Abstract
CCAAT/enhancer-binding proteins (C/EBPs) are a family of transcription factors that regulate cell growth and differentiation in numerous cell types. To identify novel C/EBP-target genes, we performed transcriptional profiling using inducible NIH 3T3 cell lines expressing 1 of 4 members of the C/EBP family. Functional analysis revealed a previously unknown link between C/EBP proteins and circadian clock genes. Our microarray data showed that the expression levels of 2 core components of the circadian network, Per2 and Rev-Erbα, were significantly altered by C/EBPs. Recent studies suggested that Per2 behaves as a tumor suppressor gene in mice. Therefore, we focused our additional studies on Per2. We showed that Per2 expression is up-regulated by C/EBPα and C/EBPϵ. Per2 levels were reduced in lymphoma cell lines and in acute myeloid leukemia (AML) patient samples. In addition, we generated stable K562 cells that expressed an inducible Per2 gene. Induction of Per2 expression resulted in growth inhibition, cell cycle arrest, apoptosis, and loss of clonogenic ability. These results suggest that Per2 is a downstream C/EBPα-target gene involved in AML, and its disruption might be involved in initiation and/or progression of AML. (Blood. 2005; 106:2827-2836)
Introduction
The precise transcriptional regulation of numerous genes is required to maintain the balance between normal cellular proliferation and terminal differentiation. Such control is achieved through specific transcription factors that act as master regulators of various cellular functions. The CCAAT/enhancer-binding protein (C/EBP) family falls into this category of transcription factors, with many physiologic and pathologic conditions associated with their activities.1,2 To date, 6 C/EBP family members have been identified, with further diversity achieved by the generation of different isoforms and extensive protein-protein interactions both within the family and with other transcription factors. All C/EBP family members contain a highly conserved basic region and a leucine zipper domain (bZIP). Tissue- and stage-specific expression, as well as variable DNA-binding specificities, contributes to the differences in the biologic functions of the C/EBP isoforms.
C/EBP proteins play a key role in regulating proliferation and differentiation of many cell types including mammary epithelia cells, neuronal cells, granulocytes, hepatocytes, and adipocytes.3 Increasing evidence now shows that deregulated activity of some C/EBPs is involved in tumorigenesis.4-7 Within the hematopoietic system, C/EBPα is crucial for granulocytic differentiation. It is expressed in hematopoietic stem cells and myeloid progenitors cells, and no mature granulocytes are found in C/EBPα-deficient mice.8-10 Inactivation of C/EBPα leads to a differentiation block in acute myeloid leukemia (AML), and conditional expression of C/EBPα results in AML growth arrest and differentiation.11,12 In addition, mutations in the CEBPA gene are found in a subclass of human myeloid leukemias,13-15 implicating it as a tumor suppressor gene. Furthermore, a variety of fusion proteins (ie, acute myeloid leukemia 1/eight-twenty-one [AML/ETO], breakpoint cluster region/Abelson murine leukemia [BCR/ABL], core binding factor beta/myosin heavy polypeptide 11 [CBFB/MYH11], and promyelocytic leukemia-retinoic acid receptor [PML/RAR]) that result from chromosomal translocations in myeloid leukemia, either directly or indirectly, have been associated with inappropriately low expression of C/EBPα.7,16-19
Circadian rhythms are generated by a set of clock genes organized in interlocking transcriptional-translational feedback loops. Circadian oscillations of clock genes are found in the suprachiasmatic nucleus (SCN), where the central pacemaker is located, and in many peripheral tissues including liver, muscle, and bone marrow.20-22 Recent studies provide evidence for molecular links between the circadian clock and cell proliferation.23,24 Many cell cycle-related genes are deregulated and cell cycle progression from S to M phase is impaired in mice lacking the Cry gene, a core component of the clock network.25 Recent studies suggest that the murine Per2 gene, another key factor of the circadian system, is involved in tumor suppression by regulating cell cycle- and apoptosis-related genes.26 In addition, disruption of circadian rhythms has been associated with cancer in humans.27 Understanding the molecular links between the cell and the circadian cycles may lead to new therapeutic approaches to cancer as well as other challenging diseases.
In the present study, we used cDNA microarray analysis to examine the composition of C/EBP target genes following induction of C/EBPα, C/EBPβ, C/EBPδ, or C/EBPϵ. Functional analysis of the data revealed a previously unknown link between C/EBP proteins and the circadian clock pathway. Further experiments focused on Per2 as a possible downstream target of C/EBPα. We showed that C/EBPα and C/EBPϵ induced Per2 expression in hematopoietic cancer cell lines. Additional studies found low levels of Per2 expression in 42% of fresh AML bone marrow mononuclear cell samples. We also showed that re-establishment of Per2 expression in leukemia cells leads to dramatic growth inhibition, cell cycle arrest, apoptosis, and reduced anchorage-independent cell growth. To our knowledge, this is the first study implicating abnormalities of Per2 associated with AML.
Patients, materials, and methods
Patient samples
Low-density mononuclear bone marrow cells from 21 patients with AML as well as from 9 healthy individuals were obtained after their informed consent; approval was obtained from Cedars-Sinai Medical Center institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. The samples were processed as previously described.28
Cell culture
NIH 3T3 (murine fibroblast), KCL22, K562 (chronic myelocytic leukemia), U937 (myelomonoblastic), and Daudi (Burkitt lymphoma) cell lines were obtained from the American Type Culture Collection (Manassas, VA) and grown in the recommended medium and conditions.
Generation of zinc-inducible stable cell lines
The zinc-inducible C/EBP expression vectors pMTα, pMTβ, pMTδ, and pMTϵ were constructed by cloning the human C/EBPα, C/EBPβ, C/EBPδ, and C/EBPϵ cDNAs, respectively, into the pMTCB6+ vector (pMT, a kind gift from F. J. Rauscher III, The Wistar Institute, Philadelphia, PA). NIH 3T3 cells were transfected with zinc-inducible C/EBP vectors as well as control empty vector using the GenePORTER transfection Reagent (GTS, San Diego, CA). Multiple polyclonal clones were obtained by selection with G418 (700 μg/mL). Clones were screened by Western blot analysis for C/EBP protein expression following induction for 16 hours with ZnSO4 (100 μM). The zinc-inducible Per2 expression vector (pMTPer2) was constructed by inserting a full-length V5-taggeted Per2 (generous gift from S. Lei, University of Massachusetts Medical School, Worcester, MA) at the KpnI and EcoRV sites of the pMT vector. To generate stable inducible K562-pMTPer2, K562-pMT, Daudi-pMTα, Daudi-pMT, KCL22-pMTα, KCL22-pMT,12 U937-pMTϵ, and U937-pMT29 cell lines, K562, Daudi, KCL22, and U937 cells were transfected with pMTPer2, pMTα, pMTϵ, or pMT vectors using an electroporation apparatus (Electro Square Porator T820; BTX, San Diego, CA). Selection with G418 at 1 mg/mL was started 48 hours after electroporation to obtain stably transfected cells. Multiple monoclonal cultures were screened for zinc-inducible Per2, C/EBPα, or C/EBPϵ expression by Western blot analysis.
Oligonucleotide array hybridization and data analysis
Triplicate clones of NIH 3T3 cells were induced by addition of ZnSO4 (100 μM) for 16 hours to the medium. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). Biotinylated cRNAs were prepared and hybridized to murine MG_74Av2 microarrays (Affymetrix, Santa Clara, CA), which contain more than 12 000 genes. The probed arrays were scanned with a Hewlett Packard Gene Array scanner (Hewlett Packard, Palo Alto, CA). The scanned output image files were analyzed using Affymetrix Microarray Suite version 5.0 (Affymetrix Microarray, Santa Clara, CA). To identify genes that were differentially expressed between the 5 sample sets (empty vector, C/EBPα, C/EBPβ, C/EBPδ, and C/EBPϵ), class compression analysis was performed using BRBArray Tools (developed by Richard Simon and Amy Peng Lam; http://linus.nci.nih.gov/BRB-ArrayTools.html). Medium normalization was applied to the arrays, and the percent absent filter was set at 80% to exclude probe sets that were unreliable. A list of 158 genes was generated with probability of 95% that they contained no more then 10% false discoveries. Of these genes, 117 were significant at the 0.001 level of univariate F-test, and the remaining 41 were significant at the 0.027 level. Functional annotation analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, MD).30
Semiquantitative RT-PCR and real-time RT-PCR
Total RNA (2 μg) was converted into cDNA using SuperScript II reverse transcriptase (Invitrogen). Semiquantitative reverse-transcriptase-polymerase chain reaction (RT-PCR) was performed to determine the expression levels of haptoglobin (Hp) and lipocalin (Lcn2). RT-PCR for 18S was used as an internal control. Reaction products were visualized on ethidium bromide-stained agarose gels. Real-time RT-PCR analysis of selected genes was performed to confirm the microarray data. The expression levels of Per2 and Rev-Erbα were also measured in human cell lines and human specimens. Levels of 18S or glyceraldehyde phosphate dehydrogenase (GAPDH) were measured for endogenous reference. Reactions were performed using HotMaster Taq DNA Polymerase (Eppendorf, Hamburg, Germany) and SYBRGreen I (Molecular Probes, Eugene, OR). Reactions were performed in triplicates using a iCycler iQ system (Biorad, Hercules, CA). For each sample, the amount of the target gene and reference gene was determined from standard curves. Comparisons between expression of Per2 in bone marrow samples from healthy patients and patients with AML were analyzed using Student t test.
Northern blot analysis
Total RNA (10 μg) was electrophoresed on a denaturing formaldehyde gel, transferred onto a nylon membrane, and hybridized with [α-32P] deoxyadenosine triphosphate (dATP)-labeled (Strip-EZ DNA; Ambion, Austin, TX) cDNA probes. To ensure equal loading of RNA, blots were stripped and rehybridized with a GAPDH probe.
Western analysis
Cells were lysed with lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]-HCl [pH 7.4], 150 mM NaCl, 0.5% nonidet P-40 [NP-40]); subsequently cell lysates were resolved on 4% to 15% gradient sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGEs) and transferred to nitrocellulose membranes (Sigma, St Louis, MO). Immunoblots were incubated with the following antibodies: anti-C/EBPα (sc-61), anti-C/EBPβ (sc-150), anti-C/EBPδ (sc-636), and anti-C/EBPϵ (sc-158) from Santa Cruz Biotechnology (Santa Cruz, CA); anti-V5 from Invitrogen; and anti-GAPDH from Research Diagnostics (Flanders, NJ). SuperSignal West Pico substrate (Pierce, Rockford, IL) was used for detection. Western blots were stripped between hybridizations with stripping buffer (10 mM Tris-HCl [pH 2.3], 150 mM NaCl).
Reporter assay
NIH 3T3 cells were cotransfected with a murine Per2 promoter reporter construct (pGL3B/mPer2 [-1670 to +53], generous gift from P. Sassone-Corsi, Louis Pasteur, France,31 0.6 μg), along with either C/EBPα, C/EBPϵ, Dbp (kind gift from M. Noshiro, Hiroshima University, Japan32 ), or empty expression vectors (0.6 μg). The total amount of DNA was kept equal in each transfection with the addition of empty vector. Lysates were harvested 24 hours after transfection and luciferase activity was measured with the Dual-Luciferase reporter 1000 assay system (Promega, Madison, WI). Transfection efficiency was normalized using pRL-TK vector (0.1 μg). Results represent the mean of 3 separate experiments done in triplicate.
Electrophoretic mobility-shift assay (EMSA)
Double-stranded oligonucleotides containing the C/EBP site (underlined: CCCAGGGCTTCTTTGGAAAGGGCTGCTGAA) from the Per2 promoter were end-labeled with γ-32P-ATP by T4 polynucleotide kinase. Nuclear extracts from NIH 3T3 cells either untransfected or transfected with a C/EBPα expression vector were prepared with the CelLytic Nuclear Extraction Kit (Sigma). Nuclear extract proteins (10 μg) were incubated with 20 000 cpm of labeled oligonucleotides. Binding reactions were incubated for 30 minutes on ice and then analyzed on 4% polyacrylamide gel. When either cold competitor (100-fold excess) or anti-C/EBPα antibody was used, they were added to the reactions 20 minutes prior to the labeled probe.
Chromatin immunoprecipitation assay
Bone marrow cells from 5 mice were suspended in Iscoves modified Dulbecco medium (IMDM) with 10% fetal bovine serum (FBS). Chromatin was prepared and immunoprecipitated according to the manufacturer's protocol (Upstate, Lake Placid, NY). Samples were immunoprecipitated with either rabbit anti-C/EBPα or rabbit anti-C/EBPϵ antibodies. As negative controls, immunoprecipitation containing either no antibody or rabbit preimmune serum was included. PCR was performed with the following primers: Per2, 5′ (-1640)CCCAGCTCTGCTCAGTGTTT and 5′(-1365)AGGCATGCAATTCCTCAGAT; β-actin, 5′ (+31)GCTTCTTTGCAGCTCCTTCGTTG and 5′ (+135)TTTGCACATGCCGGAGCCGTTGT.
Transient transfections and cell viability assays
K562 and U937 cells (8 × 106) were electroporated using a Gene Pulser electroporation apparatus (BTX, San Diego, CA), at 310 V for 35 milliseconds. For cell viability assays, cells were electroporated with either pcDNA3.1 V5-taggeted Per2 expression vector or empty pcDNA3.1 vector, together with pMSCVpuro vector (Clontech, Palo Alto, CA) and selected with puromycin (0.8 μg/mL) for 3 days. Equal numbers of transfected cells (5 × 104/mL) were then plated in fresh medium. The mean number of viable cells was determined daily using trypan blue exclusion.
Cell proliferation, cell cycle, apoptosis, and clonogenic assays
Cell proliferation was determined by methyl-thiazol tetrazolium (MTT) assays (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocol. For cell cycle analyses, cells were fixed in cold ethanol, stained with propidium iodide, and analyzed by FACScan and CELLFit program (Becton Dickinson, San Jose, CA). Apoptosis analysis was performed with annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (BD PharMingen, San Diego, CA) according to the manufacturer's instructions. For clonogenic assay, cells (1 × 103) were plated into 24-well flat-bottomed plates using a 2-layer soft agar system. After 14 days of incubation, colonies were counted and measured. Experiments were done at least twice using triplicate plates per experimental point. Statistical significance of the results was analyzed using t test.
Results
Transcriptional profiling of C/EBP-inducible NIH 3T3 cells
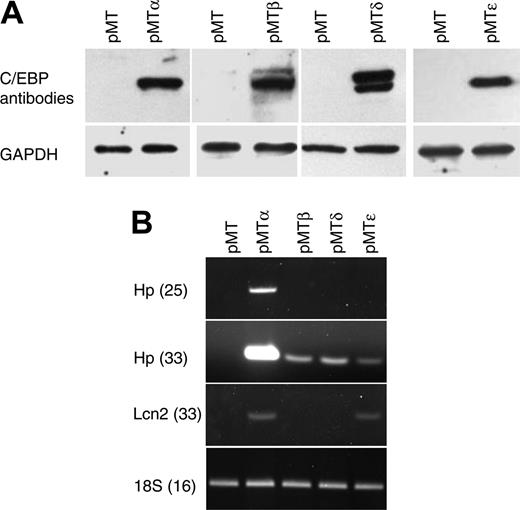
To identify C/EBP target genes, we generated a series of cell lines by stably transfecting NIH 3T3 cells with zinc-inducible vectors that express either C/EBPα (pMTα), C/EBPβ (pMTβ), C/EBPδ (pMTδ), or C/EBPϵ (pMTϵ), as well as a control empty vector (pMT). Following G418 selection and expansion, polyclonal populations of resistant cells were stimulated to express the CEBP genes, followed by harvesting of their RNA and protein. C/EBP protein expression was demonstrated by Western blot analysis using specific antibodies (Figure 1A).
Characterization of NIH 3T3 cells carrying zinc-inducible CEBPA, CEBPB, CEBPD, or CEBPE genes. (A) NIH 3T3 cell lines stably transfected with zinc-inducible cDNA coding for C/EBP proteins (pMTα, pMTβ, pMTδ, and pMTϵ) and cells transfected with empty vector (pMT) were treated with zinc for 16 hours. Protein lysates were analyzed by Western blot with C/EBPα-, C/EBPβ-, C/EBPδ-, or C/EBPϵ-specific antibodies. The blots were stripped and rehybridized with a GAPDH antibody as control for equal loading. (B) Semiquantitative RT-PCR was performed on RNAfrom the NIH 3T3 cell lines incubated with zinc for 16 hours. PCR products of haptoglobin (Hp, 25 and 33 cycles) and lipocalin (Lcn2, 33 cycles) were gel separated and stained with ethidium bromide. PCR for 18S was carried out as an internal control.
Characterization of NIH 3T3 cells carrying zinc-inducible CEBPA, CEBPB, CEBPD, or CEBPE genes. (A) NIH 3T3 cell lines stably transfected with zinc-inducible cDNA coding for C/EBP proteins (pMTα, pMTβ, pMTδ, and pMTϵ) and cells transfected with empty vector (pMT) were treated with zinc for 16 hours. Protein lysates were analyzed by Western blot with C/EBPα-, C/EBPβ-, C/EBPδ-, or C/EBPϵ-specific antibodies. The blots were stripped and rehybridized with a GAPDH antibody as control for equal loading. (B) Semiquantitative RT-PCR was performed on RNAfrom the NIH 3T3 cell lines incubated with zinc for 16 hours. PCR products of haptoglobin (Hp, 25 and 33 cycles) and lipocalin (Lcn2, 33 cycles) were gel separated and stained with ethidium bromide. PCR for 18S was carried out as an internal control.
To test the transcriptional activity of the C/EBP proteins, semiquantitative RT-PCR was performed using haptoglobin (Hp) and lipocalin (Lcn2), 2 known C/EBP target genes. Zinc treatment induced the expression of both genes (Hp, strong induction by C/EBPα and low induction by C/EBPβ, C/EBPδ, and C/EBPϵ; Lcn2, induction by C/EBPα and C/EBPϵ), demonstrating that the stably transfected C/EBP proteins could specifically activate target genes that are normally expressed in other tissues (Figure 1B).
For global gene expression profiling, the transfected NIH 3T3 cell lines (3 independent polyclones from each stably transfected C/EBP family member) were cultured in the presence of zinc for 16 hours. Oligonucleotide microarray analysis was performed with total RNA using Affymetrix U74Av2 chips. The raw expression data were processed and filtered according to the criteria described in “Patients, materials, and methods.” Using these criteria, we found that the expression of 158 genes was modulated in a statistically significantly fashion by one or more of the C/EBP proteins (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). These genes were classified into 4 functional categories (Table 1). Consistent with previous knowledge about C/EBP functions, many of the differentially regulated genes are involved in regulation of cell growth, immune response, cellular metabolism, and differentiation.3,33-37 Our analysis also showed that induction of C/EBPα led to the most significant changes in gene expression, as well as altering the largest number of genes.
Functional characterization of C/EBP target genes
GenBank accession no., by gene function . | . | . | Fold change . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | Probe set . | Gene symbol . | C/EBPα . | C/EBPβ . | C/EBPδ . | C/EBPϵ . | |||
| Immune response | |||||||||
| M63695 | 103422_at | Cd1d1 | 53.47 | 3.05 | 7.36 | 2.42 | |||
| AW124547 | 96092_at | Hp* | 28.05 | 2.72 | 3.76 | 1.85 | |||
| M27008 | 100436_at | Orm1† | 14.20 | 1.03 | 3.25 | 1.23 | |||
| M83218 | 103448_at | S100a8 | 12.99 | 0.99 | 1.72 | 2.12 | |||
| M63697 | 101897_g_at | Cd1d2 | 11.76 | 1.63 | 2.65 | 2.27 | |||
| A1786089 | 93837_at | Kng | 9.13 | 1.74 | 2.15 | 2.16 | |||
| K02782 | 95910_f_at | C3† | 8.43 | 2.18 | 3.39 | 1.62 | |||
| X81627 | 160564_at | Lcn2† | 5.84 | 0.75 | 1.16 | 3.82 | |||
| U27267 | 98772_at | Cxcl5‡ | 4.21 | 2.03 | 2.48 | 5.02 | |||
| M74123 | 94224_s_at | Ifi205‡ | 3.90 | 1.23 | 1.88 | 3.34 | |||
| X57349 | X57349_M_at | Trfr | 3.70 | 3.46 | 1.61 | 2.78 | |||
| M21050 | 100611_at | Lyzs†* | 3.69 | 1.34 | 1.00 | 1.12 | |||
| M31419 | 98465_f_at | Ifi204* | 3.07 | 1.03 | 1.50 | 2.51 | |||
| M32486 | 100477_at | M32486 | 3.06 | 1.15 | 1.30 | 2.74 | |||
| AB012693 | 103611_at | Cd47† | 2.91 | 2.09 | 1.61 | 2.52 | |||
| X83601 | 92731_at | Ptx3 | 2.82 | 1.28 | 2.44 | 4.19 | |||
| L09737 | 102313_at | Gch | 0.31 | 0.50 | 0.89 | 0.39 | |||
| X68273 | 103016_s_at | Cd68 | 0.25 | 0.50 | 0.91 | 0.36 | |||
| L39017 | 98018_at | Procr | 0.16 | 0.29 | 0.90 | 0.15 | |||
| AJ242830 | 102948_at | Hemt1 | 0.08 | 0.19 | 0.86 | 0.07 | |||
| Cell proliferation | |||||||||
| AF099973 | 92472_f_at | Slfn2 | 8.85 | 2.48 | 2.12 | 6.82 | |||
| AF036893 | 93694_at | Per2* | 4.85 | 1.73 | 1.75 | 11.40 | |||
| AF099977 | 92315_at | Slfn4 | 4.98 | 1.10 | 2.36 | 2.27 | |||
| Z22703 | 99435_at | Fgf7 | 4.28 | 1.59 | 2.46 | 4.75 | |||
| X72307 | 102898_at | Hgf* | 4.07 | 1.17 | 0.86 | 1.31 | |||
| X53929 | 93534_at | Dcn | 3.93 | 3.09 | 1.73 | 9.75 | |||
| A1834950 | 98507_at | Nr1d1* | 0.31 | 0.44 | 0.69 | 0.31 | |||
| M64292 | 101583_at | Btg2 | 0.30 | 0.49 | 0.92 | 0.28 | |||
| X61940 | 104598_at | Dusp1* | 0.27 | 0.44 | 0.98 | 0.28 | |||
| AV373612 | 161980_f_at | Bag3 | 0.25 | 0.38 | 0.74 | 0.25 | |||
| D14077 | 95286_at | Clu | 0.24 | 0.41 | 0.75 | 0.31 | |||
| sX67083 | 101429_at | Ddit3* | 0.22 | 0.42 | 0.93 | 0.22 | |||
| AA770736 | 161067_at | Ifld2* | 0.20 | 0.36 | 0.93 | 0.07 | |||
| M17298 | 102298_at | Ngfb | 0.16 | 0.49 | 0.65 | 0.16 | |||
| X54149 | 102779_at | Gadd45b*§ | 0.16 | 0.46 | 1.03 | 0.07 | |||
| U21673 | 99134_at | Tcte3 | 0.16 | 0.23 | 0.97 | 0.16 | |||
| L31958 | 100548_at | Pea15 | 0.11 | 0.46 | 1.09 | 0.28 | |||
| AF061972 | 103671_at | Htatip2 | 0.10 | 0.34 | 0.78 | 0.11 | |||
| V00727 | 160901_at | Fos* | 0.08 | 0.20 | 1.12 | 0.06 | |||
| AF055638 | 101979_at | Gadd45g*ठ| 0.05 | 0.19 | 0.94 | 0.04 | |||
| Metabolism | |||||||||
| AA726364 | 95611_at | Lp1 | 38.65 | 3.49 | 6.44 | 3.99 | |||
| Z48670 | 92913_at | Abcd2† | 13.46 | 2.52 | 2.73 | 3.44 | |||
| AI849718 | 160111_at | elF1A | 6.81 | 6.02 | 1.60 | 5.63 | |||
| AW048512 | 95426_at | Echs1 | 6.38 | 4.35 | 3.45 | 6.12 | |||
| AW049778 | 160770_at | Mvd | 5.69 | 6.38 | 1.59 | 6.09 | |||
| U15977 | 94507_at | Facl2 | 5.40 | 1.22 | 1.39 | 1.48 | |||
| L16992 | 102302_at | Bckdhb | 3.05 | 1.73 | 1.06 | 1.62 | |||
| X07888 | 99425_at | HMGCR | 2.91 | 2.39 | 1.30 | 1.68 | |||
| AW106745 | 98631_g_at | Nsdh1 | 2.74 | 2.91 | 1.39 | 3.54 | |||
| AW122523 | 103665_at | Lce-pending | 2.67 | 3.22 | 1.19 | 2.95 | |||
| AW047343 | 160841_at | Dbp | 0.69 | 2.10 | 0.91 | 0.41 | |||
| X05862 | 93833_s_at | Hist1h2bc† | 0.62 | 0.58 | 0.97 | 0.36 | |||
| M18070 | 100606_at | Prnp* | 0.36 | 0.50 | 0.76 | 0.41 | |||
| Y11092 | 92473_at | Mknk2 | 0.35 | 0.45 | 0.74 | 0.22 | |||
| M18070 | 100606_at | Prnp* | 0.36 | 0.50 | 0.76 | 0.41 | |||
| AI835630 | 96680_at | Dnajb9 | 0.34 | 0.48 | 1.33 | 0.35 | |||
| AI845732 | 101007_at | Mknk2 | 0.30 | 0.47 | 0.77 | 0.25 | |||
| L46651 | 102341_at | Gla | 0.25 | 0.37 | 0.71 | 0.07 | |||
| M74495 | 98435_at | Adss1 | 0.21 | 0.32 | 0.65 | 0.27 | |||
| J03520 | 93981_at | Plat | 0.19 | 0.37 | 0.96 | 0.31 | |||
| AB028272 | 96254_at | Dnajb1 | 0.09 | 0.25 | 0.76 | 0.10 | |||
| Differentiation/development | |||||||||
| L10385 | 96566_at | Tgm3* | 47.62 | 1.20 | 2.87 | 2.83 | |||
| U10374 | 97926_s_at | Pparg*‡ | 44.91 | 1.72 | 4.92 | 0.39 | |||
| AJ001700 | 99494_at | Serpini1 | 14.00 | 3.66 | 3.33 | 2.26 | |||
| X16490 | 92978_s_at | Serpinb2 | 10.94 | 1.31 | 1.36 | 1.54 | |||
| Y12293 | 99846_at | Foxf2* | 4.96 | 2.22 | 1.51 | 2.21 | |||
| X75129 | 97950_at | Xdh | 4.14 | 2.49 | 2.31 | 3.82 | |||
| U36760 | 161049_at | Foxg1*† | 2.80 | 3.17 | 1.36 | 2.19 | |||
| M35523 | 100127_at | Crabp2† | 0.39 | 0.59 | 0.78 | 0.26 | |||
| V00756 | 160092_at | Ifrd1 | 0.35 | 0.47 | 1.12 | 0.33 | |||
| AA41985 | 9277_at | Cyr61† | 0.34 | 1.09 | 1.02 | 0.81 | |||
| X13297 | 93100_at | Acta2† | 0.34 | 0.47 | 0.84 | 0.37 | |||
| AF077861 | 93013_at | Idb2 | 0.32 | 0.39 | 1.05 | 0.46 | |||
| M60523 | 92614_at | Idb3* | 0.25 | 0.39 | 1.14 | 0.36 | |||
| X51829 | 160463_at | Myd116‡ | 0.18 | 0.24 | 0.82 | 0.17 | |||
| AV138783 | 161666_f_at | Gadd45b†* | 0.09 | 0.22 | 1.15 | 0.06 | |||
GenBank accession no., by gene function . | . | . | Fold change . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | Probe set . | Gene symbol . | C/EBPα . | C/EBPβ . | C/EBPδ . | C/EBPϵ . | |||
| Immune response | |||||||||
| M63695 | 103422_at | Cd1d1 | 53.47 | 3.05 | 7.36 | 2.42 | |||
| AW124547 | 96092_at | Hp* | 28.05 | 2.72 | 3.76 | 1.85 | |||
| M27008 | 100436_at | Orm1† | 14.20 | 1.03 | 3.25 | 1.23 | |||
| M83218 | 103448_at | S100a8 | 12.99 | 0.99 | 1.72 | 2.12 | |||
| M63697 | 101897_g_at | Cd1d2 | 11.76 | 1.63 | 2.65 | 2.27 | |||
| A1786089 | 93837_at | Kng | 9.13 | 1.74 | 2.15 | 2.16 | |||
| K02782 | 95910_f_at | C3† | 8.43 | 2.18 | 3.39 | 1.62 | |||
| X81627 | 160564_at | Lcn2† | 5.84 | 0.75 | 1.16 | 3.82 | |||
| U27267 | 98772_at | Cxcl5‡ | 4.21 | 2.03 | 2.48 | 5.02 | |||
| M74123 | 94224_s_at | Ifi205‡ | 3.90 | 1.23 | 1.88 | 3.34 | |||
| X57349 | X57349_M_at | Trfr | 3.70 | 3.46 | 1.61 | 2.78 | |||
| M21050 | 100611_at | Lyzs†* | 3.69 | 1.34 | 1.00 | 1.12 | |||
| M31419 | 98465_f_at | Ifi204* | 3.07 | 1.03 | 1.50 | 2.51 | |||
| M32486 | 100477_at | M32486 | 3.06 | 1.15 | 1.30 | 2.74 | |||
| AB012693 | 103611_at | Cd47† | 2.91 | 2.09 | 1.61 | 2.52 | |||
| X83601 | 92731_at | Ptx3 | 2.82 | 1.28 | 2.44 | 4.19 | |||
| L09737 | 102313_at | Gch | 0.31 | 0.50 | 0.89 | 0.39 | |||
| X68273 | 103016_s_at | Cd68 | 0.25 | 0.50 | 0.91 | 0.36 | |||
| L39017 | 98018_at | Procr | 0.16 | 0.29 | 0.90 | 0.15 | |||
| AJ242830 | 102948_at | Hemt1 | 0.08 | 0.19 | 0.86 | 0.07 | |||
| Cell proliferation | |||||||||
| AF099973 | 92472_f_at | Slfn2 | 8.85 | 2.48 | 2.12 | 6.82 | |||
| AF036893 | 93694_at | Per2* | 4.85 | 1.73 | 1.75 | 11.40 | |||
| AF099977 | 92315_at | Slfn4 | 4.98 | 1.10 | 2.36 | 2.27 | |||
| Z22703 | 99435_at | Fgf7 | 4.28 | 1.59 | 2.46 | 4.75 | |||
| X72307 | 102898_at | Hgf* | 4.07 | 1.17 | 0.86 | 1.31 | |||
| X53929 | 93534_at | Dcn | 3.93 | 3.09 | 1.73 | 9.75 | |||
| A1834950 | 98507_at | Nr1d1* | 0.31 | 0.44 | 0.69 | 0.31 | |||
| M64292 | 101583_at | Btg2 | 0.30 | 0.49 | 0.92 | 0.28 | |||
| X61940 | 104598_at | Dusp1* | 0.27 | 0.44 | 0.98 | 0.28 | |||
| AV373612 | 161980_f_at | Bag3 | 0.25 | 0.38 | 0.74 | 0.25 | |||
| D14077 | 95286_at | Clu | 0.24 | 0.41 | 0.75 | 0.31 | |||
| sX67083 | 101429_at | Ddit3* | 0.22 | 0.42 | 0.93 | 0.22 | |||
| AA770736 | 161067_at | Ifld2* | 0.20 | 0.36 | 0.93 | 0.07 | |||
| M17298 | 102298_at | Ngfb | 0.16 | 0.49 | 0.65 | 0.16 | |||
| X54149 | 102779_at | Gadd45b*§ | 0.16 | 0.46 | 1.03 | 0.07 | |||
| U21673 | 99134_at | Tcte3 | 0.16 | 0.23 | 0.97 | 0.16 | |||
| L31958 | 100548_at | Pea15 | 0.11 | 0.46 | 1.09 | 0.28 | |||
| AF061972 | 103671_at | Htatip2 | 0.10 | 0.34 | 0.78 | 0.11 | |||
| V00727 | 160901_at | Fos* | 0.08 | 0.20 | 1.12 | 0.06 | |||
| AF055638 | 101979_at | Gadd45g*ठ| 0.05 | 0.19 | 0.94 | 0.04 | |||
| Metabolism | |||||||||
| AA726364 | 95611_at | Lp1 | 38.65 | 3.49 | 6.44 | 3.99 | |||
| Z48670 | 92913_at | Abcd2† | 13.46 | 2.52 | 2.73 | 3.44 | |||
| AI849718 | 160111_at | elF1A | 6.81 | 6.02 | 1.60 | 5.63 | |||
| AW048512 | 95426_at | Echs1 | 6.38 | 4.35 | 3.45 | 6.12 | |||
| AW049778 | 160770_at | Mvd | 5.69 | 6.38 | 1.59 | 6.09 | |||
| U15977 | 94507_at | Facl2 | 5.40 | 1.22 | 1.39 | 1.48 | |||
| L16992 | 102302_at | Bckdhb | 3.05 | 1.73 | 1.06 | 1.62 | |||
| X07888 | 99425_at | HMGCR | 2.91 | 2.39 | 1.30 | 1.68 | |||
| AW106745 | 98631_g_at | Nsdh1 | 2.74 | 2.91 | 1.39 | 3.54 | |||
| AW122523 | 103665_at | Lce-pending | 2.67 | 3.22 | 1.19 | 2.95 | |||
| AW047343 | 160841_at | Dbp | 0.69 | 2.10 | 0.91 | 0.41 | |||
| X05862 | 93833_s_at | Hist1h2bc† | 0.62 | 0.58 | 0.97 | 0.36 | |||
| M18070 | 100606_at | Prnp* | 0.36 | 0.50 | 0.76 | 0.41 | |||
| Y11092 | 92473_at | Mknk2 | 0.35 | 0.45 | 0.74 | 0.22 | |||
| M18070 | 100606_at | Prnp* | 0.36 | 0.50 | 0.76 | 0.41 | |||
| AI835630 | 96680_at | Dnajb9 | 0.34 | 0.48 | 1.33 | 0.35 | |||
| AI845732 | 101007_at | Mknk2 | 0.30 | 0.47 | 0.77 | 0.25 | |||
| L46651 | 102341_at | Gla | 0.25 | 0.37 | 0.71 | 0.07 | |||
| M74495 | 98435_at | Adss1 | 0.21 | 0.32 | 0.65 | 0.27 | |||
| J03520 | 93981_at | Plat | 0.19 | 0.37 | 0.96 | 0.31 | |||
| AB028272 | 96254_at | Dnajb1 | 0.09 | 0.25 | 0.76 | 0.10 | |||
| Differentiation/development | |||||||||
| L10385 | 96566_at | Tgm3* | 47.62 | 1.20 | 2.87 | 2.83 | |||
| U10374 | 97926_s_at | Pparg*‡ | 44.91 | 1.72 | 4.92 | 0.39 | |||
| AJ001700 | 99494_at | Serpini1 | 14.00 | 3.66 | 3.33 | 2.26 | |||
| X16490 | 92978_s_at | Serpinb2 | 10.94 | 1.31 | 1.36 | 1.54 | |||
| Y12293 | 99846_at | Foxf2* | 4.96 | 2.22 | 1.51 | 2.21 | |||
| X75129 | 97950_at | Xdh | 4.14 | 2.49 | 2.31 | 3.82 | |||
| U36760 | 161049_at | Foxg1*† | 2.80 | 3.17 | 1.36 | 2.19 | |||
| M35523 | 100127_at | Crabp2† | 0.39 | 0.59 | 0.78 | 0.26 | |||
| V00756 | 160092_at | Ifrd1 | 0.35 | 0.47 | 1.12 | 0.33 | |||
| AA41985 | 9277_at | Cyr61† | 0.34 | 1.09 | 1.02 | 0.81 | |||
| X13297 | 93100_at | Acta2† | 0.34 | 0.47 | 0.84 | 0.37 | |||
| AF077861 | 93013_at | Idb2 | 0.32 | 0.39 | 1.05 | 0.46 | |||
| M60523 | 92614_at | Idb3* | 0.25 | 0.39 | 1.14 | 0.36 | |||
| X51829 | 160463_at | Myd116‡ | 0.18 | 0.24 | 0.82 | 0.17 | |||
| AV138783 | 161666_f_at | Gadd45b†* | 0.09 | 0.22 | 1.15 | 0.06 | |||
Shown are the top 20 genes in each category. Mean fold changes were calculated using a simple division of the raw expression values between experimental sample and control.
Gene also involved in metabolism.
Gene also involved in cell proliferation.
Gene also involved in immune response.
Gene also involved in differentiation/development.
Confirmation of microarray data
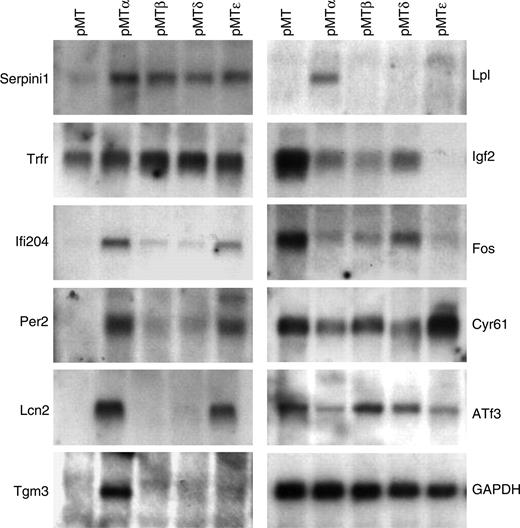
To verify the microarray results, the induction/repression of several genes was assessed by real-time PCR (data not shown) and Northern blot analysis (Figure 2). A high degree (93%) of concordance occurred between the microarray results and the confirmation studies. In addition, several known C/EBP target genes (such as Pparg, Hp, Lcn2, and Saa3) were represented on the microarrays. In several cases, a gene predicted to be selectively modulated by one or more C/EBP transcription factors by the microarray analysis was shown also to be regulated by other C/EBPs in real-time and/or Northern studies. Furthermore, with the exception of a small number of genes, the fold changes in gene expression, induced by the C/EBP proteins as calculated by the chip analysis, underestimated the level of modulation as found by quantitative real-time PCR. These studies indicate that our transcriptional profiling accurately reflected target gene expression levels in the transfected NIH 3T3 populations.
Per2 is a direct target for C/EBPα and C/EBPϵ
Functional annotation analysis showed that the expression levels of 2 core circadian genes, Per2 (Period 2) and Nr1d1 (nuclear receptor subfamily 1, group D, member 1, Rev-Erbα), as well as a circadian output gene, Dbp (albumin site d-binding protein), are significantly altered by C/EBPs, suggesting for the first time a link between C/EBPs and clock genes. These findings are particularly interesting because recent studies now link the circadian clock to cell proliferation and tumor growth. In additional experiments, we chose to focus mainly on Per2 as a possible critical target of C/EBPα-induced growth arrest and tumor suppression.
Verification of the microarray data by Northern blot analysis. Total RNA was harvested from pMT, pMTα, pMTβ, pMTδ, and pMTϵ NIH 3T3 cell lines cultured in the presence of zinc-supplemented media for 16 hours. Northern blots were performed with specific probes for selected genes as indicated.
Verification of the microarray data by Northern blot analysis. Total RNA was harvested from pMT, pMTα, pMTβ, pMTδ, and pMTϵ NIH 3T3 cell lines cultured in the presence of zinc-supplemented media for 16 hours. Northern blots were performed with specific probes for selected genes as indicated.
Analysis of the Per2 promoter region revealed that it contains several potential C/EBP binding sites between -1520 and -910 relative to the mRNA start site. We used reporter assays to examine whether C/EBPα and C/EBPϵ can regulate Per2 promoter activity. Cotransfection of NIH 3T3 cells with a Per2 promoter-luciferase construct along with either the C/EBPα or C/EBPϵ expression vector resulted in a 7- and 5-fold stimulation of Per2 promoter activity, respectively (Figure 3A). Dbp, one of the genes identified by our transcriptional profiling, is a transcription factor that recognizes DNA binding sites similar to those recognized by C/EBPs.38 We, therefore, tested the ability of DBP to activate Per2 transcription. Cotransfection of NIH 3T3 cells with the Per2 promoter-luciferase construct and a DBP expression vector led to a 3-fold increase in the reporter activity (Figure 3A). These results show that the Per2 promoter is induced by C/EBPα, C/EBPϵ, and DBP.
EMSAs were performed to examine whether C/EBPα can bind to its cognate site in the Per2 promoter. When a radiolabeled probe encompassing the putative C/EBP binding site was incubated with nuclear extracts from untransfected NIH 3T3 cells, only a weak band was detected (Figure 3B). In contrast, nuclear extracts from cells transfected with C/EBPα expression vector gave rise to an intense band. The binding was specific as it was competed with unlabeled probe and addition of a C/EBPα antibody supershifted the protein complex. This demonstrates that the transcriptional activation of Per2 is due to direct binding of C/EBPα to the Per2 promoter.
We also performed chromatin immunoprecipitation (ChIP) experiments using murine bone marrow cells to determine whether C/EBPα and C/EBPϵ can directly bind to the Per2 promoter. The C/EBPα and C/EBPϵ antibodies, but not the negative controls (no antibody and preimmune serum), precipitated the endogenous Per2 promoter sequence. The β-actin used as a control sequence was not pulled down by the C/EBP antibodies (Figure 3C). These results indicate that C/EBPα and C/EBPϵ are specifically associated with the Per2 promoter in vivo.
C/EBPα and C/EBPϵ directly regulate the Per2 promoter. (A) Reporter assays were performed with NIH 3T3 cells cotransfected with murine Per2 reporter vector (pGL3-Per2) and either C/EBPα, C/EBPϵ, or DBP expression vectors. Control transfections indicated that the expression vectors had little to no effect on the pGL3 empty reporter vector. All transfections included the pRL-TK vector that served as an internal control for transfection efficiency. Results represent the mean ± SD of triplicate transfections. (B) EMSA was done using 10 μg nuclear extract proteins from NIH 3T3 cells either untransfected (UT) or transfected with a C/EBPα expression vector. Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP site from the Per2 promoter. Unlabeled competitor oligonucleotides (× 100) or C/EBPα antibody was added as indicated. The asterisk indicates the position of the supershifted band. (C) Chromatin immunoprecipitation was performed from murine bone marrow cells using either C/EBPα or C/EBPϵ antibodies, preimmune serum, or no antibody. The samples were analyzed by PCR using primers specific for the C/EBP site in the murine Per2 promoter. Primers for the murine β-actin 5′ untranslated region were used as negative control. The input chromatin was included as a positive control. Images are inverted.
C/EBPα and C/EBPϵ directly regulate the Per2 promoter. (A) Reporter assays were performed with NIH 3T3 cells cotransfected with murine Per2 reporter vector (pGL3-Per2) and either C/EBPα, C/EBPϵ, or DBP expression vectors. Control transfections indicated that the expression vectors had little to no effect on the pGL3 empty reporter vector. All transfections included the pRL-TK vector that served as an internal control for transfection efficiency. Results represent the mean ± SD of triplicate transfections. (B) EMSA was done using 10 μg nuclear extract proteins from NIH 3T3 cells either untransfected (UT) or transfected with a C/EBPα expression vector. Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP site from the Per2 promoter. Unlabeled competitor oligonucleotides (× 100) or C/EBPα antibody was added as indicated. The asterisk indicates the position of the supershifted band. (C) Chromatin immunoprecipitation was performed from murine bone marrow cells using either C/EBPα or C/EBPϵ antibodies, preimmune serum, or no antibody. The samples were analyzed by PCR using primers specific for the C/EBP site in the murine Per2 promoter. Primers for the murine β-actin 5′ untranslated region were used as negative control. The input chromatin was included as a positive control. Images are inverted.
Per2 is regulated by C/EBPα and C/EBPϵ in human hematopoietic leukemic cell lines
We next tested whether C/EBPα and C/EBPϵ can regulate Per2 expression in human hematopoietic cancer cell lines. For these experiments, we used KCL22 (chronic myelocytic leukemia) and Daudi (Burkitt lymphoma) cell lines stably transfected with a zinc-inducible CEBPA gene, as well as a U937 (myelomonocytic leukemia) cell line stably transfected with a zinc-inducible CEBPE gene. Real-time PCR analysis showed that Per2 expression levels increased by 7- and 5-fold following induction of C/EBPα in the KCL22 and Daudi cell lines, respectively; and by 14-fold after induction of C/EBPϵ in the U937 cell line (Figure 4A). We also measured the expression levels of Nr1d1, the second clock gene identified by transcriptional profiling. Real-time PCR analysis showed that Rev-Erbα expression levels were down-regulated by 2- and 4-fold following induction of C/EBPα in the KCL22 and Daudi cell lines, respectively, and by 2-fold in the U937 cell line after induction of C/EBPϵ (Figure 4A). This is in agreement with the microarray data showing that C/EBPα and C/EBPϵ induced Per2 and repressed Rev-Erbα expression levels in NIH 3T3 cells.
C/EBPα and C/EBPϵ up-regulate Per2 expression in human leukemia cell lines. (A) KCL22, Daudi, and U937 cells stably transfected with zinc-inducible C/EBPα (pMTα), C/EBPϵ (pMTϵ), or control empty vector (pMT) were incubated in the presence of zinc for 24 hours. Total RNA was extracted and analyzed by real-time PCR with Per2- and Rev-Erbα-specific primers. (B) Per2 expression levels were measured by real-time PCR in samples from 4 normal human lymph nodes and the indicated lymphoma cell lines. The results are expressed in arbitrary units as a ratio of either Per2 (A-B) or Rev-Erbα (A) transcripts to 18S transcripts (each value represents the mean of 3 measurements of the sample).
C/EBPα and C/EBPϵ up-regulate Per2 expression in human leukemia cell lines. (A) KCL22, Daudi, and U937 cells stably transfected with zinc-inducible C/EBPα (pMTα), C/EBPϵ (pMTϵ), or control empty vector (pMT) were incubated in the presence of zinc for 24 hours. Total RNA was extracted and analyzed by real-time PCR with Per2- and Rev-Erbα-specific primers. (B) Per2 expression levels were measured by real-time PCR in samples from 4 normal human lymph nodes and the indicated lymphoma cell lines. The results are expressed in arbitrary units as a ratio of either Per2 (A-B) or Rev-Erbα (A) transcripts to 18S transcripts (each value represents the mean of 3 measurements of the sample).
Per2 expression is down-regulated in lymphoid and myeloid malignancies
A significant number of mice deficient in expression of Per2 develop lymphomas.26 We used real-time RT-PCR to measure Per2 expression in 6 human cell lines representing different lymphoma subtypes, as well as in normal human lymph nodes. Results showed that in 4 types of B-cell malignant cell lines, Burkitt (Daudi, Raji, and Ramos), pre-B-cell acute lymphoblastic leukemia (B-ALL; Naml6 and blin1), Mantle cell (Jeko1), and large B-cell lymphoma (Sudhl6 and Sudhl16), Per2 levels were extremely low in comparison with levels in cells from normal human lymph nodes (Figure 4B). These results suggest that Per2 expression is down-regulated in several subsets of lymphomas.
Inactivation of C/EBPα often occurs in AML and may subsequently result in deregulation of C/EBPα target genes. We, therefore, tested whether Per2 might be abnormally expressed in patients with AML. Total RNA from light-density mononuclear samples from patients with AML as well as from similar samples from healthy volunteers was isolated, and Per2 expression levels were determined by real-time RT-PCR. We found that in 42% of samples from patients with AML, Per2 mRNA expression levels were significantly lower (P < .05) in comparison with levels in normal bone marrow cells (Figure 5).
Expression of Per2 in AML. Expression levels of Per2 were measured in light density, mononuclear bone marrow samples (enriched for less differentiated, dividing cells) from 9 healthy donors (Healthy) and 21 patients with AML. The results are expressed in arbitrary units as a ratio of the Per2 transcripts to GAPDH transcripts (each value represents the mean of 3 measurements of the sample).
Expression of Per2 in AML. Expression levels of Per2 were measured in light density, mononuclear bone marrow samples (enriched for less differentiated, dividing cells) from 9 healthy donors (Healthy) and 21 patients with AML. The results are expressed in arbitrary units as a ratio of the Per2 transcripts to GAPDH transcripts (each value represents the mean of 3 measurements of the sample).
Expression of Per2 leads to growth arrest
Further studies explored the consequences of expressing Per2 on cell proliferation. The CML, K562, and the myelomonocytic, U937, human cell lines were transfected with either a V5-tagged Per2 expression vector or an empty vector as control. After a short period of antibiotic selection, equal numbers of polyclonal populations were cultured in fresh media, and their growth rate was measured by daily viable cell counts. In both cell lines, forced expression of Per2 led to a dramatic decrease in proliferation (Figure 6B). Expression of the Per2 protein was verified by Western analysis (Figure 6A).
To analyze additionally the role of Per2 in cell proliferation, we established an inducible cell line system. The K562 cell line was stably transfected with a zinc-inducible V5-tagged Per2 expression vector (pMTPer2) as well as control empty vector (pMT). Clones were selected on the basis of G418 resistance, and inducibility of Per2 expression was demonstrated by Western blot (Figure 6C). Induction of Per2 led to very substantial growth reduction as measured by MTT assays (Figure 6D).
Cell cycle analysis was carried out to determine which phase of the cell cycle is inhibited by Per2 expression. After 3 days of culture in the presence of zinc, the K562-pMTPer2 cells had a significantly (P < .01) increased number of cells (8%) in the G2/M phase and a decreased number of cells (30%) in the S phase of the cell cycle compared with K562-pMT cells containing the empty vector (2% G2/M and 44% S phase [Figure 7A]). Furthermore, increased apoptosis (statistically significant, P < .01) was observed in K562-pMTPer2 cells (10%) by day 5 of culture in zinc-supplemented media, in contrast to the low level of apoptosis in the K562-pMT cells (1%) cultured under identical conditions (Figure 7B).
We next examined the effect of Per2 expression on anchorage-independent clonal growth of K562 in soft agar (Figure 7C). Incubation of the K562-pMTPer2 cells in zinc-containing media led to complete inhibition of colony formation (0 colonies). Exposure of the K562-pMT control cells to zinc also inhibited colony formation compared with the wild-type untreated cells, but to a significantly lesser extent.
Discussion
As one approach for better understanding the full extent of gene expression under the control of C/EBP proteins, we performed transcriptional profiling with NIH 3T3 cells ectopically expressing either C/EBPα, C/EBPβ, C/EBPδ, or C/EBPϵ. Several C/EBP microarray studies using a number of cell types were previously reported.12,39-41 We elected to use NIH 3T3 cells because they do not express endogenous C/EBP proteins. Furthermore, these multi-potential mesenchymal stem cells have the capacity to express genes normally restricted to more differentiated cell types such as adipocytes, myocytes, granulocytes, and neuronal cells42-46 ; using theses cells allowed induction of numerous C/EBP target genes from heterologous cell types.
An interesting question is whether C/EBP family members regulate specific genes or if they regulate a common set of genes. One major finding of our study is that almost all identified genes were regulated by more than 1 of the C/EBP members, albeit at different levels. These results suggest that strict C/EBP target gene specificity is rare; rather, specificity may be conveyed by how efficiently the C/EBPs can activate a given target gene. Of the 4 C/EBPs examined, C/EBPα, was notably the strongest transcriptional modulator. In further studies, we focused mainly on C/EBPα-regulated genes.
Per2 inhibits cell proliferation. (A) K562 and U937 cells were cotransfected with either an empty expression vector (Con) or a V5-tagged Per2 expression vector (Per2-V5) along with a vector expressing the puromycin-resistant gene. Following 3 days of antibiotic selection, the surviving cells were harvested and analyzed by Western blotting with a V5 antibody. The blots were stripped and rehybridized with a GAPDH antibody. (B) Growth curve. K562 and U937 cells transfected with either empty vector (K562/Con and U937/Con) or Per2 expression vector (K562/Per2 and U937/Per2) were grown in fresh media. Aliquots were taken at 24-hour intervals for assessment of total viable cells. Data represent the mean ± SD of duplicate experiments. (C) K562 cells stably transfected with either a zinc-inducible V5-tagged Per2 expression vector (pMTPer2-V5) or an empty vector (pMT) were incubated with zinc for 16 hours and analyzed by Western blot with V5 antibody. The blot was stripped and rehybridized with a GAPDH antibody. (D) MTT assay. Equal numbers (3 × 104/mL) of K562 cells stably transfected with either empty vector (pMT) or a Per2 expression vector (pMTPer2) were grown in the presence of zinc. After 5 days, cell proliferation was determined by MTT assays. Untreated, wild-type K562 cells (wt) were included as control. Data are expressed as the mean ± SD of quadruplicate samples. The experiment was repeated twice.
Per2 inhibits cell proliferation. (A) K562 and U937 cells were cotransfected with either an empty expression vector (Con) or a V5-tagged Per2 expression vector (Per2-V5) along with a vector expressing the puromycin-resistant gene. Following 3 days of antibiotic selection, the surviving cells were harvested and analyzed by Western blotting with a V5 antibody. The blots were stripped and rehybridized with a GAPDH antibody. (B) Growth curve. K562 and U937 cells transfected with either empty vector (K562/Con and U937/Con) or Per2 expression vector (K562/Per2 and U937/Per2) were grown in fresh media. Aliquots were taken at 24-hour intervals for assessment of total viable cells. Data represent the mean ± SD of duplicate experiments. (C) K562 cells stably transfected with either a zinc-inducible V5-tagged Per2 expression vector (pMTPer2-V5) or an empty vector (pMT) were incubated with zinc for 16 hours and analyzed by Western blot with V5 antibody. The blot was stripped and rehybridized with a GAPDH antibody. (D) MTT assay. Equal numbers (3 × 104/mL) of K562 cells stably transfected with either empty vector (pMT) or a Per2 expression vector (pMTPer2) were grown in the presence of zinc. After 5 days, cell proliferation was determined by MTT assays. Untreated, wild-type K562 cells (wt) were included as control. Data are expressed as the mean ± SD of quadruplicate samples. The experiment was repeated twice.
While a number of genes identified by our screen were common to C/EBP targets identified by previous studies (eg, gadd45, Ptx3, Rgs2, Btg2, Pim1, Fos, Cyr61, and Lcn2), a substantial number of genes found in our list were not previously linked to C/EBP function. Nonetheless, many of them correlate very well with known physiologic activities of C/EBP proteins. A large number of the identified genes are implicated in the inflammatory response. A second group of genes includes those associated with cell proliferation. Several of the repressed genes (such as Fos, Junb, Atf3, Pim1, and Pea15) are genes known to promote growth, while a number of the induced genes (such as Slfn2, Slfn4, Dcn, Orm1, and Lnc2) inhibit growth. This is in accordance with known functions of C/EBP proteins in arrest of proliferation in different cell types. Two additional sets of modulated genes include those associated with differentiation/development and those involved in cellular metabolism.
Per2 induces growth arrest, apoptosis, and loss of clonogenic potential. (A, top) Cell cycle analysis. K562 cells stably transfected with either empty vector (pMT; ▧) or a Per2 expression vector (pMTPer2; □) were cultured with zinc for 3 days, harvested, stained with propidium iodide (PI), and analyzed by flow cytometry for cell cycle analysis.  indicates wt. (Bottom) Bar graphs present the means ± SD of 3 independent experiments. (B) Apoptosis analysis. Stably transfected K562 cells (pMT and pMTPer2) treated with zinc for 4 days were stained with annexin V-FITC and PI. Data represent the mean ± SD of 3 experiments. (C) Clonogenic analysis. Zinc-treated stably transfected K562 cells (pMT and pMTPer2 [1 × 104/well]) were cultured in soft agar. Colonies containing approximately 1000 cells or more were counted on day 14. Experiments were performed in triplicate and repeated twice, and the mean ± SD of a representative experiment is shown. Untreated wild-type K562 cells (wt) were included as controls.
indicates wt. (Bottom) Bar graphs present the means ± SD of 3 independent experiments. (B) Apoptosis analysis. Stably transfected K562 cells (pMT and pMTPer2) treated with zinc for 4 days were stained with annexin V-FITC and PI. Data represent the mean ± SD of 3 experiments. (C) Clonogenic analysis. Zinc-treated stably transfected K562 cells (pMT and pMTPer2 [1 × 104/well]) were cultured in soft agar. Colonies containing approximately 1000 cells or more were counted on day 14. Experiments were performed in triplicate and repeated twice, and the mean ± SD of a representative experiment is shown. Untreated wild-type K562 cells (wt) were included as controls.
Per2 induces growth arrest, apoptosis, and loss of clonogenic potential. (A, top) Cell cycle analysis. K562 cells stably transfected with either empty vector (pMT; ▧) or a Per2 expression vector (pMTPer2; □) were cultured with zinc for 3 days, harvested, stained with propidium iodide (PI), and analyzed by flow cytometry for cell cycle analysis.  indicates wt. (Bottom) Bar graphs present the means ± SD of 3 independent experiments. (B) Apoptosis analysis. Stably transfected K562 cells (pMT and pMTPer2) treated with zinc for 4 days were stained with annexin V-FITC and PI. Data represent the mean ± SD of 3 experiments. (C) Clonogenic analysis. Zinc-treated stably transfected K562 cells (pMT and pMTPer2 [1 × 104/well]) were cultured in soft agar. Colonies containing approximately 1000 cells or more were counted on day 14. Experiments were performed in triplicate and repeated twice, and the mean ± SD of a representative experiment is shown. Untreated wild-type K562 cells (wt) were included as controls.
indicates wt. (Bottom) Bar graphs present the means ± SD of 3 independent experiments. (B) Apoptosis analysis. Stably transfected K562 cells (pMT and pMTPer2) treated with zinc for 4 days were stained with annexin V-FITC and PI. Data represent the mean ± SD of 3 experiments. (C) Clonogenic analysis. Zinc-treated stably transfected K562 cells (pMT and pMTPer2 [1 × 104/well]) were cultured in soft agar. Colonies containing approximately 1000 cells or more were counted on day 14. Experiments were performed in triplicate and repeated twice, and the mean ± SD of a representative experiment is shown. Untreated wild-type K562 cells (wt) were included as controls.
The highest up-regulated gene was Cd1d1, which showed a 53-fold induction following C/EBPα expression. CD1d1, a member of the major histocompatibility complex (MHC) family, presents glycolipids to natural killer T cells and is thought to be involved in the antitumor immune response.47 Peroxisome proliferator-activated receptor gamma (PPARγ), a well-known C/EBPα target, was also strongly induced by C/EBPα (45-fold). Of interest, C/EBPϵ down-regulated PPARγ expression by 2.5-fold. Although C/EBPα and C/EBPϵ are both expressed during granulocytic differentiation, recent findings demonstrated specific roles for these 2 C/EBPs in modulating secondary granule gene expression.48 Thus, they may have unique functions in regulating common target genes in other cell types. Of note, PPARγ, C/EBPα, and C/EBPϵ are all expressed during macrophage development and play key roles in maturation and metabolic functions of macrophages.49-52 It is possible that cross-talk between PPARγ and C/EBP signaling pathways is necessary for coordinated gene expression in macrophages.
Functional annotation analysis suggested a previously unreported relationship between C/EBP transcription factors and the circadian clock. Our microarray data identified 3 circadian genes, Per2, Nr1d1, and Dbp, as novel C/EBP targets (Figure 8). This finding is especially intriguing given recent reports that link the circadian clock to cell-cycle regulation and tumor suppression. In a recent study, Fu et al26 presented strong evidence supporting a role for Per2 in tumor suppression and response to DNA damage; Per2 mutant mice showed increased sensitivity to γ radiation and tumor development; aberrant activity of cell cycle-related genes such as Myc, Cend1, and Trp53 may contribute to the cancer-prone phenotype of these mice. C/EBPα is a potent inhibitor of cell growth and an established tumor suppressor gene in acute leukemia.7 Consequently, we focused on Per2 as a possible downstream target for C/EBPα-mediated tumor suppression in leukemia. Using reporter assays, EMSA, chromatin immunoprecipitation, and conditionally expressing cell lines, we clearly demonstrated that C/EBPα and C/EBPϵ induce expression of Per2 mRNA.
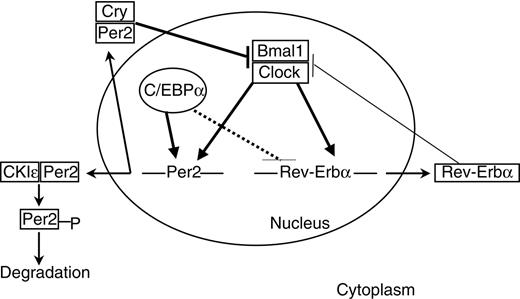
Links between C/EBPα and circadian clock gene expression. The clock mechanism involves transcriptional-translational feedback loops. The transcriptional activators Clock and Bmal1 drive the expression of Per2 and Rev-erbα genes. Per and Cry proteins dimerize in the cytoplasm and enter the nucleus to inhibit the Clock/Bmal1 complexes. Phosphorylation by CKIϵ leads to proteolysis of cytoplasmic Per2. Rev-erbα inhibits bmal1 expression. C/EBPα positively regulates Per2 promoter. Solid thick lines indicate direct regulation; solid thin lines, indirect regulation; and dashed lines, undetermined mode of regulation.
Links between C/EBPα and circadian clock gene expression. The clock mechanism involves transcriptional-translational feedback loops. The transcriptional activators Clock and Bmal1 drive the expression of Per2 and Rev-erbα genes. Per and Cry proteins dimerize in the cytoplasm and enter the nucleus to inhibit the Clock/Bmal1 complexes. Phosphorylation by CKIϵ leads to proteolysis of cytoplasmic Per2. Rev-erbα inhibits bmal1 expression. C/EBPα positively regulates Per2 promoter. Solid thick lines indicate direct regulation; solid thin lines, indirect regulation; and dashed lines, undetermined mode of regulation.
Clock genes are expressed in several peripheral tissues including bone marrow where they control cell proliferation and apoptosis by regulating genes involved in those processes.20-26,53 To investigate whether Per2 is involved in leukemia, we measured its expression in normal bone marrow and bone marrow from patients with AML. Our results show that Per2 expression is reduced in 42% of AML samples. Casein kinase I ϵ (CKIϵ), a core component of the circadian system, regulates the stability of Per by phosphorylating these proteins. A recent report showed that CKIϵ is essential for granulocytic differentiation.53 Perhaps the decreased activity of Per2 in leukemic cells, which are blocked in their terminal differentiation, occurs not only by down-regulation of its mRNA but also by a posttranscriptional mechanism such as phosphorylation.
We showed that forced expression of Per2 in K562 and U937 human myeloid leukemia cell lines leads to a marked growth inhibition. We also generated a stably transfected K562 line containing an inducible Per2 gene. Induction of Per2 in this cell system resulted in arrest of proliferation, apoptosis, and loss of clonogenic ability. Our finding that Per2 overexpression results in a significant G2/M arrest is in agreement with several earlier studies showing that the G2/M checkpoint is under circadian control.25,54 Nonetheless, genetic and molecular data point to circadian regulation of multiple stages of the cell cycle pathway.23,24 Increased expression of c-myc was suggested as a possible mechanism contributing to the cancer-prone phenotype of the Per2 mutant mice. In myeloid cells, down-regulation of c-myc is critical for terminal differentiation and growth arrest associated with C/EBPα.55 We found that c-myc expression is deregulated in the K562-pMTPer2 inducible cells (data not shown). The molecular pathways underlying Per2 effects in myeloid cells and what role c-myc, as well as other cell cycle-related genes, plays in mediating these effects remain the subject of further studies.
A significant number of Per2 mutant mice die before the age of 16 months from spontaneous lymphomas.26 We found that Per2 expression is down-regulated in several types of human B-ALL and lymphoma cell lines, suggesting that inactivation of Per2 may contribute to the development of B-cell leukemias and lymphomas in humans. We also found that Per2 inhibits the proliferation of several epithelial cell types including breast, prostate, and lung cancer cells (data not shown). C/EBPα is expressed in normal epithelial cells from a variety of tissues and was recently suggested to act as a tumor suppressor gene in those cells.56-58 Perhaps induction of Per2 is one of the pathways contributing to antitumor-genic effects of C/EBPα. These findings further support a role for Per2 as a potent growth inhibitor in a variety cell types.
Nr1d1, the second clock gene identified by our transcriptional profiling, is a transcriptional repressor that plays a key role in the circadian clock feedback loops. A recent report showed an association between Rev-Erbα and V-erb-b2 erythroblastic leukemia viral oncogene homolog (ERBB2) expression levels in breast cancer samples,59 suggesting that deregulation of Rev-Erbα plays a role in cancer. The third circadian gene that we identified, Dbp, is not one of the core clock genes but is a component of the clock output system.60 Our microarray data showed that DBP expression is induced by C/EBPβ, repressed by C/EBPϵ, and is unaffected by either C/EBPα or C/EBPδ. This is in agreement with an earlier study showing that although the DBP promoter contains C/EBP binding sites, C/EBPα does not regulate its expression.61 DBP is a member of the proline- and acidic amino acid-rich (PAR) bZIP transcription factor family that binds a subset of C/EBP sites.38 Indeed, we were able to show that DBP positively regulates the Per2 promoter. More detailed studies are needed to determine whether DBP and C/EBPs regulate Per2 expression through a common element in the Per2 promoter.
In summary, our transcriptional profile study identified C/EBP-target genes, many of which were not previously associated with the C/EBP family. These studies identified for the first time a link between C/EBPs and circadian clock genes. We showed that C/EBPα, a bona fide tumor suppressor gene in leukemias, directly up-regulates Per2 expression in hematopoietic cells. Additional studies of Per2 showed that its expression is reduced in AML samples and that forced expression of this gene inhibits leukemic cell growth. These data support a model in which Per2, and possibly other clock genes, is involved in C/EBPα-induced growth arrest and block of differentiation, the hallmarks of myeloid transformation. Further elucidating the links between circadian rhythms and malignant growth may help open new therapeutic avenues.
Prepublished online as Blood First Edition Paper, June 28, 2005; DOI 10.1182/blood-2005-01-0358.
Supported in part by National Institutes of Health grants, as well as by the Ronald Havner, Parker Hughes, and the Cindy, Alan Horn, and Inger funds. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and the Molecular Biology Institute (UCLA) and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This work is dedicated to the memory of David Golde, a mentor and friend.





![Figure 7. Per2 induces growth arrest, apoptosis, and loss of clonogenic potential. (A, top) Cell cycle analysis. K562 cells stably transfected with either empty vector (pMT; ▧) or a Per2 expression vector (pMTPer2; □) were cultured with zinc for 3 days, harvested, stained with propidium iodide (PI), and analyzed by flow cytometry for cell cycle analysis. indicates wt. (Bottom) Bar graphs present the means ± SD of 3 independent experiments. (B) Apoptosis analysis. Stably transfected K562 cells (pMT and pMTPer2) treated with zinc for 4 days were stained with annexin V-FITC and PI. Data represent the mean ± SD of 3 experiments. (C) Clonogenic analysis. Zinc-treated stably transfected K562 cells (pMT and pMTPer2 [1 × 104/well]) were cultured in soft agar. Colonies containing approximately 1000 cells or more were counted on day 14. Experiments were performed in triplicate and repeated twice, and the mean ± SD of a representative experiment is shown. Untreated wild-type K562 cells (wt) were included as controls.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/8/10.1182_blood-2005-01-0358/6/m_zh80200585230007.jpeg?Expires=1765892720&Signature=kbIkV4X6R-Z0hyMhD4PFRgv6P8bWfBwVrcHS8D0bZloc0H2c~oNtcdQycc20DRosbNbhMW7ZyQftOMj0pNpRzjY-Zufkto3UUeLPBpP03JZuMGCe7hAtDp59syR-EAtF-0APV6eV64XFZ3zVCRB2HtBOMXVDbaqpLN5gs7Osnq5CqyOMqO54yImTVq8-Bwr7mmafhLRRAAb-cIjVugEUR251OG9AEQMC4CzVtdnCqND1nGGWy6fbl3MUtw1By8HwpvnlPtnfN5SSJH~sR0Q7MxmESoQ6gYSIsoMZAokis5brcV1xBIO~0QLy-pE9KuVhCmlwFP5ngPEItVw-xAZEbg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal