Abstract
Sequence analysis of the ADAMTS13 locus of 2 patients with hereditary thrombotic thrombocytopenic purpura (TTP) revealed the homozygous presence of 4 single nucleotide polymorphisms (SNPs) (R7W, Q448E, P618A, A732V) and a rare missense mutation (R1336W). Analysis of the individual effect of any amino acid exchanges showed that several sequence variations can interact with each other, thereby altering the phenotype of ADAMTS13 deficiency. Introduction of polymorphisms R7W, Q448E, and A732V had no or only minor effects on ADAMTS13 secretion. In contrast, P618A, R1336W, and the A732V-P618A combination strongly reduced ADAMTS13-specific activity and antigen levels. Surprisingly, R7W and Q448E were positive modifiers of ADAMTS13 secretion in the context of P618A and A732V but neither could rescue the severely reduced specific activity conferred by P618A. However, in the context of R1336W, polymorphisms R7W and Q448E enhanced the detrimental effect of the missense mutation and led to undetectable enzyme activity. We show that dependent on the sequence context, the same polymorphisms might be either positive or negative modifiers of gene expression. Our results might therefore be widely relevant to understanding the influence of polymorphisms on the phenotypic expression of complex diseases.
Introduction
von Willebrand factor (VWF) is crucial for primary hemostasis by mediating extracellular matrix-platelet interactions and delivering factor VIII to the site of vascular injury. The large and heterogeneous glycoprotein circulates as a multimer composed of identical units, ranging in molecular weight from 500 kDa to more than 20 000 kDa.1 Plasma VWF is predominately secreted as high molecular weight, “unusually large,” VWF (ULVWF) multimers. Because ULVWF binds more avidly to components of the extracellular matrix2 and to various platelet receptors3,4 than low molecular weight VWF, the hemostatic activity of VWF is size dependent and much more pronounced for high molecular weight VWF than for smaller multimers. This intrinsic property of ULVWF allows for platelet attachment to sites of vascular injury under high shear stress conditions of fast-flowing blood.5 However, immediately downstream from the site of injury, the multimeric size of VWF needs to be physiologically regulated to prevent the formation of platelet and VWF-rich thrombi.
A VWF-cleaving protease, now called ADAMTS13 (a disintegrin and metalloprotease with thrombospondin type-1 motifs) has been identified.6-13 ADAMTS13 cleaves VWF between Tyr1605 and Met1606 in the A2 domain of the VWF monomer,6,7,14,15 yielding typical fragments of 176 kDa and 140 kDa and smaller VWF multimers. Severe deficiency of ADAMTS13 may result in ULVWF spontaneously interacting with platelet receptors5 and cause the life-threatening disorder thrombotic thrombocytopenic purpura (TTP).16,17 Acquired TTP has been associated with the inhibition of ADAMTS13 activity by autoimmune antibodies,18-21 whereas mutations in the ADAMTS13 gene have been causatively linked to hereditary TTP.22
More than 50 ADAMTS13 mutations, including missense, nonsense, and splice site mutations as well as nucleotide deletions and insertions, have been described.11,23-31 However, in vitro expression analyses correlating the genotype and clinical phenotype were done only for some of these mutations.23,26,29,32 Defective secretion of the mutated protein was shown to be the primary mechanism leading to ADAMTS13 deficiency.23,26,29,32 Conversely, the nonsense mutation Q448X leading to the truncation after the first thrombospondin type-1 motif almost lacked detectable ADAMTS13 activity despite normal secretion.23
In addition to ADAMTS13 mutations, evidence is growing that single nucleotide polymorphisms (SNPs) located in the ADAMTS13 coding region influence plasmatic ADAMTS13 activity levels. For example, in a healthy Japanese cohort the relatively frequent SNP variant ADAMTS13 P475S (allelic frequency of 5.1%) was shown to be secreted normally, but it has been associated with profoundly reduced VWF-cp activity, whereas another common SNP variant, Q448E (allelic frequency of 19.4%), had no obvious effect on secretion levels and ADAMTS13 activity.23
The recently described24 complex ADAMTS13 genotype of 2 brothers with familial TTP was found to consist of 4 coding SNPs,11 a single missense mutation and a nonsense mutation. Here, we report on the investigation of the effect of individual polymorphisms and missense mutations as well as combinations of polymorphisms and missense mutations on ADAMTS13 antigen and specific-activity levels.
Materials and methods
Cloning of ADAMTS13 mutant expression plasmids
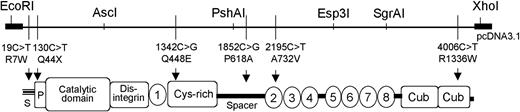
The assembly of the complete ADAMTS13 cDNA into the mammalian expression vector pcDNA 3.1 (Invitrogen, Lofer, Austria) has been described.15 Single ADAMTS13 mutations (19C>T, R7W; 1342C>G, Q448E; 1852C>G, P618A; 2195C>T, A732V; 4006C>T, R1336W) and combinations of amino acid exchanges leading to the series of 20 plasmids described in Figure 1 were introduced either individually or in combination with one another by polymerase chain reaction (PCR) techniques into wild-type (wt) ADAMTS13 cDNA using specific primers or by switching the appropriate restriction fragment between expression constructs using standard cloning techniques.33 A detailed cloning scheme is available on request.
Accuracy of DNA sequences of all generated constructs was confirmed by sequencing. The reporter vector pCMVβ expressing Escherichia coli β-galactosidase was purchased from BD Biosciences Clontech (Erembodegem, Belgium).
Transfection
Human embryonic kidney (HEK 293) cells (American Type Culture Collection [ATCC] CRL-1573; ATCC, Manassas, VA) were cultivated in D-MEM/HAM F12 (1:1) medium (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (full medium). Subconfluent HEK 293 cells grown in 150-mm dishes were transfected with 12 μg plasmid DNA and 60 μL lipofectamine 2000 in Opti-mem I according to the supplier's instructions (Invitrogen). To normalize for transfection efficiency across a range of individual transfections, the reporter plasmid pCMVβ was cotransfected as an internal reference (10:1 molar ratio of test plasmid and pCMVβ). The patient's heterozygous genotype was mimicked by cotransfection with the mutant ADAMTS13 expression plasmids corresponding to the paternal and maternal allele (or one mutant plasmid together with the control vector pcDNA3.1 to compensate total DNA input) and pCMVβ in a molar ratio of 4.5:4.5:1. Each construct was examined in 3 to 10 separate transfections. Medium was replaced by serum-free full medium (SF-FM) 24 hours after transfection, and the cells were incubated for an additional 24 hours. Conditioned medium was collected, clarified by centrifuging, and concentrated 20-fold using Centriprep YM-30 (Millipore, Bedford, MA). Cells were trypsinized and lysed in 20 mM Tris, pH 7.8, and 0.5% Triton X-100 at a concentration of 2.5 × 107 cells/mL as described.15
β-galactosidase activity
The β-galactosidase (β-gal) activity was determined in duplicates in a constant volume of cell lysate of transfected cells using the β-gal assay from Invitrogen with o-nitrophenyl-β-d-galactopyranoside as substrate according to the supplier's instructions.34 The calculated activity coefficient was used to obtain a normalized expression value for the lysate and conditioned medium of the transfected cells.
Western blot analysis
Volumes equivalent to up to 2.5 × 105 cells and 1 × 106 cells per lane for the lysate and the conditioned medium, respectively, adjusted according to the results of the β-gal assay were resolved by 8% SDS-PAGE (acrylamide/bis 29:1; 3.3% C) under reducing and denaturing conditions. Recombinant ADAMTS13 was visualized by Western blotting using the monoclonal anti-ADAMTS13 antibody 242/H1, directed against the catalytic domain.15 Alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma, St Louis, MO) was used for secondary antibody with BCIP/NBT (Promega, Madison, WI) as substrate.
Determination of ADAMTS13 antigen
The determination of ADAMTS13 antigen was done according to M.R., S.F., Johanna A. Kremer Hovinga, C.K., Andrea Herzog, Letitia Koller, Alfred Weber, Giuseppe Remuzzi, Michael Dockal, B.P., and F.S. (manuscript submitted, October 2005) with minor modifications. Briefly, microtiter plates (Nunc-Immuno Maxisorp; Nalge Nunc International, Rochester, NY) were coated with purified polyclonal anti-ADAMTS13 IgG from rabbit35 (2 μg/mL in 0.1 M bicarbonate buffer, pH 9.6) and blocked with 0.5% nonfat milk (Bio-Rad, Hercules, CA) in PBS + 0.1% Tween-20. Cell-culture supernatants and citrated plasma were diluted in blocking solution. For calibration, purified recombinant ADAMTS13 was used in final concentrations from 20 to 0.6 ng/mL added into blocking solution. ADAMTS13 antigen was detected with rabbit anti-ADAMTS13 IgG35 conjugated to horseradish peroxidase using the labeling kit from Roche (Mannheim, Germany) and Sure Blue TMB (Kirkegaard and Perry Laboratories, KPL Inc, Gaithersburg, MD) as substrate. The optical density was read at 450 nm versus 620 nm as reference after stopping the color reaction with 1 M HCl. The concentration of ADAMTS13 antigen in the conditioned medium was analyzed in duplicates in 3 independent analyses.
Assay of ADAMTS13 activity
The ADAMTS13 activity was analyzed in the concentrated conditioned medium of the separate transfections by measuring the residual collagen-binding activity (CBA) of the degraded substrate essentially as described36 with slight modifications. Purified recombinant VWF (Baxter BioScience, Vienna, Austria) was used as substrate, and heat-inactivated (incubated 30 minutes at 56°C, centrifuged 15 minutes at 10 000g) pooled normal human plasma (NHP; Baxter BioScience) was used for sample dilution. A calibration curve was established with NHP. The ADAMTS13 activity in the conditioned medium of each transfection was determined in duplicates in 3 independent analyses.
Normalization of transfection efficiency by cotransfected β-gal reporter plasmid
To monitor transfection efficiency and to obtain a normalized expression value important for comparing results of experiments carried out with different constructs, the activity of the cotransfected β-gal was analyzed in each cell lysate. ADAMTS13 activity and antigen values determined in the conditioned medium were first transcribed into units and micrograms per 106 transfected cells, respectively, and then depicted as normalized values according to the β-gal activity results obtained in the cell lysate. Values (mean of replicate transfections) are presented as a percentage of normal plus or minus SEM (wt ADAMTS13 = 100%).
Statistical analysis
Data are presented as means plus or minus SEMs. The mean ratio of ADAMTS13 activity to antigen levels for the variants and the wild type were compared by using 2-sided t tests for independent samples, and a P value less than .05 was considered to be of statistical significance.
Results
Sequence analysis of the ADAMTS13 gene locus of 2 patients with familial TTP identified 6 amino acid changes putatively involved in ADAMTS13 deficiency.24 On the paternal allele the transition 130C>T introduced a stop codon (Q44X) in the propeptide region and caused premature protein termination. On the maternal allele, 4 of the 5 amino acid substitutions R7W (signal peptide), Q448E (cysteine-rich domain), P618A (spacer domain), A732V (thrombospondin type 1 repeat -2) were identified as common SNPs,11,24 whereas the amino acid exchange R1336W (cub2 domain) was not found in 230 alleles of healthy subjects and therefore was suspected as a new ADAMTS13 deficiency causing missense mutation. Interested by the accumulation of 4 SNPs and a missense mutation on a single allele, we investigated whether these genetic variations individually or through mutual interplay or by both mechanisms would influence the function or the secretion level of the resulting ADAMTS13 molecule.
Schematic domain structure of ADAMTS13. Restriction enzymes used to facilitate cloning of the ADAMTS13 expression plasmids. Relative positions of nucleotide exchanges with the predicted amino acid substitutions in the ADAMTS13 domains indicated. S indicates signal peptide; P, propeptide; 1-8, thrombospondin-1 type 1 repeats.
Schematic domain structure of ADAMTS13. Restriction enzymes used to facilitate cloning of the ADAMTS13 expression plasmids. Relative positions of nucleotide exchanges with the predicted amino acid substitutions in the ADAMTS13 domains indicated. S indicates signal peptide; P, propeptide; 1-8, thrombospondin-1 type 1 repeats.
Expression of mutant ADAMTS13 and effect on secretion
Twenty different ADAMTS13 variants, containing either single mutations or various combinations of amino acid exchanges, were constructed (Figure 1, Table 1). Each construct was individually expressed in HEK 293 cells. Cotransfection of a plasmid coding for β-gal provided an internal reference protein, facilitating the calculation of transfection efficiency and ensuring comparability between experiments. Volumes of cell lysate and conditioned medium, adjusted to cell counts and normalized for transfection efficacy according to β-gal activity, were subjected to immunoblot analysis using a monoclonal antibody raised against the catalytic domain of ADAMTS1315 for ADAMTS13-specific detection (Figures 2, 3, and 4).
Overview of the ADAMTS13 mutants constructed containing single or multiple amino acid substitutions. Cloning strategy and mutations affecting the particular domain(s) are denoted.
Cloning strategy . | Amino acid exchange in wt ADAMTS13 . | Domain(s) affected . |
|---|---|---|
| Overlapping primer PCR | ||
| W7 | R7W | Signalpeptide (S) |
| X44 | Q44X | Propeptide (P) |
| E448 | Q448E | Cysteine-rich (Cys) |
| Inverse PCR | ||
| A618 | P618A | Spacer (Sp) |
| V732 | A732V | Tsp1-2 |
| W1336 | R1336W | Cub2 |
| A-V | P618A-A732V | Sp/Tsp1-2 |
| Switching restriction fragments | ||
| W1-A | R7W-P618A | P/Sp |
| W1-V | R7W-A732V | P/Tsp1-2 |
| E-A | Q448E-P618A | Cys/Sp |
| V-W2 | A732V-R1336W | Tsp1-2/Cub2 |
| E-A-V | Q448E-P618A-A732V | Cys/Sp/Tsp1-2 |
| W1-A-V | R7W-P618A-A732V | S/Sp/Tsp1-2 |
| A-V-W2 | P618A-A732V-R1336W | Sp/Tsp1-2/Cub2 |
| W1-E-A | R7W-Q448E-P618A | P/Cys/Sp |
| W1-E-W2 | R7W-Q448E-R1336W | P/Cys/Cub2 |
| W1-E-A-V | R7W-Q448E-P618A-A732V | S/Cys/Sp/Tsp1-2 |
| W1-A-V-W2 | R7W-P618A-A732V-R1336W | S/Sp/Tsp1-2/Cub2 |
| W1-E-A-V-W2 | R7W-Q448E-P618A-A732V-R1336W | S/Cys/Sp/Tsp1-2/Cub2 |
Cloning strategy . | Amino acid exchange in wt ADAMTS13 . | Domain(s) affected . |
|---|---|---|
| Overlapping primer PCR | ||
| W7 | R7W | Signalpeptide (S) |
| X44 | Q44X | Propeptide (P) |
| E448 | Q448E | Cysteine-rich (Cys) |
| Inverse PCR | ||
| A618 | P618A | Spacer (Sp) |
| V732 | A732V | Tsp1-2 |
| W1336 | R1336W | Cub2 |
| A-V | P618A-A732V | Sp/Tsp1-2 |
| Switching restriction fragments | ||
| W1-A | R7W-P618A | P/Sp |
| W1-V | R7W-A732V | P/Tsp1-2 |
| E-A | Q448E-P618A | Cys/Sp |
| V-W2 | A732V-R1336W | Tsp1-2/Cub2 |
| E-A-V | Q448E-P618A-A732V | Cys/Sp/Tsp1-2 |
| W1-A-V | R7W-P618A-A732V | S/Sp/Tsp1-2 |
| A-V-W2 | P618A-A732V-R1336W | Sp/Tsp1-2/Cub2 |
| W1-E-A | R7W-Q448E-P618A | P/Cys/Sp |
| W1-E-W2 | R7W-Q448E-R1336W | P/Cys/Cub2 |
| W1-E-A-V | R7W-Q448E-P618A-A732V | S/Cys/Sp/Tsp1-2 |
| W1-A-V-W2 | R7W-P618A-A732V-R1336W | S/Sp/Tsp1-2/Cub2 |
| W1-E-A-V-W2 | R7W-Q448E-P618A-A732V-R1336W | S/Cys/Sp/Tsp1-2/Cub2 |
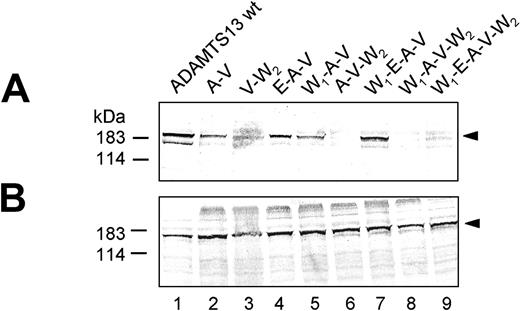
Recombinant expression of wt ADAMTS13 produced an immunoreactive band of approximately 185 kDa in the cell lysate and in the conditioned medium (Figure 2A-B, lane 2). Each ADAMTS13 mutant construct, except X44 (Figure 2A-B, lane 4), was produced with similar efficiency within the transfected cells as judged in the triton-soluble lysate fraction, implying that neither single nor multiple amino acid substitutions greatly affected ADAMTS13 synthesis (Figures 2B, 3B, and 4B). However, large differences in the secretion levels among the various ADAMTS13 mutant constructs were observed.
Introducing the polymorphic mutations W7 and E448 into wt ADAMTS13 had no detectable influence on ADAMTS13 secretion levels, as judged by Western blot analysis (Figure 2A, lanes 3 and 5) and ADAMTS13 antigen enzyme-linked immunosorbent assay (ELISA) (Figure 5, Table 2). Analysis of ADAMTS13 antigen levels in the conditioned cell-culture medium revealed a mean value (±SEM) in percentage compared with wt ADAMTS13 of 99% ± 5% for W7 and 95% ± 8% for E448 (Table 2). Transfection of the SNP mutant V732 led to a moderately lower level of V732 antigen in the cell-culture supernatant (Figure 2, lane 7), which was quantified to be 60% ± 5% of wt ADAMTS13. In contrast, protein secretion significantly decreased for the polymorphism A618 and the missense mutation W1336 (Figure 2A, lanes 6 and 8), and ADAMTS13 antigen levels were reduced to 27% ± 4% for A618 and 23% ± 3% for W1336 (Table 2).
ADAMTS13 antigen and activity levels obtained in the conditioned medium of cells expressing ADAMTS13 variants
. | Percentage of wt ADAMTS13 . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | Antigen level . | . | Activity level . | . | |||
| Construct . | Mean . | SEM . | Mean . | SEM . | |||
| Vector | ND | – | ND | – | |||
| WT | 100 | – | 100 | – | |||
| W7 | 99 | 5 | 86 | 8 | |||
| E448 | 95 | 8 | 75 | 8 | |||
| A618 | 27 | 4 | 14 | 1 | |||
| V732 | 60 | 5 | 71 | 4 | |||
| W1336 | 23 | 3 | 12 | 1 | |||
| A-V | 7 | 1 | 4 | 0 | |||
| W1-A | 35 | 2 | 9 | 2 | |||
| W1-V | 63 | 12 | 75 | 15 | |||
| E-A | 67 | 5 | 28 | 6 | |||
| V-W2 | 18 | 2 | 4 | 1 | |||
| E-A-V | 26 | 2 | 15 | 1 | |||
| W1-A-V | 25 | 2 | 12 | 1 | |||
| A-V-W2 | 4 | 0 | ND | – | |||
| W1-E-A | 63 | 13 | 32 | 2 | |||
| W1-E-W2 | 12 | 1 | 7 | 1 | |||
| W1-E-A-V | 80 | 8 | 40 | 4 | |||
| W1-A-V-W2 | ND | – | ND | – | |||
| W1-E-A-V-W2 | ND | – | ND | – | |||
| X44 | ND | – | ND | – | |||
| X44/W1-E-A-V-W2 | ND | – | ND | – | |||
. | Percentage of wt ADAMTS13 . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | Antigen level . | . | Activity level . | . | |||
| Construct . | Mean . | SEM . | Mean . | SEM . | |||
| Vector | ND | – | ND | – | |||
| WT | 100 | – | 100 | – | |||
| W7 | 99 | 5 | 86 | 8 | |||
| E448 | 95 | 8 | 75 | 8 | |||
| A618 | 27 | 4 | 14 | 1 | |||
| V732 | 60 | 5 | 71 | 4 | |||
| W1336 | 23 | 3 | 12 | 1 | |||
| A-V | 7 | 1 | 4 | 0 | |||
| W1-A | 35 | 2 | 9 | 2 | |||
| W1-V | 63 | 12 | 75 | 15 | |||
| E-A | 67 | 5 | 28 | 6 | |||
| V-W2 | 18 | 2 | 4 | 1 | |||
| E-A-V | 26 | 2 | 15 | 1 | |||
| W1-A-V | 25 | 2 | 12 | 1 | |||
| A-V-W2 | 4 | 0 | ND | – | |||
| W1-E-A | 63 | 13 | 32 | 2 | |||
| W1-E-W2 | 12 | 1 | 7 | 1 | |||
| W1-E-A-V | 80 | 8 | 40 | 4 | |||
| W1-A-V-W2 | ND | – | ND | – | |||
| W1-E-A-V-W2 | ND | – | ND | – | |||
| X44 | ND | – | ND | – | |||
| X44/W1-E-A-V-W2 | ND | – | ND | – | |||
Values were converted to micrograms per 106 cells and units per 106 cells, synchronized according to the calculated β-gal activity, and correlated to wt ADAMTS13 taken as 100%. The mean of ADAMTS13 antigen and activity including SEM (n = 3-10) is depicted. ND indicates not detectable.
– indicates not applicable.
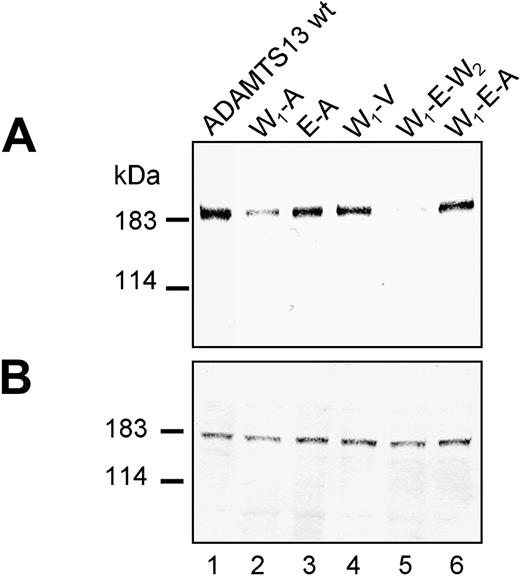
Potential synergistic interactions of individual mutations with each other were analyzed in a series of mutants containing several combinations of the 5 amino acid exchanges R7W, Q448E, P618A, A732V, and R1336W (Figure 1). For example, expression of W7 and V732 together in construct W7-V732 (W1-V), did not markedly alter protein secretion (Figure 4A, lane 4), and secreted ADAMTS13 was detected to 63% ± 12% of wt ADAMTS13, similar to the concentration obtained in the single V732 expression. However, although V732 when expressed individually in the wt ADAMTS13 background had only a moderate effect on ADAMTS13 secretion, its combination with the polymorphism A618 in the ADAMTS13 double mutant A618-V732 (A-V) decreased protein secretion (Figure 3A, lane 2) to about 7% ± 1% of wt ADAMTS13 levels (Table 2). Surprisingly, adding a further polymorphism, either W7 or E448, to the combination A-V, giving rise to the triple polymorphism variants W7-A618-V732 (W1-A-V; Figure 3A, lane 5) and E448-A618-V732 (E-A-V; Figure 3A, lane 4), recovered protein secretion to 25% ± 2% for W1-A-V and 26% ± 2% for E-A-V (Table 2). However, the secretion-recovering effect of the individual SNPs leading to W7 and E448 was more pronounced when both polymorphisms were combined with A-V, as seen with the construct W7-E448-A618-V732 (W1-E-A-V; Figure 3A, lane 7). Secretion of W1-E-A-V was reestablished to 80% ± 8% of wt ADAMTS13 levels (Table 2). This argues for a highly beneficial and synergistic effect of W7 and E448 on ADAMTS13 secretion in the context of polymorphisms A618 and V732.
Expression of ADAMTS13 containing single point mutations. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing single mutant ADAMTS13 variants (abbreviations correspond to Figure 1) were normalized according to β-gal activity and analyzed by immunoblotting using an antibody against ADAMTS13 catalytic domain. The representative of at least 3 repetitive experiments is shown.
Expression of ADAMTS13 containing single point mutations. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing single mutant ADAMTS13 variants (abbreviations correspond to Figure 1) were normalized according to β-gal activity and analyzed by immunoblotting using an antibody against ADAMTS13 catalytic domain. The representative of at least 3 repetitive experiments is shown.
The individual effect of W7 and E448 on A618 was investigated in more detail with double mutants W7-A618 (W1-A) and E448-A618 (E-A), and triple mutant W7-E448-A618 (W1-E-A). Secretion was slightly improved (Figure 4A, lane 2) in W1-A to 35% ± 2% but was strongly intensified (Figure 4A, lane 3) in E-A to 67% ± 5% of wt ADAMTS13 (compared with 27% ± 4% for A618). Similar ADAMTS13 secretion of 63% ± 13% was observed when both W7 and E448 were introduced as in W1-E-A (Figure 4A, lane 6), suggesting that in context with A618 mainly E448 contributes to an improvement of protein secretion (Table 2).
In contrast, neither W7 nor E448 or both were able to compensate for the adverse effect of mutation W1336 on protein secretion as shown in the construct W7-E448-W1336 (W1-E-W2; Figure 4A, lane 5), and ADAMTS13 secretion was decreased to 12% ± 1% (compared with 23% ± 3% for W1336), suggesting rather a detrimental than a beneficial effect of W7 or E448 in this context. Expressing R1336W together with polymorphism V732, as in construct V732-W1336 (V-W2; Figure 3A, lane 3), reduced ADAMTS13 secretion to about 18% ± 2% (Table 2) of wild type. Addition of A618 as in the triple mutant A618-V732-W1336 (A-V-W2; Figure 3A, lane 6) further compromised secretion to 4% of wt ADAMTS13 levels (Table 2). Neither W7 nor W7 and E448 together introduced into the A-V-W2 context as in the mutants W7-A618-V732-W1336 (W1-A-V-W2; Figure 3A, lane 8) and W7-E448-A618-V732-W1336 (W1-E-A-V-W2; Figure 3A, lane 9) improved protein secretion (Figure 5). For both constructs, ADAMTS13 antigen became undetectable in the cell-culture supernatant, suggesting that W1336, in the context of all 4 amino acid polymorphisms, has a strongly detrimental effect on ADAMTS13 secretion.
Expression of ADAMTS13 containing multiple point mutations. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing ADAMTS13 variants (abbreviations corresponding to Table 1) were normalized according to β-gal activity and subjected to immunoblot analysis using anti-ADAMTS13 antibody. The representative of at least 3 repetitive experiments is shown.
Expression of ADAMTS13 containing multiple point mutations. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing ADAMTS13 variants (abbreviations corresponding to Table 1) were normalized according to β-gal activity and subjected to immunoblot analysis using anti-ADAMTS13 antibody. The representative of at least 3 repetitive experiments is shown.
Expression of ADAMTS13 variants in combination with W7 located in the signal peptide. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing ADAMTS13 variants (abbreviations corresponding to Table 1) were normalized according to β-gal activity and subjected to immunoblot analysis using anti-ADAMTS13 antibody. The representative of at least 3 repetitive experiments is shown.
Expression of ADAMTS13 variants in combination with W7 located in the signal peptide. Conditioned medium (A) and cell lysate (B) of transfected HEK 293 cells expressing ADAMTS13 variants (abbreviations corresponding to Table 1) were normalized according to β-gal activity and subjected to immunoblot analysis using anti-ADAMTS13 antibody. The representative of at least 3 repetitive experiments is shown.
ADAMTS13-specific activity
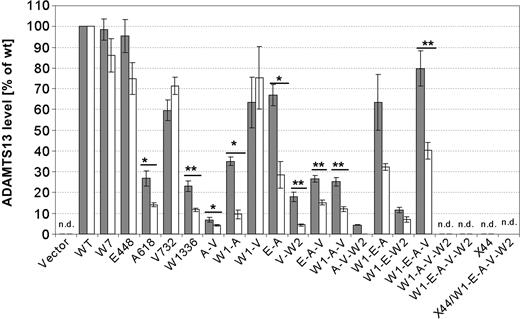
ADAMTS13 antigen and activity was analyzed in the culture medium, and results were converted to micrograms per 106 cells and units per 106 cells prior to the normalization according to the intracellular β-gal activity and then related to wt ADAMTS13 taken as 100%. The correlation between the mean plus or minus SEM levels of ADAMTS13 antigen and activity of the different constructs is depicted in Figure 5 and Table 2.
ADAMTS13 antigen and activity levels obtained in the conditioned medium of cells expressing ADAMTS13 variants. ADAMTS13 antigen (▪) and activity (□) ± SEM. P values were calculated by 2-sided t tests for independent samples (*P < .05, **P < .001).
ADAMTS13 antigen and activity levels obtained in the conditioned medium of cells expressing ADAMTS13 variants. ADAMTS13 antigen (▪) and activity (□) ± SEM. P values were calculated by 2-sided t tests for independent samples (*P < .05, **P < .001).
Single variants containing W7, E448, and V732 had an ADAMTS13 activity level similar to the ADAMTS13 antigen level, resulting in unchanged specific activities. Accordingly, the observed slight difference between antigen and activity levels for the variants W7, E448, and V732 was not statistically significant. In contrast, the isolated variants A618 and W1336 yielded not only a molecule with a much less efficient secretion level (27% ± 4% for A618, 23% ± 3% for W1336) but also with a significantly reduced protease activity (14% ± 1% for A618, 12% ± 1% for W1336). Apparently, both amino acid substitutions affected the secretion pathway and also induced a functional defect in the ADAMTS13 protein by themselves, resulting in reduced specific activities. Moreover, constructs containing A618 and W1336 either individually or together in any combination with the other 3 polymorphisms W7, E448, and V732 yielded secreted ADAMTS13 molecules with significantly impaired proteolytic activity (Figure 5). Antigen and activity levels were significantly (P < .05) different in the A618-containing double mutants A-V, W1-A, and E-A, exhibiting residual ADAMTS13 activity of 4%, 9%, and 28%, respectively. In constructs V-W2, E-A-V, and W1-A-V residual activities of 4%, 15%, and 12%, respectively, were detected, and the observed difference between antigen and activity levels became statistically highly significant (P < .001).
Although ADAMTS13 secretion was almost fully regained when all 4 polymorphic amino acids were present together as in mutant W1-E-A-V, only approximately 40% of wt enzyme activity was recovered, implying that the simultaneous presence of SNPs resulting in E448, A618, and V732 has a detrimental effect on ADAMTS13 proteolytic activity. The amino acid polymorphism W7 is located in the signal peptide and is therefore not retained within the mature protein.
No ADAMTS13 activity was detected in the conditioned medium of cells expressing combinations of polymorphisms and the mutation W1336, such as constructs A-V-W2,W1-A-V-W2, and W1-E-A-V-W2, confirming the dominant-negative effect of the secretion defect which could not be rescued by any of the amino acid polymorphisms.
Reassembling the genotype X44/W1-E-A-V-W2 of the patient with hereditary TTP
To mimic the patient's compound heterozygous genotype, we did cotransfection experiments using 2 plasmids, encoding the paternal stop mutation (Q44X) and the defective maternal allele (W1-E-A-V-W2). As expected, no ADAMTS13 polypeptide was detected in cells transfected with mutant X44 (Figure 2, lane 4). Failure to secrete ADAMTS13 when both mutant ADAMTS13 constructs were coexpressed (X44/W1-E-A-V-W2) and undetectable enzyme activity levels (Table 1) were consistent with the low plasma level of the VWF-cleaving protease (< 62.5 ng ADAMTS13 antigen/mL and < 5% of normal activity18 ) observed in the patient's plasma.
Discussion
Awareness is increasing of the critical role of multiple common polymorphisms responsible for phenotypic variability of diseases and the implication of SNPs as genetic risk factors contributing to disease development and disease susceptibility. The specific role of nonsynonymous SNPs as genetic risk factors for disease development has been associated with several multifactorial diseases such as Alzheimer disease,37,38 systemic lupus erythematosus,39 or cardiovascular disease.40 The potential role of sequence polymorphisms in coagulation factor genes and their influence on thrombosis has been reviewed recently in detail, including factor V R506Q Leiden and the prothrombin mutation G20210A as risk factors for the development of venous thrombosis.41 More recently, a single polymorphism in the VWF gene resulting in the amino acid substitution Y1584C in the A2 domain was correlated with an increased susceptibility to proteolysis by ADAMTS13.42 In addition to the function of SNPs as disease-specific markers, there is a rapidly growing and widespread interest in SNPs or SNP profiles that are associated with drug metabolism and individual drug responses.43
Our interest in a potential role of SNPs in the development of hereditary ADAMTS13 deficiency initially arose from the analysis of a case of familial TTP in which the maternal allele showed an accumulation of 4 nonsynonymous SNPs (in addition to a missense mutation) on a single allele. We report here the detailed investigation of the individual effect of SNPs and combinations of SNPs as well as the effect of the missense mutation and the combination of the missense mutation with various SNPs on the level of ADAMTS13 secretion and proteolytic function.
Individual expression analysis of the 4 amino acid polymorphisms and the rare mutation revealed variable importance of the particular amino acid substitutions for ADAMTS13 secretion and resultant ADAMTS13 activity. Although W7 and E448 polymorphisms did not notably influence ADAMTS13 secretion and proteolytic activity, and V732 polymorphism only influenced it to some minor degree, polymorphism A618 and mutation W1336 were major modifiers. We could clearly show that A618 and W1336 by themselves strongly affected protein secretion and also proteolytic activity.
The amino acid change W1336, located in the second cub domain, and the fairly common (allelic frequency of 9.2%) SNP resulting in A618, located in the spacer region, led to a reduced secretion of the mutated protein (23% for W1336, 27% for A618) with a significantly lower residual activity (12% for W1336, 14% for A618). So far, the only amino acid polymorphism recombinantly analyzed and associated with a decline in the proteolytic activity of ADAMTS13 (5%-10% of wild type) despite normal secretion is P475S located in the cysteine-rich domain.23,44 Polymorphism Q448E, also in the cysteine-rich domain, has been reported as a variant11,23,26,32 with an allele frequency of 19.4% in the healthy Japanese population23 (compared with 42.5% in the European population24 ), and recombinant Q448E had no effect on ADAMTS13 secretion and VWF-cleaving activity23 concordant with our results. The central contribution of cysteine-rich and spacer domain in VWF binding and substrate cleavage has been shown previously with C-terminal ADAMTS13 truncations.45-47 Both domains have been identified as major targets of the autoimmune response in patients with acquired TTP,35,45,48 supporting the particular importance of these domains for ADAMTS13 function in vivo. Strongly impaired secretion of a cub domain mutant (∼ 14% of wild type), however, with only little effect on enzyme activity (∼ 85% activity) has recently been proposed as the cause of hereditary TTP.28,32 It has been shown that experimentally cub-truncated ADAMTS13 mutants retain functional activity under static and under flow conditions45,46,49 and may also be dispensable in vivo in mice.50 However, cub domains have been suggested to contribute to interactions with VWF and to increase VWF-binding affinity.47,51 In conclusion, spacer and cub are probably important domains involved in substrate recognition, binding, and cleavage. Amino acid changes in these domains may significantly affect overall protein function and interaction with binding partners.
Results changed dramatically when we evaluated the functional crosstalk of individual mutations. Combining the 2 amino acid polymorphisms with a demonstrable effect on ADAMTS13-specific activity (A618 and V732), as exemplified in the mutant A-V, led to a further reduction of the secretion level, suggesting an additive effect of the polymorphisms. However, amino acid polymorphisms W7 and E448 were clear positive modifiers of the negative effects of A618 and V732. Either combination of A-V with W7 or with E448, such as W7-A618-V732 (W1-A-V) or E448-A618-V732 (E-A-V), led to an improvement in secretion; however, functional activity of ADAMTS13 was not restored. Moreover, ADAMTS13 activity was strongly improved (mean activity, 40% ± 4%) and expression of ADAMTS13 antigen reached almost wt level (mean antigen, 80% ± 8%) when all 4 amino acid polymorphisms (W7-E448-A618-V732, W1-E-A-V) were combined on the same ADAMTS13 polypeptide. This unequivocally points to the corrective modulatory role of both polymorphisms W7 and E448 and implicates a complex, synergistic interaction of polymorphisms located in different ADAMTS13 domain regions. Taken together, amino acid polymorphisms W7 and E448 function clearly as positive modifiers of ADAMTS13 expression in the context of polymorphisms A618 and V732.
The reported allelic frequency of P618A is 9.2%,24 indicating that theoretically 1 in about 125 people is homozygous for A618, which would, according to our data, translate into a strongly reduced ADAMTS13 activity. It is therefore notable that in 120 healthy alleles analyzed, A618 was detected 11 times, and almost always (10 of 11; 91%) in association with E448 and to a lesser percentage (7 of 11; 64%) in association with both W7 and E448. Although we do not currently know whether A618 and W7 (E448) are present on the same allele, it is tempting to speculate that the detrimental effect of A618 might be attenuated by the presence of either W7 or E448 in vivo. We have provided experimental evidence that particularly E448 is the main positive modulator of the negative effect of A618, at least in terms of secretion. As exemplified in construct W1-E-A, antigen and thereby the total activity levels could be raised to 63% and 32% of wild type, respectively, compared with antigen and activity levels of 27% and 14% for A618. The genetic defect leading to roughly 50% specific activity in all the A618-containing constructs is not corrected by any of the polymorphisms.
Although we could show that W7 and E448 increased the secretion rate of 2 other amino acid polymorphisms, substantially different results were obtained when we combined the missense mutation R1336W with various polymorphisms. The adverse effect of W1336 could not be rescued by the presence of any amino acid polymorphism, in particular W7 or E448. Moreover, the detrimental effect of R1336W on protein secretion and functionality was enhanced in the context of the polymorphisms leading to absent ADAMTS13 activity (eg, W1-A-V-W2,W1-E-A-V-W2). These data highlight the complexity of interactions between amino acid changes and the limitations of the analysis of single mutant proteins.
The positioning of R7W on the signal peptide of ADAMTS13, together with its modulatory influence only becoming evident when it is put into context with polymorphisms E448, A618, and V732, makes the apparent long-reaching effect of W7 on secretion levels difficult to explain.
Signal peptides have a significant role in the targeting of soluble proteins to the secretory pathway and contain specific information, such as for the interaction with membrane phospholipids, with protein components of the secretory machinery, and for cleavage by the signal peptidase.52,53 At the moment, the molecular mechanism by which the polymorphism R7W exerts its clear-cut effect on the process of ADAMTS13 secretion cannot be adequately explained. Possible explanations are an enhancing effect of W7 on the electrostatic interaction of the signal peptide with anionic phospholipids, a positive effect on the interaction with components of the secretion machinery or an effect on the translocation competency of the unfolded protein by a signal peptide-mature protein interaction as reported by Beena et al.54 Alternatively, the signal peptide might have some influence on the stability and the folding of the mature protein, as has been shown for the maltose-binding protein.54 We also cannot entirely rule out the possibility that the observed effect of R7W is only apparent in cell-culture systems using a single cell type without any physiologically relevant consequence.
Mutual functional influence of a rare mutation and a nearby single amino acid polymorphism has only been reported to our knowledge in the gene coding for the cardiac Na+ channel (SCN5A).55 Our study provides new evidence that the positive or negative biologic effect, and therefore the clinical relevant expression, of a mutation or of multiple polymorphisms or any combination of mutation and polymorphism might be dependent on the presence or absence of additional amino acid changes. The important general application of our findings is that they promote the concept that nonsynonymous polymorphisms can have profound modulatory effects which need to be analyzed in concert, rather than on the level of single amino acid changes.
In conclusion, we provide evidence that the ADAMTS13 missense mutation R1336W, found in patients with hereditary TTP, by itself reduces ADAMTS13 activity (12%) but most probably not to the extent necessary (< 5%) to induce disease. We also show that any combination of R1336W with 4 amino acid polymorphisms entirely abolishes ADAMTS13 activity, explaining the lack of ADAMTS13 activity in the patient's plasma. Through detailed analysis of intermediate genotypes we were able to show that polymorphism P618A severely reduces protein secretion and specific activity but that the deleterious effect of this polymorphism could partly be counteracted by the coexpression of other polymorphisms (particularly E448 and W7) that were shown to be functionally benign.
These observations are indicative of a genetic-predisposing effect of common polymorphisms (1) in causatively contributing to an increased potential of ADAMTS13 deficiency and (2) perhaps in increasing the risk of thrombotic diseases. They also support the importance of including SNPs into the risk management of disease susceptibility.
Prepublished online as Blood First Edition Paper, September 13, 2005; DOI 10.1182/blood-2005-06-2482.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Elise Langdon-Neuner for expert editorial assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal