We analyzed 2502 patients with acute myeloid leukemia at diagnosis for NRAS mutations around the hot spots at codons 12, 13, and 61 and correlated the results to cytomorphology, cytogenetics, other molecular markers, and prognostic relevance of these mutations. Two hundred fifty-seven (10.3%) of 2502 patients had NRAS mutations (NRASmut). Most mutations (112 of 257; 43.6%) were found at codon 12, mostly resulting in changes from glycine to asparagine. The history of AML did not differ significantly in association with NRAS mutations. The subgroups with inv(16)/t(16;16) and inv(3)/t(3;3) showed a significantly higher frequency of NRASmut (50 of 133, 37.6% [P < .001], and 11 of 41, 26.8% [P = .004], respectively) than the total cohort. In addition, in these 2 subgroups, mutations of codon 61 were significantly overrepresented (both P < .001). In contrast, NRAS mutations were significantly underrepresented in t(15;17) (2 of 102; 2%; P = .005) in the subgroup with MLL/11q23 rearrangements (3 of 77; 3.9%; P = .061) and in the complex aberrant karyotype (4 of 258; 1.6%; P < .001). Overall, we did not find a significant prognostic impact of NRASmut for overall survival, event-free survival, and disease-free survival. However, there was a trend to better survival in most subgroups, especially when other molecular markers (FLT3-LM, MLL-PTD, and NPM) were taken into account.
Introduction
Acute myelogenous leukemia (AML) is a clinically and molecularly heterogeneous disease.1 The WHO classification2 subdivides AML predominantly according to karyotype because recurrent chromosomal abnormalities identify distinct leukemia entities and have a major impact on prognosis. In addition, molecular analyses indicate that many types of AML have cytogenetically undetectable mutations that have an impact on prognosis in different cytogenetic subgroups. Some of these mutations affect genes encoding for transcription factors (AML1, CEBPA) (2%-3% and 5%-15% of patients, respectively),3-5 and others affect the receptor tyrosine kinases FLT3 and KIT (25%-30% and 1% of patients, respectively)6-9 or have partial tandem duplications within the MLL gene (MLL-PTD), which has many different functions (5%-10% of patients).10-12 For some molecular markers, such as FLT3 length mutation (FLT3-LM),6-8 and for partial tandem duplication within the MLL gene,10,11 the unfavorable prognostic impact in AML is well known. In contrast, mutations in CEBPA and NPM are of favorable prognostic impact.4,11,13,14 In addition, RAS genes are frequently affected by mutations in AML (10%-15% of patients),15-19 but thus far the prognostic impact is still under discussion.
RAS oncogenes encode a family of membrane-associated proteins that regulate signal transduction on binding to a variety of membrane receptors. They play important roles in the regulatory processes of proliferation, differentiation, and apoptosis.17 There are 3 functional RAS genes—NRAS, KRAS, and HRAS—and all homologs carry mutations nearly exclusively in codons 12, 13, and 61, conferring constitutive activation of the RAS protein, which subsequently is held in GTP-bound status. These mutations have been described in various solid tumors and in hematologic malignancies.16,20 NRAS mutations seem to be the most prominent RAS mutations in patients with AML and have been reported in 11% to 30% of patients.15-22 They lead to increased activity of the RAS pathway, resulting in increased proliferation and decreased apoptosis rates.18 Activation of the RAS pathway can also be mediated by mutations of other more upstream genes.18,20
Although in AML NRAS mutations were first reported 18 years ago,23,24 the prognostic impact of NRAS mutations is still under discussion and seems to vary from disease to disease.16-19,22,25-26 Several studies indicate an association with poor outcome,25,26 but Kiyoi et al16 found a negative prognostic impact of NRAS mutations only in AML with favorable karyotype. In contrast, Neubauer et al15 found NRAS mutations associated with a favorable prognosis, whereas in some studies a prognostic impact of NRAS mutations could not be defined at all.18,19,21 In addition, whether NRAS mutations are associated with specific morphology, karyotypes, or other molecular markers in AML must still be determined. Some studies did not find a significant association of NRAS mutations with certain cytogenetic subgroups.15,17 In contrast, others found a higher percentage of AML M4 subtypes in patients with NRAS15,21 or even demonstrated an association especially with AML M4eo/inv(16).27,28 However, many historical data sets are of insufficient size to distinguish prognostic differences or to show correlations to certain biologic subgroups. To further clarify the biologic and prognostic impact of NRAS mutations, we analyzed a large cohort of 2502 patients with AML who were well characterized with respect to cytomorphology, cytogenetics, and other molecular mutations.
Patients, materials, and methods
Patients
Included in this study were 2502 patients with AML who were between 18.3 and 91.8 years of age (median age, 63.4 years) at diagnosis, and who were screened for NRAS mutations. Patient samples were referred for investigation between January 1998 and March 2005. All patients gave informed consent before entering the study. The study design adhered to the principles of the Declaration of Helsinki and was approved by the ethics committees of the participating institutions.
Samples were evaluated by cytomorphology, cytochemistry, multiparameter flow cytometry, cytogenetics, fluorescence in situ hybridization (FISH), and molecular genetics in parallel.7,29-31 Table 1 shows the characteristics of the patients. The cytomorphologic classification of AML was performed according to the French-American-British (FAB) classification.32,33
Characterization of the patient cohort (n = 2502 patients)
. | No. (%) . |
|---|---|
| Type of AML | |
| De novo | 2037 (81.4) |
| s-AML | 281 (11.2) |
| t-AML | 184 (7.4) |
| Total | 2502 (100.0) |
| Cytogenetic subgroup | |
| Normal karyotype | 1198 (48.5) |
| t(15;17) | 102 (4.1) |
| t(8;21) | 132 (5.3) |
| inv(16) | 133 (5.4) |
| 11q23/MLL | 77 (3.1) |
| inv(3)/t(3;3) | 41 (1.7) |
| Complex karyotype,37 typical | 258 (10.4) |
| Complex karyotype,37 atypical | 56 (2.3) |
| Trisomy 8 | 122 (4.9) |
| 5q-/-5 | 24 (1.0) |
| 7q-/-7 | 58 (2.3) |
| Other aberrations | 301 (12.2) |
| Total | 2502 (100.0) |
| FAB subtype | |
| M0 | 94 (4.6) |
| M1 | 365 (17.9) |
| M2 | 750 (36.8) |
| M3 | 62 (3.0) |
| M3v | 32 (1.6) |
| M4 | 343 (16.8) |
| M4eo | 116 (5.7) |
| M5a | 77 (3.7) |
| M5b | 81 (4.0) |
| M6 | 84 (4.1) |
| M7 | 8 (0.4) |
| Not classifiable | 26 (1.3) |
| Total | 2038 (100.0) |
. | No. (%) . |
|---|---|
| Type of AML | |
| De novo | 2037 (81.4) |
| s-AML | 281 (11.2) |
| t-AML | 184 (7.4) |
| Total | 2502 (100.0) |
| Cytogenetic subgroup | |
| Normal karyotype | 1198 (48.5) |
| t(15;17) | 102 (4.1) |
| t(8;21) | 132 (5.3) |
| inv(16) | 133 (5.4) |
| 11q23/MLL | 77 (3.1) |
| inv(3)/t(3;3) | 41 (1.7) |
| Complex karyotype,37 typical | 258 (10.4) |
| Complex karyotype,37 atypical | 56 (2.3) |
| Trisomy 8 | 122 (4.9) |
| 5q-/-5 | 24 (1.0) |
| 7q-/-7 | 58 (2.3) |
| Other aberrations | 301 (12.2) |
| Total | 2502 (100.0) |
| FAB subtype | |
| M0 | 94 (4.6) |
| M1 | 365 (17.9) |
| M2 | 750 (36.8) |
| M3 | 62 (3.0) |
| M3v | 32 (1.6) |
| M4 | 343 (16.8) |
| M4eo | 116 (5.7) |
| M5a | 77 (3.7) |
| M5b | 81 (4.0) |
| M6 | 84 (4.1) |
| M7 | 8 (0.4) |
| Not classifiable | 26 (1.3) |
| Total | 2038 (100.0) |
Two thousand thirty-seven patients had diagnoses of de novo AML, 281 patients had secondary AML (s-AML) following myelodysplastic syndrome (MDS), and 184 patients had therapy-related AML (t-AML). One hundred seven patients were further compared with respect to NRAS mutation status at diagnosis and at relapse.
Mutation analysis
Mononucleated cells were isolated by standard Ficoll density gradient centrifugation. Nucleic acid isolation, cDNA synthesis, and screening for NRAS mutations using a melting curve–based LightCycler (Roche Applied Science, Indianapolis, IN) assay was performed as described.7,34 All positive results for NRAS mutations were confirmed by sequence analysis. Approximately 100 ng purified polymerase chain reaction (PCR) products were directly sequenced with 3.3 pmol of each forward and reverse primer using the Big Dye Terminator Cycle Sequencing Kit (Applera, Darmstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and at 60°C for 4 minutes were performed. Sequence analysis was performed on an ABI 3100 Avant sequence detection system (Applied Biosystems, Foster City, CA).
With this assay, clone levels as low as 2% of the entire population were detectable as estimated by limited-dilution experiments. Patients with 2% to 10% of mutated cells could not be sequenced because the sensitivity of direct sequencing was only up to around 10%.
Statistical analysis
Overall survival (OS) and event-free survival (EFS) were determined according to the Kaplan-Meier method.35 The correlation of OS and EFS with other parameters was assessed by Cox regression analysis. Survival curves were compared using the double-sided log rank test. Comparisons of dichotomous variables between different groups were performed with the use of 2-sided Fisher exact test. For statistical analysis, SPSS (version 12.4) software (SPSS, Chicago, IL) was used.
Results
Frequency of NRAS mutations in AML samples
A total of 2502 AML patients were screened for NRAS mutations around the mutational hotspots at codons 12, 13, and 61. Of all 2502 patients, 257 (10.3%) had NRAS mutations (NRASmut), whereas 2245 had only the NRAS wild-type allele (NRASwt). In 3 (1.2%) of the 257 patients with mutations, 2 different mutations were detected (compound heterozygote). None of the patients with mutations was homozygous for the mutation. This could be assessed by the melting curve assay, which clearly showed 2 peaks in each patient: a peak at 65°C (indicating the wild-type allele) and a peak at 56°C (indicating the mutation). Neither the loss of the wild-type peak nor a shift to higher intensity of the wild-type peak was observed in any patient. Thus, uniparental disomy, as described in other AML-specific mutations such as FLT3-LM and CEBPA, could be excluded.36
Characterization of NRAS mutations
Most mutations (112 of 257; 43.6%) were found at codon 12 (Table 2). Mutations at codon 13 were found in 54 (21%) of 257 patients. In both cohorts, the most frequent changes were from glycine to asparagine (codon 12: 76 of 112, 29.6%; codon 13: 41 of 54, 16%) (Table 3). Mutations at codon 61 were detected in 64 (24.9%) of 257 patients, mostly from glycine to arginine. A subset of 27 mutations remained uncharacterized because they were detected by LightCycler (Roche Applied Science) assay in a subclone of less than 10%, which was below the resolution of direct sequencing.
Distribution of mutations in codons 12, 13, and 61 in all patients with NRAS mutations and in patients with inv(16)/t(16;16) and inv(3)/t(3;3)
Codon . | All NRAS mutations, no. (%) . | NRASmut inv(16)/t(16;16), no. (%) . | NRASmut inv(3)/t(3;3), no. (%) . |
|---|---|---|---|
| 12 | 112 (43.6) | 14 (28.0) | 4 (36.4) |
| 13 | 54 (21.0) | 10 (20.0) | 1 (9.1) |
| 61 | 64 (24.9) | 21 (42.0) | 6 (54.5) |
| Not sequenced | 27 (10.5) | 5 (10.0) | 0 (0.0) |
| Total | 257 (100.0) | 50 (100.0) | 11 (100.0) |
Codon . | All NRAS mutations, no. (%) . | NRASmut inv(16)/t(16;16), no. (%) . | NRASmut inv(3)/t(3;3), no. (%) . |
|---|---|---|---|
| 12 | 112 (43.6) | 14 (28.0) | 4 (36.4) |
| 13 | 54 (21.0) | 10 (20.0) | 1 (9.1) |
| 61 | 64 (24.9) | 21 (42.0) | 6 (54.5) |
| Not sequenced | 27 (10.5) | 5 (10.0) | 0 (0.0) |
| Total | 257 (100.0) | 50 (100.0) | 11 (100.0) |
Frequency of involvement of codon 61 in comparison with involvement of codons 12 and 13, P < .001.
Mutation analysis of all patients with NRAS mutations compared with patients with inv(16) and other NRAS mutations
. | All NRAS mutations, no. (%) . | NRASmut with inv(16), no. (%) . | All other NRAS mutations, no. (%) . |
|---|---|---|---|
| Codon 12 | |||
| Gly > Ala | 7 (2.7) | 1 (2.0) | 6 (2.9) |
| Gly > Arg | 2 (0.8) | 0 (0.0) | 2 (1.0) |
| Gly > Asp | 76 (29.6) | 11 (22.0) | 65 (31.4) |
| Gly > Cys | 10 (3.9) | 2 (4.0) | 8 (3.9) |
| Gly > Phe | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gly > Ser | 12 (4.7) | 0 (0.0) | 12 (5.8) |
| Gly > Val | 4 (1.6) | 0 (0.0) | 4 (1.9) |
| Total | 112 (43.6) | 14 (28.0) | 98 (47.3) |
| Codon 13 | |||
| Gly > Ala | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gly > Arg | 5 (1.9) | 2 (4.0) | 3 (1.4) |
| Gly > Asp | 41 (16.0) | 7 (14.0) | 34 (16.4) |
| Gly > Cys | 2 (0.8) | 0 (0.0) | 2 (1.0) |
| Gly > Val | 5 (1.9) | 1 (2.0) | 4 (1.9) |
| Total | 54 (21.0) | 10 (20.0) | 44 (21.3) |
| Codon 61 | |||
| Gln > Arg | 26 (10.1) | 7 (14.0) | 19 (9.2) |
| Gln > Glu | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gln > His | 5 (1.9) | 1 (2.0) | 4 (1.9) |
| Gln > Leu | 9 (3.5) | 0 (0.0) | 9 (4.3) |
| Gln > Lys | 23 (8.9) | 13 (26.0) | 10 (4.8) |
| Total | 64 (24.9) | 21 (42.0) | 43 (20.8) |
| Not sequenced | 27 (10.5) | 5 (10.0) | 22 (10.6) |
| Overall total | 257 (100.0) | 50 (100.0) | 207 (100.0) |
. | All NRAS mutations, no. (%) . | NRASmut with inv(16), no. (%) . | All other NRAS mutations, no. (%) . |
|---|---|---|---|
| Codon 12 | |||
| Gly > Ala | 7 (2.7) | 1 (2.0) | 6 (2.9) |
| Gly > Arg | 2 (0.8) | 0 (0.0) | 2 (1.0) |
| Gly > Asp | 76 (29.6) | 11 (22.0) | 65 (31.4) |
| Gly > Cys | 10 (3.9) | 2 (4.0) | 8 (3.9) |
| Gly > Phe | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gly > Ser | 12 (4.7) | 0 (0.0) | 12 (5.8) |
| Gly > Val | 4 (1.6) | 0 (0.0) | 4 (1.9) |
| Total | 112 (43.6) | 14 (28.0) | 98 (47.3) |
| Codon 13 | |||
| Gly > Ala | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gly > Arg | 5 (1.9) | 2 (4.0) | 3 (1.4) |
| Gly > Asp | 41 (16.0) | 7 (14.0) | 34 (16.4) |
| Gly > Cys | 2 (0.8) | 0 (0.0) | 2 (1.0) |
| Gly > Val | 5 (1.9) | 1 (2.0) | 4 (1.9) |
| Total | 54 (21.0) | 10 (20.0) | 44 (21.3) |
| Codon 61 | |||
| Gln > Arg | 26 (10.1) | 7 (14.0) | 19 (9.2) |
| Gln > Glu | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Gln > His | 5 (1.9) | 1 (2.0) | 4 (1.9) |
| Gln > Leu | 9 (3.5) | 0 (0.0) | 9 (4.3) |
| Gln > Lys | 23 (8.9) | 13 (26.0) | 10 (4.8) |
| Total | 64 (24.9) | 21 (42.0) | 43 (20.8) |
| Not sequenced | 27 (10.5) | 5 (10.0) | 22 (10.6) |
| Overall total | 257 (100.0) | 50 (100.0) | 207 (100.0) |
Correlation with the history of AML
We also analyzed the distribution of NRAS mutations in patients with de novo AML, s-AML, and t-AML. NRAS mutations developed in 10% (203 of 2037) of all patients with de novo AML, 12.1% (34 of 281) of all patients with s-AML, and 10.9% (20 of 184) of all patients with t-AML. Thus, the history of AML did not differ significantly with regard to NRAS mutations.
Comparison of biologic parameters of patients with NRASmut and NRASwt
Median age of the patients with NRASmut was 63.3 years (range, 22.8-90.2 years). Median age of the patients with NRASwt was 63.4 years (range, 18.3-91.8 years). Thus, age did not differ significantly between the 2 cohorts.
The median leukocyte count was 28.0 × 109/L (range, 0.4-514.0 G/L) in the patients with NRAS mutations and 63.4 × 109/L in the patients without NRAS mutations (range, 18.3-91.8 × 109/L) (P < .001). In the subgroup with inv(16), the median leukocyte count was lower in patients with mutations (NRASmut, 30 × 109/L [range, 4.3-169 × 109/L]; NRAS wt, 47.5 × 109/L [range, 1.2-250.0 × 109/L]).
Correlation of NRASmut with cytomorphology
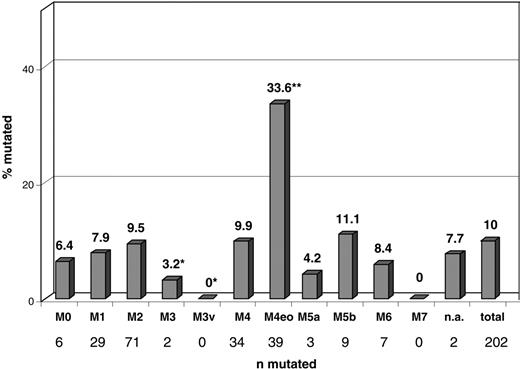
Cytomorphology findings were available for 2038 patients. Of these, 202 (10%) had NRAS mutations. In the FAB subtype M4eo, NRAS mutations were represented more frequently (33.6%, 39 of 116) than they were in all other subtypes (P < .001). In the M3v subtype (n = 32), no NRASmut was detected, making NRAS mutations highly underrepresented in this subtype (P = .058). In all other FAB subtypes, the distribution of NRAS mutations did not differ from that of the total cohort. A detailed distribution of NRAS mutations in the respective FAB subtypes is presented in Figure 1.
Correlation of NRASmut with cytogenetics
Cytogenetic data were available from all analyzed patients (n = 2502). Patients were grouped into 12 categories as follows: normal karyotype, t(15;17)/PML-RARA, t(8;21)/AML1-ETO, inv(3)/t(3;3), inv(16)/t(16;16)/CBFB-MYH11, 11q23/MLL, typical and atypical complex aberrant karyotype,37 trisomy 8, 5q– or loss of chromosome 5, 7q– or loss of chromosome 7, and so-called other aberrations.
Complex aberrant karyotype was defined by 3 or more chromosomal abnormalities. This subgroup was further divided into complex typical karyotype, which was defined by loss of at least one of the chromosomal regions of 5q, 7q, or 17p, and by loss of at least one additional area of the regions 18q21-22, 12p13, or 16q22-24 or by gain of 11q23-25, 1p33-36, 8q22-24, or 21q11-22. Complex atypical karyotype was defined by 3 or more chromosomal abnormalities without these characteristic chromosomal lesions.37
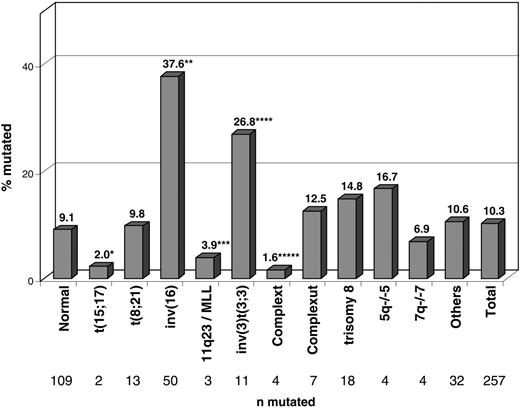
Figure 2 shows the frequency of NRAS mutations in the respective cytogenetic subgroups. In the cytogenetic subgroups with inv(16)/t(16;16) and inv(3)/t(3;3), NRAS mutations occurred at higher frequencies—37.6% (50 of 133; P < .001) and 26.8% (11 of 41; P = .004)—than in all others. In contrast, NRAS mutations were significantly underrepresented in the groups with t(15;17) (2%; 2 of 102; P = .005), 11q23/MLL (3.9%; 3 of 77; P = .061), and complex aberrant karyotype (1.6%; 4 of 258; P < .001).
In accordance with Bowen,19 we further explored the frequency of NRAS mutations in patients with t(3;5). In total, 14 patients were found with this specific aberration—13 had the complex aberrant karyotype and 1 had an isolated translocation. None of these 14 patients was NRAS mutated.
Mutation analysis in AML with inv(16)
We compared the involved codons in the 50 patients with inv(16)/t(16;16) and NRASmut and in the 11 patients with inv(3)/t(3;3) with the 257 patients with NRASmut (Table 2). Although mutations of codon 12 were found in 112 (43.6%) of 257 of all patients with mutations, they were significantly underrepresented in patients with inv(16) (14 of 50; 28%). In the subgroup with inv(3), mutations of codon 12 accounted for 4 (36.4%) of 11, respectively. The percentage of mutations of codon 13 was similar in the patients with inv(16) and in all patients (10 of 50 [20%] and 54 of 257 [21%], respectively). The inv(3) group showed codon 13 mutations in 9.1% (1 of 11) of all patients with mutations. Mutations of codon 61 were detected in 64 (24.9%) of 257 patients and thus were significantly more frequent in patients with inv(16) and with inv(3) (21 of 50; 42%, and 6 of 11; 54.5%, respectively) (P < .001).
Distribution of NRAS mutations within morphologic subgroups. Significant differences compared with the total cohort are labeled with asterisks (*underrepresentation; **overrepresentation). AML subtypes: M0, 6 of 94 (6.4%); M1, 29 of 365 (7.9%); M2, 71 of 750 (9.5%); M3, 2 of 62 (3.2%); M3v, 0 of 32 (0%) (P = .058); M4, 34 of 343 (9.9%); M4eo, 39 of 116 (33.6%) (P < .001); M5a, 3 of 77 (4.2%); M5b, 9 of 81 (11.1%); M6, 7 of 84 (8.3%); M7, 0 of 8 (0%); n.a. (no subtype documented), 2 of 26 (7.7%); total, 203 of 2037 (10%).
Distribution of NRAS mutations within morphologic subgroups. Significant differences compared with the total cohort are labeled with asterisks (*underrepresentation; **overrepresentation). AML subtypes: M0, 6 of 94 (6.4%); M1, 29 of 365 (7.9%); M2, 71 of 750 (9.5%); M3, 2 of 62 (3.2%); M3v, 0 of 32 (0%) (P = .058); M4, 34 of 343 (9.9%); M4eo, 39 of 116 (33.6%) (P < .001); M5a, 3 of 77 (4.2%); M5b, 9 of 81 (11.1%); M6, 7 of 84 (8.3%); M7, 0 of 8 (0%); n.a. (no subtype documented), 2 of 26 (7.7%); total, 203 of 2037 (10%).
Distribution of the 257 patients with NRAS mutations within cytogenetic subgroups. Significant differences compared with the cohort of 2245 patients with NRASwt and available cytogenetics are labeled with asterisks. Normal karyotype, 109 of 1198 (9.1%); t(15;17), 2 of 102 (2%) (*P = .005); t(8;21), 13 of 132 (9.8%); inv(16), 50 of 133 (37.6%) (**P < .001); 11q23/MLL, 3 of 77 (3.9%) (***P = .062); inv(3)t(3;3), 11 of 41 (26.8%) (****P = .001); complex typical karyotype37 , 4 of 258 (1.6%) (*****P = .001); complex atypical karyotype37 , 7 of 56 (12.5%); +8, 18 of 122 (14.8%); 5q–/–5, 4 of 24 (16.7%); 7q–/–7, 4 of 58 (6.9%); other aberrations, 32 of 301 (10.6%); total, 257 of 2502 (10.3%).
Distribution of the 257 patients with NRAS mutations within cytogenetic subgroups. Significant differences compared with the cohort of 2245 patients with NRASwt and available cytogenetics are labeled with asterisks. Normal karyotype, 109 of 1198 (9.1%); t(15;17), 2 of 102 (2%) (*P = .005); t(8;21), 13 of 132 (9.8%); inv(16), 50 of 133 (37.6%) (**P < .001); 11q23/MLL, 3 of 77 (3.9%) (***P = .062); inv(3)t(3;3), 11 of 41 (26.8%) (****P = .001); complex typical karyotype37 , 4 of 258 (1.6%) (*****P = .001); complex atypical karyotype37 , 7 of 56 (12.5%); +8, 18 of 122 (14.8%); 5q–/–5, 4 of 24 (16.7%); 7q–/–7, 4 of 58 (6.9%); other aberrations, 32 of 301 (10.6%); total, 257 of 2502 (10.3%).
Analysis of paired samples at diagnosis and at relapse
We compared NRAS mutation status at diagnosis and at relapse in 107 patients (Table 4). Overall, when taking all patients into account, including those who were negative at diagnosis, NRAS mutation status was constant in 102 of 107 (95.3%) patients at both time points. However, of 8 patients with NRAS mutations at diagnosis, 4 had lost the mutation at relapse. In 1 patient, the mutation was gained at relapse. From these data, we conclude that NRAS mutations are not good markers for follow-up control.
Analysis of NRAS mutation status in 107 paired samples at diagnosis and at relapse
. | Status . | . | . | . | |
|---|---|---|---|---|---|
| Diagnosis . | Relapse . | No. (%) . | Constant . | No. (%) . | |
| Positive | Positive | 4 (3.7) | Yes | 4 (3.7) | |
| Positive | Negative | 4 (3.7) | No | NA | |
| Negative | Positive | 1 (0.9) | No | NA | |
| Negative | Negative | 98 (91.6) | Yes | 98 (91.6) | |
| Total | NA | 107 (100.0) | NA | 102 (95.3) | |
. | Status . | . | . | . | |
|---|---|---|---|---|---|
| Diagnosis . | Relapse . | No. (%) . | Constant . | No. (%) . | |
| Positive | Positive | 4 (3.7) | Yes | 4 (3.7) | |
| Positive | Negative | 4 (3.7) | No | NA | |
| Negative | Positive | 1 (0.9) | No | NA | |
| Negative | Negative | 98 (91.6) | Yes | 98 (91.6) | |
| Total | NA | 107 (100.0) | NA | 102 (95.3) | |
NA indicates not applicable.
Analysis of prognostic value
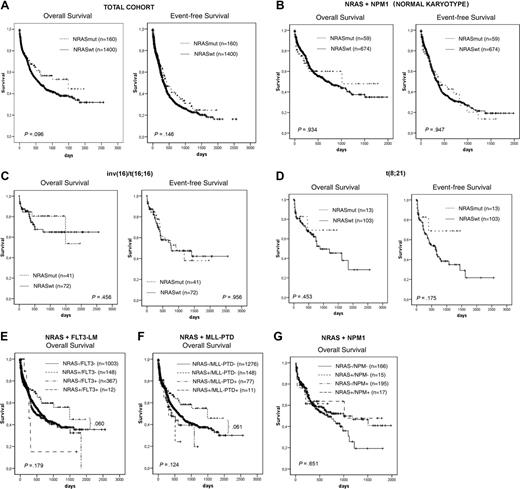
Based on the previous findings, we analyzed the influence of NRAS mutations on prognosis in 1560 patients for whom clinical follow-up data were available. In the total group, there was no difference with regard to complete remission (CR) rate (NRASmut, 60.6%; NRASwt, 55.7%; P = .34) or to OS (1460 vs 604 days; P = .096) and EFS (424 vs 335 days, P = .146) (Figure 3A). In the detailed analyses within single cytogenetic subgroups with inv(16) inv(3), t(8;21), normal karyotype, and other aberrations, we found no statistically significant impact of NRAS mutations on these 3 end points (Table 5).
CR, OS, and EFS in 1560 patients for whom clinical data are available
. | Total no. patients . | No. patients . | . | CR . | . | . | Median OS . | . | . | Median EFS . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | NRASmut . | NRASWT . | NRASmut, % . | NRASWT, % . | P . | NRASmut, d . | NRASWT, d . | P . | NRASmut, d . | NRASWT, d . | P . | |||||||
| All patients | 1560 | 160 | 1400 | 60.6 | 55.7 | .340 | 1490 | 604 | .096 | 424 | 335 | .146 | |||||||
| Normal karyotype | 733 | 59 | 674 | 65.9 | 64.2 | .620 | 1012 | 737 | .934 | 368 | 352 | .947 | |||||||
| All intermediate karyotypes | 299 | 27 | 272 | 62.7 | 60.9 | .748 | 310 | 436 | .672 | 187 | 303 | .138 | |||||||
| inv(16) | 113 | 41 | 72 | 66.7 | 71.8 | .429 | NR | NR | .456 | 918 | 769 | .956 | |||||||
| inv(3) | 20 | 4 | 16 | NA | NA | NA | 100 | 752 | .320 | 100 | 296 | .835 | |||||||
| t(8;21) | 116 | 13 | 103 | 80 | 62.8 | .250 | NR | 882 | .453 | NR | 648 | .175 | |||||||
. | Total no. patients . | No. patients . | . | CR . | . | . | Median OS . | . | . | Median EFS . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | NRASmut . | NRASWT . | NRASmut, % . | NRASWT, % . | P . | NRASmut, d . | NRASWT, d . | P . | NRASmut, d . | NRASWT, d . | P . | |||||||
| All patients | 1560 | 160 | 1400 | 60.6 | 55.7 | .340 | 1490 | 604 | .096 | 424 | 335 | .146 | |||||||
| Normal karyotype | 733 | 59 | 674 | 65.9 | 64.2 | .620 | 1012 | 737 | .934 | 368 | 352 | .947 | |||||||
| All intermediate karyotypes | 299 | 27 | 272 | 62.7 | 60.9 | .748 | 310 | 436 | .672 | 187 | 303 | .138 | |||||||
| inv(16) | 113 | 41 | 72 | 66.7 | 71.8 | .429 | NR | NR | .456 | 918 | 769 | .956 | |||||||
| inv(3) | 20 | 4 | 16 | NA | NA | NA | 100 | 752 | .320 | 100 | 296 | .835 | |||||||
| t(8;21) | 116 | 13 | 103 | 80 | 62.8 | .250 | NR | 882 | .453 | NR | 648 | .175 | |||||||
NR indicates not reached; NA, not available.
However, detailed analyses of Kaplan-Meier plots in the NRASmut cohort showed a trend to reach a plateau after 1 year of follow-up in normal karyotype and inv(16) according to OS analysis and in t(8;21) according to OS and EFS analyses (Figure 3B-D). The plateau is disturbed by only one event each.
In further analysis, we focused on the association with other molecular markers. Taking other molecular markers (FLT3-LM, MLL-PTD, NPM1) into account in this group indicated an even better trend for a more favorable prognosis (Figure 3E-G).
In the total cohort without FLT3 mutations, EFS was slightly improved for patients with NRAS mutations (426 ± 73 days) compared with patients negative for NRAS mutations (378 ± 21 days) (P = .060).
Discussion
Characterizing molecular mutations in AML is of growing importance for better understanding of the biology of the disease, better risk assessment for individual patients, and better detection of minimal residual disease (MRD). NRAS mutations occur at high incidence in patients with AML and were first reported 18 years ago.23,24 However, whether they influence prognosis in patients with AML is not been determined. We analyzed 2502 AML patients for NRAS mutations and performed analyses with respect to cytomorphology, cytogenetics, and other biologic aspects and addressed their prognostic relevance.
In the total cohort, NRAS mutations were found in 10.3% (257 of 2502) of patients. This corresponded to findings in the literature that show a range of 11.4% to 13.9% NRAS mutations.15-19 We did not observe different proportions of NRAS mutations in de novo, secondary, or therapy-associated AML. This was in contrast to the results of Illmer et al,18 who found a higher percentage of NRAS mutations in patients with s-AML (43%) and t-AML (75%) than in those with de novo AML (17%).
The highest frequency (37.6%) of NRAS mutations in our cohort compared with the total cohort was detected in patients with inv(16)/t(16;16) (P < .001). The high incidence of NRAS mutations in inv(16) in our study corresponded with most of the previously published studies reporting frequencies of 26% to 33%.18,28,38 Only Panagopoulos et al39 found NRAS mutations in a small subset of 1 of 8 patients with inv(16), perhaps because of the small case numbers.
Surprisingly, the NRAS mutation rate was also increased in patients with inv(3)/t(3;3), another subtype of AML with an inversion and reciprocal translocation of homologous chromosomes. For the first time, our study could show a cooperative mutation for the 3q21q26 leukemia subtype. It may be speculated that a common yet unknown mechanism underlies these NRAS mutations and the formation of inversions and reciprocal translocations of homologous chromosomes. We found a different distribution of codon involvement in patients with inv(16)/t(16;16) and inv(3)/t(3;3) than in all other patients with NRASmut. In patients with inv(16)/t(16;16) and inv(3)/t(3;3), codon 61 was significantly more frequently mutated than it was in all other patients, who most frequently showed involvement of codon 12 (P < .001). The different mutation patterns suggest that different types of NRAS mutations might cooperate specifically better with certain primary mutations in leukemogenesis. Alternatively, the mechanisms leading to inv(16)/t(16;16) and inv(3)/t(3;3) may better trigger mutations in codon 61 than in codon 12 or 13. Thus, common mutational mechanisms might exist in inv(16)/t(16;16) and in inv(3)/t(3;3).
Kaplan-Meier analysis. Total cohort (A) in different cytogenetic (B-D) and molecular genetic (E-G) subgroups.
Kaplan-Meier analysis. Total cohort (A) in different cytogenetic (B-D) and molecular genetic (E-G) subgroups.
In contrast, in t(15;17) or in complex aberrant karyotypes, NRAS mutations were underrepresented. It is well known that FLT3-LM is a highly frequent accompanying mutation in patients with acute promyelocytic leukemia (APL)40,41 and that patients with t(8;21) tend to have KIT mutations.9,42 This also supports the association of certain cytogenetic alterations with molecular mutations. According to the 2-hit hypothesis of Knudson et al and Gilliland et al,43-45 leukemogenesis in AML is the result of 2 genetic events: mutations of class I, which mediate a proliferative signal, and mutations of class II, which impair hematopoietic differentiation. CBFB gene fusion is supposed to act as a class II mutation.44 It was demonstrated in animal models that CBFB gene fusion is insufficient to induce leukemogenesis without the interaction of other genetic events.28,46 NRAS mutations are supposed to function as class I mutations according to this model and, according to our results, seem to be the optimal cooperators of inv(16)/t(16;16) and inv(3)/t(3;3).
In the study of Bowen et al,19 there was an association between KRAS mutations and t(3;5), with 62.5% of the patients with this karyotype positive for this aberration. We did not find a prevalence for NRAS in our 14 patients with t(3;5), all of whom were negative for NRAS mutations. However, 13 of 14 of our patients with t(3;5) had complex aberrant karyotypes, a subgroup in which we found an overall underrepresentation of NRAS mutations.
We further correlated FAB subtypes and NRAS mutations. As suspected, patients with AML M4eo had a significantly higher frequency (33.6%) of NRAS mutations than did the entire patient cohort (10%) (P < .001). In contrast, we did not find a significant correlation of AML M4 with NRAS mutations. Our results were in accordance with those of Illmer et al,18 who also did not find an association between NRAS mutations and the FAB subtype M4 (14.8% in the cohort with NRASmut vs 16.2% in patients with NRASwt). Others found an association between NRAS mutations and the FAB subtype M421 or between KRAS and AML M419 ; however, in these studies, M4 and M4eo were not analyzed separately, and the high percentage may thus depend on M4eo patients.
NRAS mutations were significantly associated with lower peripheral leukocyte counts, in accordance with findings by Kiyoi et al.16 The same was true in our study for the subgroup with the inv(16) mutation.
In our study, NRAS mutations did not influence prognosis significantly. This was true for all patients and especially for the subgroups with normal karyotype, inv(16), and t(8;21). These results were in agreement with those of others,18,19,21 who also were unable to establish a clear prognostic impact of NRAS mutations in AML. However, when looking at the Kaplan-Meier plots in detail, there was a trend in the NRASmut groups to reach a plateau after 1 year.
In total, our results were in contrast to those of Kiyoi et al,16 who found NRAS mutations to represent an unfavorable prognostic parameter in patients with AML with favorable karyotype. AML with intermediate or unfavorable karyotype prognosis was not influenced by NRAS mutations. In contrast, our study showed a slightly improved prognostic impact in the subgroup with normal karyotype without FLT3 mutations in comparison with patients with NRAS wild-type (P = .060). In summary, it must be stated that most studies were relatively small and that large cohorts and other cytogenetic and molecular aberrations must be considered to definitively answer the question of prognostic impact of NRAS mutations.
However, in the study of Ilmer et al,18 increased NRAS activity, even in the absence of known NRAS mutations, was associated with improved prognosis in patients younger than 61 years receiving high-dose cytarabine, indicating that NRAS activity occurs regardless of direct NRAS mutation or that alternative activating mechanisms are more important than the NRAS mutation itself. It may be speculated that highly redundant mechanisms induce NRAS activation, leading to undetectable impact on prognosis in the NRASmut patients.
In conclusion, we found a clear pattern of NRAS mutation in defined subgroups of patients with AML. For the first time, an association between NRASmut and inv(3)/t(3;3) was detected in addition to the high frequency in the inv(16)/(t16;16) subgroup. This supports the hypothesis of a 2-hit mutation model in AML. Overall the prognostic impact of NRAS mutations was weak and not significant in most AML subgroups. Only in the subgroup with normal karyotype and nonmutated FLT3 status did NRAS mutations account for an improved prognosis. However, because of the lack of clear prognostic value of the NRAS mutations, mutation analysis for this gene cannot be recommended for inclusion within the routine diagnostic setting.
Prepublished online as Blood First Edition Paper, January 24, 2006; DOI 10.1182/blood-2005-08-3522.
Supported in part by a grant from the NGFN (BMBF Nationales Genomforschungsnetz Förderkennzeichen 01GSS0105-Teilprojekt 2).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Gudrun Mellert, Claudia Tschulik, Madlen Fuchs, Nina Leopold, and Theresa Förster for their excellent technical work.
We would like to thank all participants of the AMLCG study group for sending bone marrow or blood samples to our laboratory for reference diagnosis, and for submitting clinical data, as part of the patients were treated within the AMLCG study.
Prof Dr med W. Hiddemann serves as head of the Department of Internal Medicine III of the Ludwig-Maximilians University, Munich, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal