper panel). Sfpi1 encodes an ETS1-related transcription factor essential for normal myeloid and lymphoid development. Recent studies have shown that unlike mice heterozygous or homozygous for an Sfpi1 null allele, mice carrying a combination of hypomorphic Sfpi1 alleles that reduce Sfpi1 expression by 80% develop myeloid leukemia.17 Similarly, mice that carry an Sfpi1 deletion in combination with an Sfpi1 hypomorphic allele also develop myeloid leukemia.18 These results suggest that Sfpi1 encodes a dose-sensitive myeloid tumor-suppressor gene. The probability of finding a 3-hit common integration site (CIS), assuming random integration given that only 50 integrations were cloned, is extremely low (2.52 × 10-4).
Integrations at genes identified in MSCV-Sox4 virus-induced leukemias
Tumor no., gene . | Protein family . | Location and distance . | Orientation . | Mouse chromosome . | Human chromosome . | Accession no. . |
|---|---|---|---|---|---|---|
| 0137 | ||||||
| Mef2c* | Transcription factor | Intron 3 | Same | 13 | 5q14 | AK_009139 |
| EKI | Ethanolamine kinase | 5′, 10.5 kb | Same | 6 | 12p12.2 | AK_044502 |
| BC060648 | Enzyme | 5′, 0.5 kb | Same | 4 | ND | BC_060648 |
| Arhh | GTPase | 5′, 2.2 kb | Same | 5 | 4p13 | AK_017885 |
| AK016052 | ND | Intron 5 | Same | 17 | ND | AK_016052 |
| AF397014 | ND | Intron 11 | Inverse | 13 | ND | AF_397014 |
| 0841 | ||||||
| Sfpi 1* | Transcription factor | Intron 1 | Same | 2 | 11p11.2 | BC003815 |
| CB520747 | ND | Intron 6 | Inverse | 4 | ND | CB_520747 |
| Cish | Signal transduction | Intron 1 | Same | 9 | 3p21.3 | BC_003783 |
| Wrnip1 | Helicase | 3′, 10 kb | Same | 13 | 6p25.2 | AK_050368 |
| Ifitm31 | Transmembrane protein | 5′, 11 kb | Inverse | 7 | ND | BC_002160 |
| Tde1 | Membrane protein | Intron 1 | Inverse | 2 | 20q13.1-13.3 | AK_031101 |
| 0935 | ||||||
| Hcph* | Tyrosine phosphatase | Intron 3 | Inverse | 6 | 12p13 | BC_012660 |
| 1494 | ||||||
| Rnf8 | Ring finger protein | Intron 1 | Inverse | 17 | ND | AJ_242721 |
| Rnf11 | Ring finger protein | Intron 1 | Same | 4 | 1pter-p22.1 | BC_010299 |
| Golga5 | Golgi protein | 3′, 24 kb | Same | 12 | 14q32.12-32.13 | BC_016883 |
| AK042193 | ND | exon 2 | Same | 1 | ND | AK_042193 |
| 1668 | ||||||
| AK054410 | ND | Intron 20 | Same | 9 | ND | AK_054410 |
| Soat1 | Sterol O-acyltransferase | Intron 1 | Same | 1 | 1q25 | L_42293 |
| AK017935 | ND | 5′, 17 kb | Inverse | 19 | ND | AK_017935 |
| 1865 | ||||||
| Plekha2* | pH domain | Intron 1 | Same | 8 | 8p11 | BC_010215 |
| Scm14 | Polycomb protein | Intron 1 | Inverse | 10 | ND | BC_043310 |
| Aif1 | Calcium binding | 5′, 0.5 kb | Inverse | 17 | 6p21.3 | AK_006184 |
| AK089567 | ND | 5′, 2 kb | Inverse | ND | ND | AK_089567 |
| Nfat5* | Transcription factor | 5′, 40 kb | Inverse | 8 | 16q22.1 | AF_200687 |
| 2208 | ||||||
| Elf4* | Transcription factor | 5′, 7 kb | Inverse | X | Xq26 | AF_016714 |
| Kif21b | Cytoskeleton binding | Intron 1 | Inverse | 1 | ND | AF_202893 |
| Runx1* | Transcription factor | Intron 1 | Inverse | 16 | 21q22.3 | AK_051758 |
| AU043488 | ND | Intron 1 | Inverse | 10 | ND | AU_043488 |
| 2938 | ||||||
| AK129203* | EST1 telomerase binding | Intron 7 | Inverse | 11 | ND | AK_129203 |
| Selp1 | Selectin ligand | 3′, 6 kb | Inverse | 5 | 12q24 | AK_089214 |
| AK039624 | ND | 5′, 66 kb | Inverse | 8 | ND | AK_039624 |
| 2987 | ||||||
| Dusp2 | Phosphatase | 5′, 14 kb | Same | 2 | 2q11 | L_11330 |
| AK016300 | tRNA deacylase | Intron 2 | Same | 12 | ND | AK_016300 |
| Irak3 | Signal transduction | Intron 1 | Same | 10 | 12q14.2 | AK_029057 |
| Ern1 | Signaling | Intron 1 | Inverse | 11 | 16p12 | AB_031332 |
| AK020256 | ND | Intron 1 | Inverse | 10 | ND | AK_020256 |
| 4564 | ||||||
| Cgef | GEF | Intron 3 | Inverse | 2 | 2q31-32 | AB_021132 |
| Nrf1 | Transcription factor | 5′, 2 kb | Inverse | 6 | 7q32 | AK_037697 |
| 0249 | ||||||
| Mef2o* | Transcription factor | Intron 3 | Same | 13 | 5q14 | AK_009139 |
| Hspa9a* | Heat shock protein | 5′, 0.7 kb | Inverse | 18 | 5q31.1 | D_17556 |
| BC042709 | ND | 5′, 26 kb | Same | 4 | ND | BC_042709 |
| 5413 | ||||||
| Sfpi1* | Transcription factor | Intron 1 | Same | 2 | 11p11.2 | BC_003815 |
| AK014882 | ND | 5′, 81 kb | Same | 10 | ND | AK_014882 |
| Tuba4 | Cytoskeleton | Intron 1 | Same | 1 | ND | BC_019959 |
| Vdac2 | Anion channel | Intron 2 | Inverse | 14 | 10q22 | U_30838 |
| BC042901 | ND | Exon 9 | Same | 7 | ND | BC_042901 |
| 6577 | ||||||
| Sfpi1* | Transcription factor | Intron 1 | Inverse | 2 | 11p11.2 | BC_003815 |
| AK008545 | ND | 3′, 4 kb | Same | X | ND | AK_008545 |
| Sbf1 | Antiphosphatase | Intron 1 | Same | 15 | ND | AK_129485 |
Tumor no., gene . | Protein family . | Location and distance . | Orientation . | Mouse chromosome . | Human chromosome . | Accession no. . |
|---|---|---|---|---|---|---|
| 0137 | ||||||
| Mef2c* | Transcription factor | Intron 3 | Same | 13 | 5q14 | AK_009139 |
| EKI | Ethanolamine kinase | 5′, 10.5 kb | Same | 6 | 12p12.2 | AK_044502 |
| BC060648 | Enzyme | 5′, 0.5 kb | Same | 4 | ND | BC_060648 |
| Arhh | GTPase | 5′, 2.2 kb | Same | 5 | 4p13 | AK_017885 |
| AK016052 | ND | Intron 5 | Same | 17 | ND | AK_016052 |
| AF397014 | ND | Intron 11 | Inverse | 13 | ND | AF_397014 |
| 0841 | ||||||
| Sfpi 1* | Transcription factor | Intron 1 | Same | 2 | 11p11.2 | BC003815 |
| CB520747 | ND | Intron 6 | Inverse | 4 | ND | CB_520747 |
| Cish | Signal transduction | Intron 1 | Same | 9 | 3p21.3 | BC_003783 |
| Wrnip1 | Helicase | 3′, 10 kb | Same | 13 | 6p25.2 | AK_050368 |
| Ifitm31 | Transmembrane protein | 5′, 11 kb | Inverse | 7 | ND | BC_002160 |
| Tde1 | Membrane protein | Intron 1 | Inverse | 2 | 20q13.1-13.3 | AK_031101 |
| 0935 | ||||||
| Hcph* | Tyrosine phosphatase | Intron 3 | Inverse | 6 | 12p13 | BC_012660 |
| 1494 | ||||||
| Rnf8 | Ring finger protein | Intron 1 | Inverse | 17 | ND | AJ_242721 |
| Rnf11 | Ring finger protein | Intron 1 | Same | 4 | 1pter-p22.1 | BC_010299 |
| Golga5 | Golgi protein | 3′, 24 kb | Same | 12 | 14q32.12-32.13 | BC_016883 |
| AK042193 | ND | exon 2 | Same | 1 | ND | AK_042193 |
| 1668 | ||||||
| AK054410 | ND | Intron 20 | Same | 9 | ND | AK_054410 |
| Soat1 | Sterol O-acyltransferase | Intron 1 | Same | 1 | 1q25 | L_42293 |
| AK017935 | ND | 5′, 17 kb | Inverse | 19 | ND | AK_017935 |
| 1865 | ||||||
| Plekha2* | pH domain | Intron 1 | Same | 8 | 8p11 | BC_010215 |
| Scm14 | Polycomb protein | Intron 1 | Inverse | 10 | ND | BC_043310 |
| Aif1 | Calcium binding | 5′, 0.5 kb | Inverse | 17 | 6p21.3 | AK_006184 |
| AK089567 | ND | 5′, 2 kb | Inverse | ND | ND | AK_089567 |
| Nfat5* | Transcription factor | 5′, 40 kb | Inverse | 8 | 16q22.1 | AF_200687 |
| 2208 | ||||||
| Elf4* | Transcription factor | 5′, 7 kb | Inverse | X | Xq26 | AF_016714 |
| Kif21b | Cytoskeleton binding | Intron 1 | Inverse | 1 | ND | AF_202893 |
| Runx1* | Transcription factor | Intron 1 | Inverse | 16 | 21q22.3 | AK_051758 |
| AU043488 | ND | Intron 1 | Inverse | 10 | ND | AU_043488 |
| 2938 | ||||||
| AK129203* | EST1 telomerase binding | Intron 7 | Inverse | 11 | ND | AK_129203 |
| Selp1 | Selectin ligand | 3′, 6 kb | Inverse | 5 | 12q24 | AK_089214 |
| AK039624 | ND | 5′, 66 kb | Inverse | 8 | ND | AK_039624 |
| 2987 | ||||||
| Dusp2 | Phosphatase | 5′, 14 kb | Same | 2 | 2q11 | L_11330 |
| AK016300 | tRNA deacylase | Intron 2 | Same | 12 | ND | AK_016300 |
| Irak3 | Signal transduction | Intron 1 | Same | 10 | 12q14.2 | AK_029057 |
| Ern1 | Signaling | Intron 1 | Inverse | 11 | 16p12 | AB_031332 |
| AK020256 | ND | Intron 1 | Inverse | 10 | ND | AK_020256 |
| 4564 | ||||||
| Cgef | GEF | Intron 3 | Inverse | 2 | 2q31-32 | AB_021132 |
| Nrf1 | Transcription factor | 5′, 2 kb | Inverse | 6 | 7q32 | AK_037697 |
| 0249 | ||||||
| Mef2o* | Transcription factor | Intron 3 | Same | 13 | 5q14 | AK_009139 |
| Hspa9a* | Heat shock protein | 5′, 0.7 kb | Inverse | 18 | 5q31.1 | D_17556 |
| BC042709 | ND | 5′, 26 kb | Same | 4 | ND | BC_042709 |
| 5413 | ||||||
| Sfpi1* | Transcription factor | Intron 1 | Same | 2 | 11p11.2 | BC_003815 |
| AK014882 | ND | 5′, 81 kb | Same | 10 | ND | AK_014882 |
| Tuba4 | Cytoskeleton | Intron 1 | Same | 1 | ND | BC_019959 |
| Vdac2 | Anion channel | Intron 2 | Inverse | 14 | 10q22 | U_30838 |
| BC042901 | ND | Exon 9 | Same | 7 | ND | BC_042901 |
| 6577 | ||||||
| Sfpi1* | Transcription factor | Intron 1 | Inverse | 2 | 11p11.2 | BC_003815 |
| AK008545 | ND | 3′, 4 kb | Same | X | ND | AK_008545 |
| Sbf1 | Antiphosphatase | Intron 1 | Same | 15 | ND | AK_129485 |
Viral integrations at Sfpi1 are located in the first intron based on mRNA (BC003815) and EST (BU698148, BU698834) sequencing results. Similarly, the viral integration at Runx1 is located in the first intron (AK051758, BF135151, U19601).
Genes known CISs for replication-competent murine leukemia viruses.14
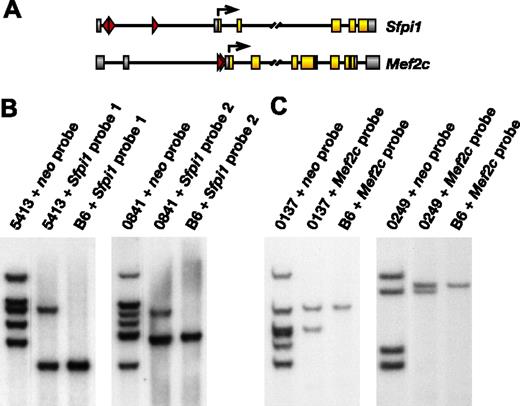
Two other leukemias contained MSCV-Sox4 integrations in the second intron of Mef2c (Table 1; Figure 2A, lower panel). Mef2c is a frequent target of replication-competent retroviruses in murine B-cell lymphomas and myeloid leukemias.13 The probability of finding 2 MSCV-Sox4 integrations near the same gene in 2 leukemias is also low (1.01 × 10-3) and supports the hypothesis that these leukemias were induced in part by the insertional mutagenesis of Mef2c.
Four other leukemias also contained MSCV-Sox4 integrations that were located at or near cancer genes (Table 1). The single MSCV-Sox4 integration in leukemia 0935 (Figure 1E) is integrated in Hcph intron 3. Hcph is expressed primarily in hematopoietic cells, where it behaves as a key regulator controlling phosphotyrosine levels.19 Aberrant HCPH splicing occurs in human acute myeloid leukemia (AML) cells, and Hcph mutant mice overexpand and inappropriately activate their myelomonocytic-cell populations,20 suggesting that Hcph is a haploinsufficient myeloid tumor-suppressor gene. An integration in leukemia 1865 is located in NFAT5 intron 1, a gene that is expressed in invasive human ductal breast carcinomas, where it promotes carcinoma invasion.21 Another integration in leukemia 2208 is located in RUNX1 intron 1, a gene that is also thought to function as a haploinsufficient human myeloid tumor-suppressor gene.22 Finally, an integration in leukemia 1494 is located 24 kb 3′ of GOLGA5, a gene that is fused to the RET tyrosine kinase gene in human sigmoid colon cancer23 and human papillary thyroid carcinoma.24
To determine whether leukemias containing Sfpi1 or Mef2c integrations are derived from single leukemic-cell clones, we designed locus-specific probes and examined the clonality of these integrations by Southern blot analysis (Figure 2B-C). Two identical leukemia DNA samples were digested with HindIII and were run side by side along with HindIII-digested C57BL/6 control DNA. One leukemia sample was then probed with a neo-specific probe, and the other identical leukemia sample and control sample were probed with a locus-specific probe. Locus-specific probes were designed to recognize the same mutant band recognized by the neo probe. Intensities of mutant and wild-type bands were then quantitated, and the percentages of spleen cells with integrations at the locus were calculated as follows: 2 × [mutant band intensity/(mutant band intensity + wild-type band intensity)]. In leukemia 5413, which represents 1 of the 2 leukemias with integrations at Sfpi1 for which RNA was available, we estimated 71% of the cells in spleen to have MSCV-Sox4 integrations at Sfpi1 (Figure 2B). Leukemia 5413 contains 5 MSCV-Sox4 integrations of similar intensity. Therefore, leukemia 5413 appears to be derived from a single leukemia-cell clone that harbors 5 integrations, one of which is located at Sfpi1. In leukemia 0841, the Sfpi1 integration is present in 41% of the spleen cells (Figure 2B). This leukemia contains 6 integrations that can be grouped into 2 groups based on hybridization intensity, suggesting that it is derived from 2 leukemia clones, with the less representative clone containing the integration at Sfpi1. Similarly, the integrations at Mef2c in leukemias 0137 and 0249 were estimated to be present in 94% and 96% of the respective splenic populations. The multiple integrations in both leukemias have similar intensities, suggesting that they are derived from single leukemia-cell clones that harbor multiple MSCV-Sox4 integrations (Figure 2C).
Frequent MSCV integrations at Sfpi1 and Mef2c in Sox4-induced leukemias. (A) MSCV integrations at Sfpi1 (top panel) or Mef2c (bottom panel). Exon-coding regions are highlighted in yellow and noncoding exons in gray. Arrows indicate translation start sites. The location and orientation of viral integrations are depicted by red triangles. (B) Southern blot analysis of spleen DNA of leukemic animals 5413 and 0841 and control C57BL/6 genomic DNA probed with neo or Sfpi1 probes. (C) Similar Southern blot analysis of spleen DNA from leukemic animals 0137 and 0249 probed with neo or Mef2c probe.
Frequent MSCV integrations at Sfpi1 and Mef2c in Sox4-induced leukemias. (A) MSCV integrations at Sfpi1 (top panel) or Mef2c (bottom panel). Exon-coding regions are highlighted in yellow and noncoding exons in gray. Arrows indicate translation start sites. The location and orientation of viral integrations are depicted by red triangles. (B) Southern blot analysis of spleen DNA of leukemic animals 5413 and 0841 and control C57BL/6 genomic DNA probed with neo or Sfpi1 probes. (C) Similar Southern blot analysis of spleen DNA from leukemic animals 0137 and 0249 probed with neo or Mef2c probe.
Real-time RT-PCR showed that leukemias 0841 and 5413, which have MSCV-Sox4 integrations at Sfpi1, express less than 50% of the Sfpi1 mRNA detected in normal bone marrow (Figure 3A). These leukemias also express lower levels of Sfpi1 mRNA than 6 of 8 other leukemias that lack Sfpi1 integrations. One exception is leukemia 0249, which harbors an uncharacterized rearrangement at the Sfpi1 locus. This rearrangement could possibly cause the low level of Sfpi1 expression seen in this leukemia (Figure 3A; data not shown). The other exception is leukemia 0137, which lacks a detectable rearrangement at the Sfpi1 locus. It is possible in this leukemia that an upstream regulator of Sfpi1 is mutated by MSCV-Sox4 integration. The choice of reference RNA is, however, critical in interpreting the effect of MSCV-Sox4 integration on Sfpi1 expression. AML is composed of a mixture of cells at varying differentiation stages, all derived from a small population of leukemia stem cells.25 Normal sorted Sca1+Lin- hematopoietic stem cells thus do not appear to represent an appropriate control for these experiments, nor do other sorted normal myeloid progenitor-cell populations. For these reasons, we used RNA from other MSCV-Sox4-induced leukemias that lacked integrations at Sfpi1 as a reference, though this is obviously not a perfect control either.
Northern blot analysis of the leukemias with integrations at Sfpi1 did not reveal any differences in message size compared with control RNA (data not shown). Fusion transcripts were also not detected between the MSCV-Sox4 virus and Sfpi1 in leukemias 0841 and 5413, making it unlikely that viral integrations at Sfpi1 affect Sfpi1 splicing. Removal of an Sfpi1 enhancer element located upstream of Sfpi1 has been shown to dramatically down-regulate Sfpi1 expression, resulting in the development of myeloid leukemia in affected animals.17 Two of the integrations at Sfpi1 are located within the region that contains this enhancer element. MSCV-Sox4 integration might, therefore, down-regulate Sfpi1 expression by disrupting this critical enhancer element. These results are consistent with the hypothesis that Sfpi1 is a myeloid tumor-suppressor gene that cooperates with Sox4 overexpression in leukemia induction.
In contrast, Northern blot analysis identified numerous smaller than normal Mef2c transcripts in leukemias 0137 and 0249, which contain MSCV-Sox4 integrations at Mef2c (data not shown). Fusion transcripts between MSCV-Sox4 LTR and Mef2c exon 3 could also be detected by RT-PCR and verified by sequencing (data not shown). Real-time RT-PCR showed that Mef2c transcripts were greatly elevated in the leukemias with integrations at Mef2c compared with normal spleen or bone marrow or other MSCV-Sox4-induced leukemias that lacked viral integrations at Mef2c (Figure 3B). These results suggest that Mef2c functions as an oncogene that cooperates with Sox4 overexpression in leukemia induction.
Viral integrations decrease Sfpi1 expression and activate Mef2c expression. (A) Real-time RT-PCR analysis of total RNA isolated from spleens of leukemic animals in addition to spleen and bone marrow of normal mice using primers specific for mouse Sfpi1. Relative gene expression levels were calculated by normalizing to β-actin mRNA levels in the same sample and in normal spleen. (B) Real-time RT-PCR analysis of the same samples using primers specific for mouse Mef2c.
Viral integrations decrease Sfpi1 expression and activate Mef2c expression. (A) Real-time RT-PCR analysis of total RNA isolated from spleens of leukemic animals in addition to spleen and bone marrow of normal mice using primers specific for mouse Sfpi1. Relative gene expression levels were calculated by normalizing to β-actin mRNA levels in the same sample and in normal spleen. (B) Real-time RT-PCR analysis of the same samples using primers specific for mouse Mef2c.
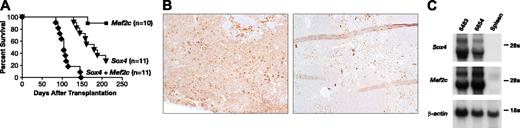
To confirm that Mef2c and Sox4 are cooperating oncogenes, we cloned Mef2c cDNA into an MSCV vector carrying a puro-selection marker and asked whether cotransduction of bone marrow cells with MSCV-Mef2c plus MSCV-Sox4 would accelerate the leukemia in mice that underwent transplantation. Two experiments were performed with bone marrow cells singly transduced with either virus alone as a control (Figure 4A). Gene delivery efficiencies determined by colony assays were 41% and 50% for Sox4 and 33% and 50% for Mef2c in single-transduction experiments. To maintain a similar exposure to virus in double-transduction experiments, only half the stable packaging cells were used for cocultivation. Double-transduction efficiencies of 13% and 5% were observed in the 2 experiments, and approximately 600 double-transduced CFCs were transplanted into each recipient (Table 2). As shown in Figure 4A, 73% of the mice that received transplanted MSCV-Sox4-infected marrow developed myeloid leukemia at an average age of onset of 163 ± 28 days, whereas none of the mice that received transplanted MSCV-Mef2c-infected marrow developed leukemia by 250 days after transplantation, although one mouse died 164 days after transplantation of an unrelated cause. Importantly, 100% of the mice that received transplanted double-infected marrow developed MPO-positive myeloid leukemia (Figure 4B) at a much reduced latency (109 ± 20 days; P < .0001, Mantel-Haenszel test) than mice that received transplanted MSCV-Sox4-infected marrow alone (Figure 4A). Mef2c thus does not induce leukemia on its own but instead accelerates the leukemia induced by Sox4. As expected, the leukemias overexpress Sox4 and Mef2c compared with normal spleen (Figure 4C). Southern blot analysis of 2 double-transduced leukemias (6290 and 6654) with neo- and puro-specific probes showed that multiple MSCV-Sox4 and MSCV-Mef2c integrations of varying intensities are present in these leukemias (Figure 5A), suggesting they are oligoclonal.
Numbers of infected CFCs transplanted in cotransduction experiments
. | . | No. injected per recipient . | . | . | ||
|---|---|---|---|---|---|---|
| Virus . | No. recipients . | G418-resistant CFCs . | Puromycin-resistant CFCs . | Double-resistant CFCs . | ||
| Experiment 1 | ||||||
| Sox4 | 6 | 2.7 × 103 | 0 | 0 | ||
| Mef2c | 5 | 0 | 1.7 × 103 | 0 | ||
| Sox4 + Mef2c | 6 | 2.1 × 103 | 1.1 × 103 | 6 × 102 | ||
| Experiment 2 | ||||||
| Sox4 | 5 | 4.1 × 103 | 0 | 0 | ||
| Mef2c | 5 | 0 | 3.6 × 103 | 0 | ||
| Sox4 + Mef2c | 5 | 4.3 × 103 | 1.4 × 103 | 6 × 102 | ||
. | . | No. injected per recipient . | . | . | ||
|---|---|---|---|---|---|---|
| Virus . | No. recipients . | G418-resistant CFCs . | Puromycin-resistant CFCs . | Double-resistant CFCs . | ||
| Experiment 1 | ||||||
| Sox4 | 6 | 2.7 × 103 | 0 | 0 | ||
| Mef2c | 5 | 0 | 1.7 × 103 | 0 | ||
| Sox4 + Mef2c | 6 | 2.1 × 103 | 1.1 × 103 | 6 × 102 | ||
| Experiment 2 | ||||||
| Sox4 | 5 | 4.1 × 103 | 0 | 0 | ||
| Mef2c | 5 | 0 | 3.6 × 103 | 0 | ||
| Sox4 + Mef2c | 5 | 4.3 × 103 | 1.4 × 103 | 6 × 102 | ||
Deregulated Mef2c expression accelerates the development of Sox4-induced leukemia. (A) Survival curve of lethally irradiated mice receiving MSCV-Mef2c-infected, MSCV-Sox4-infected, or double-infected bone marrow cells. (B) MPO staining of a representative leukemic spleen from an animal receiving double-transduced cells (top) or normal spleen (bottom). Original magnification, 100 ×. (C) Northern blot analysis of total RNA prepared from the spleens of 2 leukemic mice receiving double-infected bone marrow and normal mice using probes specific for Sox4 and Mef2c. β-Actin was used as a control for RNA loading.
Deregulated Mef2c expression accelerates the development of Sox4-induced leukemia. (A) Survival curve of lethally irradiated mice receiving MSCV-Mef2c-infected, MSCV-Sox4-infected, or double-infected bone marrow cells. (B) MPO staining of a representative leukemic spleen from an animal receiving double-transduced cells (top) or normal spleen (bottom). Original magnification, 100 ×. (C) Northern blot analysis of total RNA prepared from the spleens of 2 leukemic mice receiving double-infected bone marrow and normal mice using probes specific for Sox4 and Mef2c. β-Actin was used as a control for RNA loading.
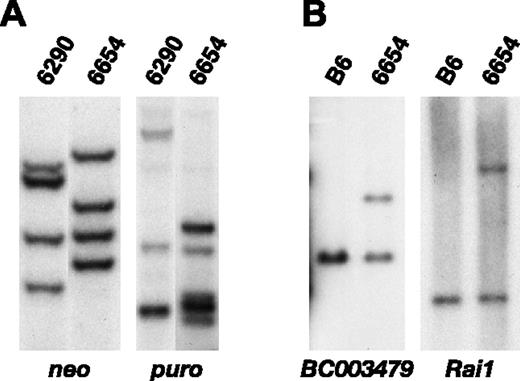
The reduced leukemia latency in double-transduction experiments confirms that Sox4 and Mef2c cooperate to induce leukemia, but it does not indicate whether Sox4 and Mef2c cooperate in a cell-autonomous or -nonautonomous manner. If cooperation is cell autonomous, then both genes should be expressed in the same tumor cell. If, however, cooperation is cell nonautonomous, cooperativity could result if Sox4 were expressed in one population of tumor cells and Mef2c in another. To address this question, the clonality of the integrations in a representative double-transduced leukemia was examined by Southern blot analysis. Leukemia 6654 contains 4 MSCV-Sox4 integrations of equal intensity detected with the neo probe, suggesting this leukemia contains one population of MSCV-Sox4-infected cells that harbors 4 integrations (Figure 5A, left panel). Leukemia 6654, in contrast, appears to contain 2 populations of MSCV-Mef2c-infected cells detected with the puro probe, a dominant population that contains 2 MSCV-Mef2c integrations and a minor population that contains 3 additional MSCV-Mef2c integrations (Figure 5A, right panel). The intensity of the 2 MSCV-Mef2c integrations in the dominant population is similar to the intensity of the 4 MSCV-Sox4 integrations, suggesting that the tumor cells in the dominant population contain 4 MSCV-Sox4 integrations and 2 MSCV-Mef2c integrations. Therefore, the 3 additional MSCV-Mef2c integrations in the minor population were presumably acquired during the 2-day infection period after division of the cell representing the major leukemia population. If this were correct, the minor population would carry 4 MSCV-Sox4 integrations in addition to 5 MSCV-Mef2c integrations.
To confirm these results, the MSCV-Sox4 and MSCV-Mef2c integrations were cloned and sequenced. Individual 3′ viral-cell junctions were then amplified using integration site-specific anti-sense primers in combination with neo- or puro-specific sense primers, respectively. PCR products were sequenced to confirm the identity of the provirus. One of the MSCV-Sox4 integrations in leukemia 6654 is located 5 kb upstream of BC003479, which encodes a putative member of the short-chain dehydrogenase/reductase (SDR) family. Southern blot analysis using a probe specific to this locus showed that it was present in 78% of the spleen cells (Figure 5B, left panel). Similarly, one of the MSCV-Mef2c integrations in leukemia 6654 is located in the first intron of Rai1. Southern blot analysis showed that this integration was present in 74% of the spleen cells (Figure 5B, right panel). Leukemia 6654 cells thus contain both the MSCV-Sox4 and the MSCV-Mef2c integrations. Similarly, the 2 leukemias with MSCV-Sox4 integrations at Mef2c (leukemias 0137, 0249) were shown by Southern blot analysis to be derived from only one population of leukemia cells, providing independent confirmation that Sox4 and Mef2c cooperate in a cell-autonomous manner.
Discussion
Here we have investigated the possibility that replication-defective oncogenic retroviruses induce leukemia in part through the insertional mutagenesis of cooperating cancer genes. Among 13 myeloid leukemias induced by transplanting in mice bone marrow cells infected in vitro with MSCV-Sox4, 9 contained integrations within or near known or suspected cancer genes. Cooperation between Sox4 and another gene, Mef2c, was subsequently confirmed in transplantation studies in which deregulated Mef2c expression was shown to accelerate the myeloid leukemia induced by deregulated Sox4 expression. Sox4 is mutated by replication-competent retroviral integration in more than 100 independent murine myeloid leukemias and B-cell lymphomas examined12,13 and is thus a candidate cancer gene.14 Our studies, presented here, confirm a role for Sox4 in leukemia transformation. SOX4 has not yet been validated in human leukemia, but it is overexpressed in human salivary adenoid cystic carcinoma26 and classic medulloblastoma though, interestingly, not in desmoplastic medulloblastoma.27,28 Sox4 is also overexpressed in ApcMin-induced mouse intestinal tumors and human colorectal cancer.29 Sox4 may, therefore, contribute to many types of human and mouse cancer.
MSCV-Sox4 and MSCV-Mef2c integrations are present in the same cell in leukemias induced by transplantation of double-transduced cells. (A) Southern blot analysis of HindIII-digested spleen DNA of leukemic animals 6290 and 6654 probed with a neo-specific (left) or a puro-specific (right) probe. (B) Southern blot analysis of EcoRI-digested spleen DNA of leukemic animal 6654 and normal C57BL/6 DNA probed with a BC003479 locus-specific probe (left) or the same DNA digested with HindIII and probed with a Rai1 locus-specific probe (right).
MSCV-Sox4 and MSCV-Mef2c integrations are present in the same cell in leukemias induced by transplantation of double-transduced cells. (A) Southern blot analysis of HindIII-digested spleen DNA of leukemic animals 6290 and 6654 probed with a neo-specific (left) or a puro-specific (right) probe. (B) Southern blot analysis of EcoRI-digested spleen DNA of leukemic animal 6654 and normal C57BL/6 DNA probed with a BC003479 locus-specific probe (left) or the same DNA digested with HindIII and probed with a Rai1 locus-specific probe (right).
Human and mouse genomes have more than 20 Sox family genes that can be allocated to 1 of 7 different subgroups.30 Overexpression or amplification of Sox genes is associated with a large number of tumor types (reviewed in Dong et al31 ). Hence, Sox4 might not be the only Sox gene involved in cancer. Consistent with this hypothesis, several Sox genes are encoded at CISs in mouse hematopoietic cancer. Sox3 is a CIS for replication-competent retroviruses in murine T-cell lymphomas.32 Ectopic SOX3 expression induces oncogenic transformation of chicken embryonic fibroblasts, and this effect is dependent on the HMG box and the transformation domains of Sox3.33 Sox5 and Sox10 are also CISs in brain tumors induced by intracranial inoculation of a defective oncogenic retrovirus carrying the PDGF B-chain (Pdgfb) gene.7
Mef2c is 1 of 4 members of the Mef2 family of myogenic basic helix-loop-helix genes. These genes link calcium-dependent signaling pathways to the genes responsible for cell division, differentiation, and death (reviewed in McKinsey et al34 ). Mef2 genes are classically associated with muscle and neuronal development but not with cancer. MEF2D, however, has recently been shown to form a fusion transcript with the RNA-binding protein gene DAZAP1 in a human pre-B acute lymphoblastic leukemia carrying a t(1;19)(q23;p13) translocation.35 MEF2D-DAZAP1 and the reciprocal DAZAP1-MEF2D proteins are both located in the nucleus. MEF2D-DAZAP1 can also form dimers with MEF2D and HDAC4, and exogenous expression of either fusion protein can promote the growth of HeLa cells. These studies confirm a role for MEF2 family genes in human cancer. Mef2c13 and Mef2b (Takeshi Suzuki, N.A.J., and N.G.C., unpublished observations, January 2003) are also CISs in murine myeloid leukemias and B-cell lymphomas. MEF2D may, therefore, not be the only MEF2 gene involved in cancer.
In mouse transplantation studies, Mef2c does not induce myeloid leukemia on its own but accelerates the leukemia induced by Sox4. This is similar to what occurs in Meis1 and Hoxa9, where Meis1 does not induce myeloid leukemia in mouse transplantation studies but is able to accelerate the myeloid leukemia induced by Hoxa9.9 That Sox4 and Mef2c double-transduced cells produce myeloid leukemia more quickly in transplant recipients than do Sox4 single-transduced cells suggests additional mutations are required for leukemic transformation. In Sox4 single-transduced cells, only rare cells in the infected population experience Mef2c expression activated by Sox4 integration, whereas in double-transduced cells many more cells overexpress both genes. Thus, the chance of accumulating additional mutations in cells that overexpress Sox4 and Mef2c is much higher in double-transduction experiments, which could result in the earlier onset of leukemia. This is also consistent with the studies reported here for leukemia 6654, which suggest that this leukemia, produced in double-transduction experiments, is derived from only 2 leukemia clones.
Three MSCV-Sox4-induced leukemias also had insertional mutations located in the first intron of Sfpi1. Sfpi1 normally suppresses myeloid leukemia development by promoting differentiation. Reducing the Sfpi1 level to 20% of that found in wild-type mice impairs the ability of Sfpil to bind DNA and is leukemogenic.17,18 Viral integration at Sfpi1 also appears to reduce Sfpi1 expression. Quantitation of Sfpi1 mRNA levels in leukemias with Sfpi1 integration is problematic, however, because it is difficult to identify an appropriate wild-type reference control. AML is composed of a mixture of cells at varying differentiation stages, and different AMLs can have different compositions. Although we compared Sfpi1 levels in leukemias with integrations at Sfpi1 to similar leukemias without Sfpi1 integrations, this variation in AML composition makes it an imperfect control. Additional studies are required to confirm the effect of MSCV-Sox4 integration on Sfpi1 mRNA levels.
Quantitation experiments suggested that Sfpi1 mRNA levels were reduced to less than 50% of normal levels after MSCV-Sox4 integration, despite that fact that viral integration occurred in only one Sfpi1 allele. Sequence analysis of the Sfpi1 mRNA expressed in leukemias with integrations at Sfpi1 showed that the transcripts are normal and do not contain any of the Sfpi1 point mutations recently identified by Cook et al18 in mouse radiation-induced myeloid leukemias (data not shown). Southern blot analysis also failed to identify any Sfpi1 genomic alterations (data not shown). As far as we could determine, the unrearranged allele in both leukemias with Sfpi1 integrations was normal but was expressed at reduced levels. Consistent with our findings, Mueller et al36 have identified SFPI1 mutations in 9 of 126 human AMLs, and 7 of them retained the wild-type allele. In the leukemias we studied and those reported by Mueller et al,36 the wild-type allele could be silenced epigenetically (eg, by methylation) or by other oncogenic changes in the tumor that act to silence the wild-type allele. This raises the intriguing possibility that Sox4, or one of its downstream targets, might function as a negative regulator of Sfpi1 expression. Sox4 overexpression alone might be insufficient to reduce Sfpi1 expression to levels that would induce leukemia, but the combination of Sox4 overexpression and a Sox4 insertional mutation at Sfpi1 might be sufficient.
Four other leukemias also contained MSCV-Sox4 integrations at known cancer genes. However, the role of these integrations in Sox4-induced leukemia remains unclear because only one integration in each gene was identified. More types of leukemia must be characterized to determine whether these genes are CISs in Sox4-induced leukemias. Three leukemias also have MSCV-Sox4 integrations at CISs for replication-competent viruses in murine leukemias. These CISs include Plekha2, Elf4, and the ETS1 telomerase-binding protein AK129203 (Table 1). In addition, 3 leukemias were identified that had 2 separate integrations in genes that are known CISs in murine leukemias (Table 1), raising the possibility that more than one insertionally mutated gene in each leukemia cooperates with Sox4 in leukemia transformation.
Although it is unclear how frequently Sox4-induced tumors contain insertional mutations in cooperating cancer genes, our data suggest that it probably occurs frequently. It is also unclear how frequently this will occur with other defective oncogenic retroviruses, though studies showing that a defective IL2RG-containing Moloney murine leukemia virus can induce T-cell leukemia after retroviral gene therapy4,5 or a similar virus carrying Pdgfb can induce brain tumors after intracranial inoculation7 suggest that it will occur. Our results also provide a note of caution for human gene therapy trials. In rare instances in which the virus used for therapy unexpectedly carries an oncogenic sequence, leukemia might occur at a much higher frequency than previously predicted. Because the preponderance of insertional mutagenesis by retroviruses involves the activation of cellular proto-oncogenes,12 insertional mutagenesis may be reduced through the use of self-inactivating retroviral vectors, which carry mutations in the viral LTR that remove the enhancers and the promoter. The animal model described here provides a means for assessing the safety of these vectors.
Prepublished online as Blood First Edition Paper, June 16, 2005; DOI 10.1182/blood-2004-12-4840.
Supported by the Intramural Research Program of the National Institutes of Health (NIH) National Cancer Institute Center for Cancer Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank D. Logsdon and C. Perella for tail-vein injection of bone marrow cells.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal