Mutations of the nucleophosmin (NPM1) gene have recently been described in patients with acute myeloid leukemia (AML). To clarify the prevalence as well as the clinical impact of this mutation, we investigated 1485 patients with AML for NPM1 exon 12 mutations using fragment analysis. A 4 bp insert was detected in 408 of 1485 patients (27.5%). Sequence analysis revealed known mutations (type A, B, and D) as well as 13 novel alterations in 229 analyzed cases. NPM1 mutations were most prevalent in patients with normal karyotype (NK) (324 of 709; 45.7%) compared with 58 of 686 with karyotype abnormalities (8.5%; P < .001) and were significantly associated with several clinical parameters (high bone marrow [BM] blasts, high white blood cell [WBC] and platelet counts, female sex). NPM1 alterations were associated with FLT3-ITD mutations, even if restricted to patients with NK (NPM1-mut/FLT3-ITD: 43.8%; versus NPM1-wt/FLT3-ITD: 19.9%; P < .001). The analysis of the clinical impact in 4 groups (NPM1 and FLT3-ITD single mutants, double mutants, and wild-type [wt] for both) revealed that patients having only an NPM1 mutation had a significantly better overall and disease-free survival and a lower cumulative incidence of relapse. In conclusion, NPM1 mutations represent a common genetic abnormality in adult AML. If not associated with FLT3-ITD mutations, mutant NPM1 appears to identify patients with improved response toward treatment.
Introduction
Acute myeloid leukemia (AML) describes a group of hematopoietic stem cell disorders characterized by the expansion of undifferentiated myeloid progenitors.1 On the molecular level, several specific changes have been identified. Reciprocal chromosomal abnormalities like the t(15;17) or the inv(16) are associated with a particular morphology and clinical behavior. Based on the results in core binding factor (CBF) leukemias, a model has been defined recently.2 It suggests that AML results from the acquisition of 2 major molecular lesions, one leading to a block of differentiation by inactivating a master regulator of myeloid differentiation and the other inducing enhanced proliferation and diminished apoptosis due to activating mutations of protooncogenes like RAS. Although the picture might be more complex, a number of recent findings in mouse models as well as in patients with CBF leukemias support this hypothesis.3-5 However, in AML patients with normal karyotype, the mechanism of leukemia development was less clear, because few molecular changes had been identified.
During the last years, several novel abnormalities have been described that are predominantly found in this patient group. Constitutive activation of the FLT3 receptor tyrosine kinase, either by internal tandem duplication (ITD) mutations of the juxtamembrane domain or point mutations clustering in the second tyrosine kinase domain (TKD mutations), has been found in 20% to 30% of patients with AML and in 30% to 45% of patients with normal karyotype (reviewed by Stirewalt and Radich6 ). ITD mutations have been associated with an increased risk of treatment failure after conventional chemotherapy,7-11 whereas the prognostic relevance of FLT3 point mutations is less evident.9,11,12 In contrast to FLT3-ITD mutations, alterations of the myeloid transcription factor CEBPα, detectable in about 10% to 15% of patients with AML and normal karyotype, have been associated with a better outcome after treatment.13,14 However, in approximately 50% of AML patients with normal karyotype the molecular basis of leukemic development is still poorly understood.
More recently, an aberrant cytoplasmic localization of the nucleophosmin protein (NPM1) has been described in 35% of patients with acute myeloid leukemia.15 NPM1, also called B23 or numatrin, is a nucleocytoplasmic shuttling protein that constantly exchanges between nucleus and cytoplasm.16 Several functions for this protein have been described, including binding of nucleic acids,17 regulation of centrosome duplication,18 and ribosomal function.19 In addition, NPM1 binds to several proteins, including p53 itself20 as well as proteins interacting with and regulating p53 (eg, Rb,21 p19ARF,22 HDM223 ). Through these interactions, NPM1 is thought to be a major stress-induced regulator of p53 function in response to hypoxia,24 UV irradiation,25-27 or cytotoxic drugs.20 In patients with AML, mutations in exon 12 of the NPM1 gene on chromosome 5q35 have been described, leading to frameshift and an elongated protein, which is retained in the cytoplasm.15 Falini and coworkers showed that NPM1 mutations are associated with several clinical features, including a normal karyotype, low or absent CD34+ expression, and an increased prevalence of FLT3-ITD mutations.15 In addition, their results indicated that patients with NPM1 mutations have a significantly higher rate of complete remissions (CRs) after standard induction chemotherapy.15 However, the role of NPM1 mutations for the long-term outcome of patients is currently unclear. In addition, the relevance of potential modulating abnormalities like FLT3-ITD mutations needs to be clarified. To study the prevalence and prognostic role of NPM1 mutations in adult patients with AML, we retrospectively analyzed almost 1500 patients with newly diagnosed AML or advanced myelodysplastic syndrome (MDS) treated in a large multicenter trial. Because all NPM1 mutations described so far induced a 4 bp insertion,15 we reasoned that high-resolution fragment analysis should be able to detect these mutations, and used this method for patient screening. With an overall prevalence of 27.5% NPM1-mutated cases, our data confirm that this abnormality is among the most common genetic abnormalities in AML. In addition, our results clearly show that patients with NPM1 mutations have a better long-term outcome if not associated with an FLT3-ITD mutation.
Patients, materials, and methods
Patients
The data set reported here is derived from the multicenter AML96 protocol of the Deutsche Studieninitiative Leukämie (DSIL), formerly known as Süddeutsche Hämoblastose Gruppe (SHG). A list of the participating study centers is given in the “Appendix.” Between 1996 and 2003, 2458 patients were registered to the study, 1684 of which were included into the clinical protocol. Of the remaining patients, 108 had AML M3 and were treated separately,28 whereas 666 cases were not included. For the analysis of NPM1, we retrospectively investigated 1504 cases for which material for molecular studies was available. Of these, 1485 patients (98.7%) had already been typed for FLT3-ITD mutations previously. Because a major focus of this study was the analysis of the impact of FLT3-ITD mutations, we restricted the data analysis to those 1485 patients. No significant differences existed between the entire cohort of patients analyzed and the subgroup with known FLT3-ITD status. Of these cases, 1221 presented with de novo AML, 189 with AML and prior history of MDS, and 55 with therapy-related AML or refractory anemia with excess blasts-1/2 (RAEB1/2) (n = 20). Most patients (n = 1328) were treated in the AML96 protocol. Details of the treatment regimen have been published previously.11,29 In this protocol, postinduction therapy was stratified according to cytogenetic risk groups as defined in Table 1 for patients 60 years old or younger. In these patients, first induction therapy consisted of MAV: mitoxantrone 10 mg/m2 (days 4 to 8), cytosine arabinoside (ara-C) 100 mg/m2 (days 1 to 8), and VP16 100 mg/m2 (days 4 to 8). Second induction consisted of MAMAC: ara-C 2 × 1000 mg/m2 (days 1 to 5) and m-AMSA (4′[9-acridinylamino]methansulfon-m-anisidide) 100 mg/m2 (days 1 to 5). Patients with intermediate cytogenetic risk were referred to allogeneic hematopoietic stem cell transplantation (HSCT) from HLA-identical sibling donors if possible. Intermediate-risk patients without a sibling donor and low-risk patients were randomized to receive intermediate (2 × 1000 mg/m2 every 12 hours on days 1 to 6) (I-MAC) or high-dose (2 × 3000 mg/m2 every 12 hours on days 1 to 6) (H-MAC) ara-C plus mitoxantrone (10 mg/m2 days 4 to 6), which was followed by autologous peripheral blood stem cell transplantation (PBSCT) (intermediate risk) or MAMAC (low risk). Patients with high-risk cytogenetics were referred to allogeneic HSCT, including the option of unrelated HSCT. Patients without a donor were treated with either I-MAC or H-MAC and referred to autologous PBSCT.
Cytogenetic risk groups
Risk group . | Abnormalities . |
|---|---|
| Low risk | t(8;21) with or without additional abnormalities |
| Intermediate risk | All other abnormalities |
| High risk | -5/del(5q); -7/del(7q); other monosomies; inv(3q); t(3;3); abnl 12p; abnl 11q; +11; +1 +21; +22; t(6;9); t(9;22); multiple aberrations (3 or more structural or numerical abnormalities) |
Risk group . | Abnormalities . |
|---|---|
| Low risk | t(8;21) with or without additional abnormalities |
| Intermediate risk | All other abnormalities |
| High risk | -5/del(5q); -7/del(7q); other monosomies; inv(3q); t(3;3); abnl 12p; abnl 11q; +11; +1 +21; +22; t(6;9); t(9;22); multiple aberrations (3 or more structural or numerical abnormalities) |
Patients older than 60 years of age received 2 induction cycles containing DA: daunorubicin 45 mg/m2 (days 3 to 5) and ara-C 100 mg/m2 (days 1 to 7). Patients in CR received MAMAC. CR was defined as the presence of fewer than 5% blasts cells in a standardized bone marrow (BM) aspirate after the second course of induction therapy. Only patients with fully regenerated peripheral blood counts were considered to be in CR.
This study was approved by the ethical board of the Technical University Dresden. Each patient gave written informed consent to participate in the study.
Patient samples
All materials investigated were obtained at diagnosis. Bone marrow was used whenever available. In all other cases, peripheral blood samples were examined. Genomic DNA or RNA was extracted from mononuclear cells using standard procedures.11,30 Cells for laser scanning confocal microscopy were prepared from DMSO-cryopreserved leukemic samples.
Polymerase chain reaction for NPM1 exon 12
Polymerase chain reaction (PCR) was performed on genomic DNA using either the published primer molecules NPM1-F and NPM1-R15 or primers NPM-I11f (5′-CTGGTAGAATGAAAAATAGAT-3′) and NPM-E12r (5′-CTTGGCAATAGAACCTGGAC-3′). Primers NPM1-F and NPM-I11f were labeled with 6-FAM or Hex (TIB MolBiol, Berlin, Germany). PCR conditions were as outlined in detail recently.11 In brief, 5 ng DNA was amplified in a volume of 50 μL containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 μM dNTPs, the oligonucleotides (0.5 μM each), and 1 unit of AmpliTaq Gold DNA-polymerase (Perkin-Elmer, Norwalk, CT). The PCR consisted of an initial incubation step at 94°C for 11 minutes followed by 27 cycles at 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 60 seconds and a final elongation step at 94°C for 30 seconds and 60°C for 45 minutes. Reverse transcriptase (RT)–PCR was performed for those 38 samples for which no DNA was available. The RT reaction was performed as outlined recently.31 One microliter of the RT reaction was used for the PCR. PCR products were then analyzed by Genescan analysis. The run conditions were identical to those described recently for FLT3-ITD mutations,11 but the ILS600 size standard (Promega, Mannheim, Germany) was used. Analyses were performed on ABI377XL or ABI310 instruments (Applied Biosystems, Darmstadt, Germany).
Sequence analysis
PCR-amplified mutant samples were purified and sequenced directly using Big Dye Terminator cycle sequencing chemistry (Applied Biosystems). When the sequence could not be identified unambiguously, mutant samples were cloned and sequenced.11 Sequences were compared with the wild-type (wt) NPM1 cDNA (accession no. NM_002520). Numbering of nucleotide positions refers to the coding sequence.
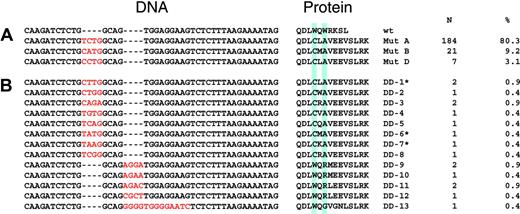
Mutations of NPM1 found in 229 sequenced patients. Comparison of the nucleotide and deduced amino acid sequences of mutations identified in AML patients. Amino acids are given in single-letter code. The blue boxes denote the 2 tryptophan residues at amino acid positions 288 and 290 important for nuclear transport.34 (A) Mutations described by Falini et al.15 (B) Novel mutations. *Also described by Suzuki et al.35
Mutations of NPM1 found in 229 sequenced patients. Comparison of the nucleotide and deduced amino acid sequences of mutations identified in AML patients. Amino acids are given in single-letter code. The blue boxes denote the 2 tryptophan residues at amino acid positions 288 and 290 important for nuclear transport.34 (A) Mutations described by Falini et al.15 (B) Novel mutations. *Also described by Suzuki et al.35
Confocal laser scanning microscopy
MV4-11 cells and blast cells from 6 patients (5 with NPM1 mutations and 1 NPM1-wt) were spun on poly-l-lysine–coated coverslips at 4°C. The cells were washed with ice-cold PBS and fixed with 4% paraformaldehyde (PFA) for 15 minutes at 37°C. Excess of PFA was quenched by 5 mM ammonium chloride. After permeabilization with 0.2% Triton X-100 and blocking with 10% FCS, cells were incubated with 8 μg/mL mouse monoclonal antibody (clone FC82291) to nucleophosmin (abcam; Biozol Diagnostica, Eching, Germany) for 1 hour. Samples were rinsed in PBS and incubated with an anti–mouse IgG antibody Cy3 conjugate (diluted 1:200; Sigma, Taufkirchen, Germany) for 30 minutes, followed by washing, and incubated for 5 minutes in equilibration buffer (Component C of SLOWFade Light Antifade Kit; Molecular Probes, Leiden, The Netherlands) and mounted in antifade reagent in glycerol buffer (Component A) containing DAPI (Sigma). The samples were scanned on a confocal laser scanning microscope system (FV-1000; Olympus, Hamburg, Germany) consisting of an Olympus IX81 microscope equipped with an oil-immersion Plan-Apo 60 ×/1.1 objective lens and a three-channel photomultiplier transmission detector using 4 × digital magnification. Images were processed with Olympus FV10-ASW 1.3 software.
Statistical analysis
Clinical variables across groups were compared using the χ2 or a 2-sided Fisher exact test for categorical variables, and the nonparametric Mann-Whitney U test was applied for continuous variables. P values below .05 were considered to be significant.
Overall and disease-free survivals were calculated only for those patients who had been included into the AML96 study using the methods of Kaplan and Meier,32 and the log-rank test was used to assess differences between survival curves. The median follow-up for all patients alive (n = 679) was 20.2 months (range, 1-87 months).
Cumulative incidence of relapse (CIR) was analyzed only for patients achieving a CR. It was measured from the CR date until date of relapse, death, or the last follow-up, where death in CR was considered a competing risk. Estimates of CIR were calculated using the Gray k-sample test.33
For multivariate analysis of prognostic factors, a Cox proportional hazard regression model was used, and stepwise forward selection was performed. Different models were tested (including continuous variables as absolute values or dichotomized, log transformation). These models obtained comparable results. Missing values were substituted by the median. Variables were added at a P value below .01 and deleted at a P value above .05.
All calculations were performed using the SPSS software package, version 12 (SPSS, Chicago, IL). CIR was calculated using the Gray algorithm with the S-Plus software package (version 6.2; Insightful, Reinach, Switzerland).
Results
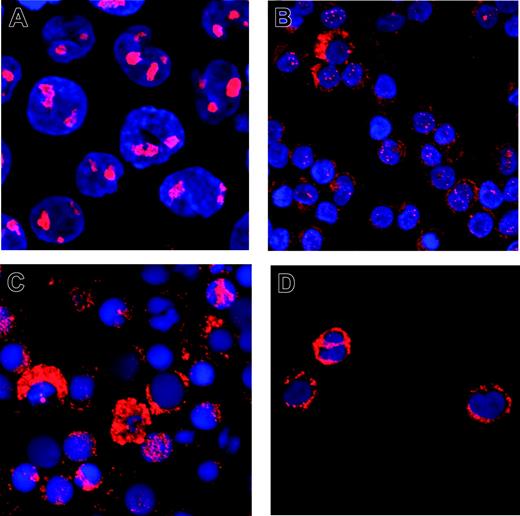
An additional, 4 bp–longer PCR fragment was detectable in 408 of 1485 patients (27.4%). In 229 randomly selected positive samples, sequence analysis verified the NPM1 mutations. Most of the patients had previously described mutations A (80.3%), B (9.2%), and D (3.1%).15 In the other 17 cases (7.4%), 13 novel mutations (Figure 1) were found. All mutations consisted of an insertion of 4 bases, either between nucleotides 960-961 or between nucleotides 964-965. Only in one case did we observe an insertion of 13 bases associated with a deletion of 9 nucleotides (965-973), which also resulted in a net addition of 4 bases. The predicted amino acid changes induced elongated proteins with very similar structures (Figure 1). Confocal laser scanning microscopy confirmed a predominant cytoplasmic localization of the NPM1 protein in 5 cases with novel mutations (Figure 2), although in 2 patients there was still some nuclear NPM1 protein detectable, potentially due to normal cells present in the sample.
NPM1 and FLT3-ITD mutations
Because FLT3-ITD mutations have previously been shown to be the most important abnormality in AML patients with normal karyotype, we correlated these 2 aberrations. The frequency of FLT3-ITD mutations was 312 of 1485 (21%). FLT3-ITD mutations were found in 164 of 408 (40.2%) NPM1-mutant cases compared with 148 of 1077 (13.7%) of the NPM1-wt cases (P < .001). Similar results were found in cases with normal karyotype (142 of 324 [43.8%] NPM1-mut were FLT3-ITD positive versus 76 of 385 [19.7%] NPM1-wt cases [P < .001]).
Confocal laser scanning microscopy of NPM1 wt and mutant cells. Cells of the NPM1-wt cell line MV4-11 (A) or mononuclear cells from patient samples with mutations DD-3 (B), DD-4 (C), or DD-1 (D) were stained with a monoclonal antibody against NPM1, Cy-3 labeled with a secondary antibody, and analyzed using confocal laser scanning microscopy. Nuclei were stained with DAPI; original magnification, × 240.
Confocal laser scanning microscopy of NPM1 wt and mutant cells. Cells of the NPM1-wt cell line MV4-11 (A) or mononuclear cells from patient samples with mutations DD-3 (B), DD-4 (C), or DD-1 (D) were stained with a monoclonal antibody against NPM1, Cy-3 labeled with a secondary antibody, and analyzed using confocal laser scanning microscopy. Nuclei were stained with DAPI; original magnification, × 240.
These data confirm that NPM1 and FLT3-ITD mutations characterize major, partially overlapping subgroups in AML, especially in patients without karyotype abnormalities. Therefore, all further analyses were performed according to 4 groups: NPM1-mut/FLT3-ITDneg (group A); NPM1-mut/FLT3-ITDpos (group B); NPM1-wt/FLT3-ITDpos (group C); and NPM1-wt/FLT3-ITDneg (group D). The clinical characteristics of these 4 groups are summarized in Table 2. Compared with patients without NPM1 and FLT3-ITD mutations (group D), patients showing only NPM1 mutations (group A) had significantly higher median white blood cell (WBC) counts (26.3 × 109/L versus 7.7 × 109/L; P < .001) and BM blasts (67.3% versus 56%; P < .001). Interestingly, patients in group A also had significantly higher median platelet counts (68 × 109/L; range, 7 × 109/L-302 × 109/L) who had compared with patients who had FLT3-ITD mutations only (group C) (median, 47 × 109/L; P < .001) and patients negative for both mutations (group D) (median, 4 × 109/L; range, 4 × 109/L-1.4 × 109/L; P < .001); a trend existed for group B (median, 56 × 109/L; range, 3 × 109/L-514 × 109/L; P = .073). As a further novel finding, NPM1 mutations were predominantly observed in female patients, where NPM1 mutations were 1.5 times as frequent as in males (mutant NPM1 females: 237 of 712 [33.3%]; males: 171 of 773 [22.1%]; P < .001). Significant differences were seen in the distribution of these mutations in different French-American-British (FAB) subgroups. Patients with NPM1 mutations were predominantly found in FAB M2, M5a, and M5b, whereas these mutations were never found in FAB M3 and less common in FAB M0, M4eo, M6, and M7. As shown previously,15 lower CD34 expression was found on blasts of patients with NPM1 mutations (median NPM1-wt, 40%; range, 0%-99%; median NPM1-mutant, 3%; range, 0%-93%; P < .001; n = 1120).
Clinical features and NPM1 and FLT3 mutations in AML and RAEB-t patients at diagnosis
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 244 | 164 | 148 | 929 |
| Median BM blasts, % (range) | 67.3 (6-95)* | 75.5 (29.5-100)* | 73 (11-96) | 56 (0.5-98.5) |
| Median WBC count, × 109/L (range) | 26.3 (0.5-380)* | 50 (1.1-372)* | 36 (0.8-465) | 7.7 (0.3-450) |
| Median platelet count, × 109/L (range) | 68 (7-302)* | 56 (3-514) | 47 (5-372) | 46 (4-1430) |
| Median LDH level, U/mL (range) | 449 (27-4065)* | 544 (23-7250)* | 607 (16-7096) | 369 (4.5-5274) |
| Median age, y (range) | 60 (18-83) | 57.5 (19-81) | 54 (17-83) | 58 (15-87) |
| Female, % | 55.3 | 62.2 | 49.3 | 43.3 |
| De novo AML, no. (%) | 223 (91.4)* | 150 (91.5) | 129 (87.2) | 719 (77.4) |
| Prior MDS, no. (%) | 16 (6.6)* | 12 (7.3) | 12 (8.1) | 149 (16.0) |
| TAML, no. (%) | 2 (0.8)* | 2 (1.2) | 5 (3.4) | 46 (5.0) |
| FAB‡ | ||||
| M0, no. (%) | 0 (0)* | 2 (1.2) | 2 (1.4) | 57 (6.1) |
| M1, no. (%) | 50 (20.5) | 47 (28.7) | 43 (29.1) | 180 (19.4) |
| M2, no. (%) | 79 (32.4) | 50 (30.5) | 37 (25) | 290 (19.4) |
| M3, no. (%) | 0 (0)* | 0 (0) | 12 (8.1) | 43 (4.6) |
| M4, no. (%) | 37 (15.2) | 20 (12.2) | 27 (18.2) | 87 (9.4) |
| M4eo, no. (%) | 1 (0.4) | 0 (0) | 4 (2.7) | 66 (7.1) |
| M5a, no. (%) | 41 (16.8)† | 25 (15.2) | 11 (7.4) | 78 (8.4) |
| M5b, no. (%) | 21 (8.6)* | 14 (8.5) | 1 (0.7) | 14 (1.5) |
| M6, no. (%) | 2 (0.8)† | 0 (0) | 2 (1.4) | 46 (5.0) |
| M7, no. (%) | 0 (0) | 2 (1.2) | 0 (0) | 10 (1.1) |
| RAEB-t, no. (%) | 7 (2.9) | 1 (0.6) | 2 (1.4) | 37 (4.0) |
| RAEB1, no. (%) | 1 (0.4) | 0 (0) | 0 (0) | 5 (0.5) |
| RAEB2, no. (%) | 2 (0.8) | 0 (0) | 2 (1.4) | 10 (1.1) |
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 244 | 164 | 148 | 929 |
| Median BM blasts, % (range) | 67.3 (6-95)* | 75.5 (29.5-100)* | 73 (11-96) | 56 (0.5-98.5) |
| Median WBC count, × 109/L (range) | 26.3 (0.5-380)* | 50 (1.1-372)* | 36 (0.8-465) | 7.7 (0.3-450) |
| Median platelet count, × 109/L (range) | 68 (7-302)* | 56 (3-514) | 47 (5-372) | 46 (4-1430) |
| Median LDH level, U/mL (range) | 449 (27-4065)* | 544 (23-7250)* | 607 (16-7096) | 369 (4.5-5274) |
| Median age, y (range) | 60 (18-83) | 57.5 (19-81) | 54 (17-83) | 58 (15-87) |
| Female, % | 55.3 | 62.2 | 49.3 | 43.3 |
| De novo AML, no. (%) | 223 (91.4)* | 150 (91.5) | 129 (87.2) | 719 (77.4) |
| Prior MDS, no. (%) | 16 (6.6)* | 12 (7.3) | 12 (8.1) | 149 (16.0) |
| TAML, no. (%) | 2 (0.8)* | 2 (1.2) | 5 (3.4) | 46 (5.0) |
| FAB‡ | ||||
| M0, no. (%) | 0 (0)* | 2 (1.2) | 2 (1.4) | 57 (6.1) |
| M1, no. (%) | 50 (20.5) | 47 (28.7) | 43 (29.1) | 180 (19.4) |
| M2, no. (%) | 79 (32.4) | 50 (30.5) | 37 (25) | 290 (19.4) |
| M3, no. (%) | 0 (0)* | 0 (0) | 12 (8.1) | 43 (4.6) |
| M4, no. (%) | 37 (15.2) | 20 (12.2) | 27 (18.2) | 87 (9.4) |
| M4eo, no. (%) | 1 (0.4) | 0 (0) | 4 (2.7) | 66 (7.1) |
| M5a, no. (%) | 41 (16.8)† | 25 (15.2) | 11 (7.4) | 78 (8.4) |
| M5b, no. (%) | 21 (8.6)* | 14 (8.5) | 1 (0.7) | 14 (1.5) |
| M6, no. (%) | 2 (0.8)† | 0 (0) | 2 (1.4) | 46 (5.0) |
| M7, no. (%) | 0 (0) | 2 (1.2) | 0 (0) | 10 (1.1) |
| RAEB-t, no. (%) | 7 (2.9) | 1 (0.6) | 2 (1.4) | 37 (4.0) |
| RAEB1, no. (%) | 1 (0.4) | 0 (0) | 0 (0) | 5 (0.5) |
| RAEB2, no. (%) | 2 (0.8) | 0 (0) | 2 (1.4) | 10 (1.1) |
RAEB-t indicates refractory anemia with excess of blasts in transformation; t-AML, therapy-related AML.
P < .001.
P < .01.
Patients with FAB M6.
NPM1 mutations and cytogenetics
The association with specific cytogenetic features was studied in 1395 patients in whom the karyotype analysis was available. As shown in Table 3, NPM1 mutations were found in 324 of 709 patients (45.7%) with normal karyotype but only in 58 of 686 patients (8.5%) with karyotype abnormalities (P < .001). Within the group of patients with aberrant karyotype, NPM1 mutations were mostly associated with single genetic abnormalities like trisomies (eg, +8 = 11; 19%; +4 = 4; 6.9%), monosomies (–Y = 7; 12.1%), or chromosomal deletions (eg, del 9q = 4; 6.9%). NPM1 mutations were only rarely found in cases with reciprocal translocations like t(8;21) or inv(16). No mutations were found in the 47 cases with t(15;17) (P = .002). A significantly lower incidence of NPM1 mutations was also found in cases with a complex karyotype (4 of 185; 2.2%; P < .001).
Comparison of cytogenetic aberrations and NPM1 and FLT3-ITD mutations in patients with AML (n = 1485)
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 244 | 164 | 148 | 929 |
| Unknown, no. (%) | 14 (5.7) | 12 (7.3) | 7 (4.7) | 57 (6.1) |
| Normal, no. (%) | 182 (79.1)† | 142 (93.4)† | 76 (53.9)† | 309 (35.4) |
| Abnormal, no. (%) | 48 (20.9) | 10 (6.6) | 65 (46.1) | 563 (64.6) |
| t(8;21), no. | 2 | 0 | 3 | 52 |
| inv(16)/t(16;16), no. | 2 | 0 | 5 | 60 |
| t(15;17), no. | 0† | 0 | 11 | 36 |
| +8, no. | 11 | 1 | 12 | 100 |
| -7/7q-, no. (%) | 5 | 0 | 3 | 114 |
| -5/5q-, no. (%) | 3 | 0 | 0 | 91 |
| t(6;9), no. | 0 | 0 | 7 | 0 |
| Complex, no.* | 4‡ | 0 | 5 | 176 |
| -X/-Y, no. | 7 | 2 | 2 | 48 |
| FLT3D835/836, no. (%)∥ | 31/204 (15.2)† | 8/133 (6.0) | 9/124 (7.3) | 42/772 (5.4) |
| MLL-PTD, no. (%)∥ | 0/126 (0)† | 0/81 (0)§ | 11/73 (15.1)§ | 28/476 (5.9) |
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 244 | 164 | 148 | 929 |
| Unknown, no. (%) | 14 (5.7) | 12 (7.3) | 7 (4.7) | 57 (6.1) |
| Normal, no. (%) | 182 (79.1)† | 142 (93.4)† | 76 (53.9)† | 309 (35.4) |
| Abnormal, no. (%) | 48 (20.9) | 10 (6.6) | 65 (46.1) | 563 (64.6) |
| t(8;21), no. | 2 | 0 | 3 | 52 |
| inv(16)/t(16;16), no. | 2 | 0 | 5 | 60 |
| t(15;17), no. | 0† | 0 | 11 | 36 |
| +8, no. | 11 | 1 | 12 | 100 |
| -7/7q-, no. (%) | 5 | 0 | 3 | 114 |
| -5/5q-, no. (%) | 3 | 0 | 0 | 91 |
| t(6;9), no. | 0 | 0 | 7 | 0 |
| Complex, no.* | 4‡ | 0 | 5 | 176 |
| -X/-Y, no. | 7 | 2 | 2 | 48 |
| FLT3D835/836, no. (%)∥ | 31/204 (15.2)† | 8/133 (6.0) | 9/124 (7.3) | 42/772 (5.4) |
| MLL-PTD, no. (%)∥ | 0/126 (0)† | 0/81 (0)§ | 11/73 (15.1)§ | 28/476 (5.9) |
The number of patients with individual aberrations adds to a higher number than the absolute number of patients with aberrations because several patients had more than one aberration.
Three or more structural or numerical chromosomal aberrations.
P < .01.
P < .001.
P < .05.
No. of positive cases/total cases analyzed.
Of the 1485 patients, 756 had previously been analyzed for MLL-PTD mutations,36 another abnormality frequently observed in patients with normal karyotype. Interestingly, we did not find any case showing both mutant NPM1 and an MLL-PTD (MLL-PTD in NPM1-mutant: 0 of 207; in NPM1-wt: 39 of 549; P < .001). FLT3-TKD mutations, which had been analyzed in 1233 patients, were twice as common in group A compared with the other groups (group A: 15.2% [31 of 204]; group B: 6%, P = .02; group C: 7.3%, P = .04; and group D: 5.4%, P < .001, for pairwise comparison).
NPM1 and FLT3-ITD: ratio of mutant to wt
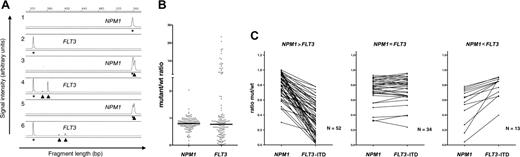
The ratio of the FLT3-ITD mutation to the wt-FLT3 allele has significant impact on the prognostic information of FLT3-ITD mutations.11 Patients showing a loss of the wt-FLT3 allele37 and, as a consequence, showing an increased ratio of mutant to wt, have a very poor prognosis.11 Because FLT3-ITD and NPM1-mutation data were generated on the same sample set using a similar method, we also asked for the mut/wt ratio in patients with NPM1 mutations. The median ratio of the 370 patients with NPM1 mutation investigated using DNA as starting material was 0.77 (range, 0.04-2.02). There were only 15 patients with a ratio above 1, indicating a heterozygous state of the mutation in all cases. In 149 cases with double mutation, which had been analyzed using fragment analysis for both mutations, the NPM1 mutation/NPM1-wt ratio was 0.8 compared with 0.77 for the FLT2-ITD/wt ratio (P = .98; Wilcoxon signed rank test). There was an obvious difference in the variance of the NPM1 ratio (coefficient of variation 28%) compared with the FLT3-ITD mutant cases (197%), indicating that NPM1 was uniformly present in most of the blasts. Because very high values for the FLT3-ITD mutation are a consequence of loss of wt-FLT3,11,37 we investigated the set of patients where both mutations had a ratio below 1. In these 99 cases, the median NPM1-mut/wt ratio was 0.78, and the FLT3-ITD/wt ratio was 0.59. Wilcoxon matched-pair analysis indicated a significantly higher NPM1-mut/wt ratio than the FLT3-ITD/wt value (P < .001). As shown in Figure 3, in most cases the NPM1-mut/wt ratio exceeded the corresponding FLT3-ITD/wt ratio. Taken together, these data indicate that NPM1 mutations were present in a higher percentage of blasts in most patients, suggesting that NPM1 mutations have occurred prior to the FLT3-ITD mutations in these cases. To further investigate the sequential acquisition of both mutations, we looked for cases showing several FLT3-mutant bands, which can be found in about 15% to 20% of patients with FLT3-ITD mutations.9,11,37 As shown in Figure 3A, several FLT3-ITD mutations were present, but only a single NPM1 mutation was found in these cases, indicating that these FLT3-ITD mutations evolved from a single NPM1-positive clone.
Analysis of NPM1 and FLT3-mutant/wt ratio in patient samples. (A) Genescan analysis for NPM1 and FLT3-ITD. Lanes 1 and 2 show examples of an NPM1-wt– and FLT3-ITD–negative case. Lanes 3 through 6 illustrate two patients with mutant NPM1 and FLT3-ITD mutations. Note that in both cases, two independent FLT3-ITD mutations were found. (B) Comparison of NPM1 and FLT3-ITD mutant/wt ratio in 149 samples analyzed for both abnormalities. (C) Comparison of NPM1 and FLT3 in 3 different groups according to the difference between NPM1-mut/wt and FLT3-ITD mut/wt ratio.
Analysis of NPM1 and FLT3-mutant/wt ratio in patient samples. (A) Genescan analysis for NPM1 and FLT3-ITD. Lanes 1 and 2 show examples of an NPM1-wt– and FLT3-ITD–negative case. Lanes 3 through 6 illustrate two patients with mutant NPM1 and FLT3-ITD mutations. Note that in both cases, two independent FLT3-ITD mutations were found. (B) Comparison of NPM1 and FLT3-ITD mutant/wt ratio in 149 samples analyzed for both abnormalities. (C) Comparison of NPM1 and FLT3 in 3 different groups according to the difference between NPM1-mut/wt and FLT3-ITD mut/wt ratio.
Presence of NPM1 mutations and response to treatment
The presence of NPM1 and FLT3 mutations (groups A to D) was correlated with the clinical outcome in those 1328 patients treated in the AML96 protocol of the DSIL. Patients with AML-M3 were not analyzed, because these patients were treated in a different protocol (APL 199328 ) and did not show NPM1 mutations. When patients of all ages and karyotype subgroups were analyzed, NPM1-mutant cases had a significantly better CR rate (group A: 58.6%; group B: 53.7%; group C: 49.3%; and group D: 41.6%; P < .001). NPM1 mutations were still associated with an improved CR rate when only cases with normal karyotype were analyzed (group A: 61%; group B: 52.8%; group C: 50%; and group D: 42.1%; P < .001; n = 709). However, no statistical significant difference was observed in cases with de novo AML in patients aged 60 years or younger and with normal karyotype (group A: 68.8%; group B: 57.7%; group C: 68.9%; and group D: 60.2%; P < .329; n = 352).
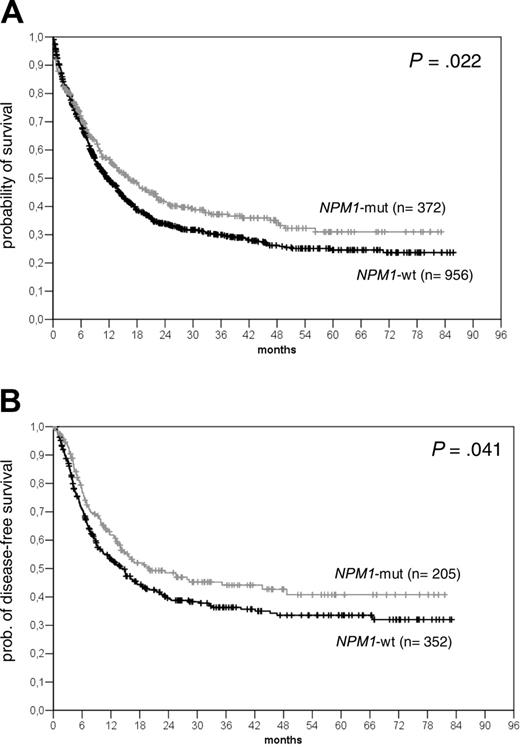
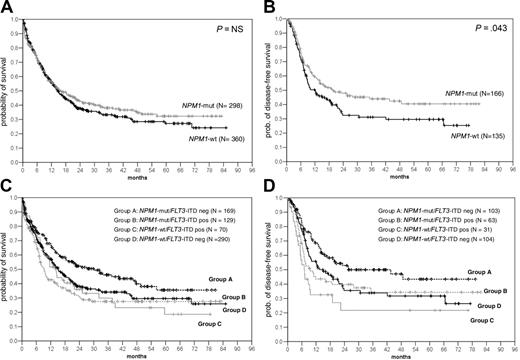
Kaplan-Meier analysis of the actuarial probability of OS and DFS in all patients analyzed. Comparison of OS (A) and DFS (B) in all patients according to the presence or absence of NPM1 mutations.
Kaplan-Meier analysis of the actuarial probability of OS and DFS in all patients analyzed. Comparison of OS (A) and DFS (B) in all patients according to the presence or absence of NPM1 mutations.
Kaplan-Meier analysis of NPM1 and FLT3-ITD mutations in patients with AML and normal karyotype. Kaplan-Meier analysis of OS (A) and DFS (B) in AML patients with normal karyotype according to NPM1 mutations; OS (C) and DFS (D) according to NPM1 and FLT3-ITD groups. Analysis was done in the 4 groups defined in the text. Patients in group A (NPM1-mut alone) had a significantly higher probability of OS than group B (double mutants; P = .001), group C (FLT3-ITDpos only; P = .032), and group D (wt for both; P = .03). Also, the DFS was significantly higher in group A than group B (P = .04), group C (P < .001), and group D (P = .04).
Kaplan-Meier analysis of NPM1 and FLT3-ITD mutations in patients with AML and normal karyotype. Kaplan-Meier analysis of OS (A) and DFS (B) in AML patients with normal karyotype according to NPM1 mutations; OS (C) and DFS (D) according to NPM1 and FLT3-ITD groups. Analysis was done in the 4 groups defined in the text. Patients in group A (NPM1-mut alone) had a significantly higher probability of OS than group B (double mutants; P = .001), group C (FLT3-ITDpos only; P = .032), and group D (wt for both; P = .03). Also, the DFS was significantly higher in group A than group B (P = .04), group C (P < .001), and group D (P = .04).
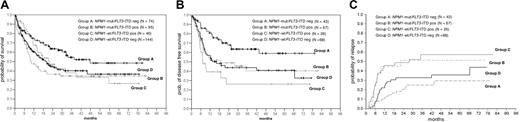
As shown in Figure 4, the presence of an NPM1 mutation was associated with an improved survival when all patients were analyzed (n = 1328) (median overall survival [OS] in the NPM1-mutant was 16.24 months [95% confidence interval (CI): 12.03 to 20.45 months] versus 11.54 months in NPM1-wt cases [95% CI: 9.96 to 13.12 months]; P = .022) (Figure 4). This was also seen for the disease-free survival (DFS) (median NPM1-mut: 19.69 months [95% CI: 6.17 to 33.22 months] versus NPM1-wt: 14.46 months [95% CI: 10.83 to 18.09 months]; P = .432). We were especially interested in patients with normal karyotype; however, no difference was seen in the OS between NPM1-mut and NPM1-wt cases and only a marginal difference for the DFS. In this group of patients, we and others have shown that FLT3-ITD mutations represent a prognostic factor. In univariate analysis, the presence of an FLT3-ITD mutation had a significant impact on survival (median OS FLT3-ITDneg: 18.28 months [95% CI: 14.46 to 22.09 months]; FLT3-ITDpos: 10.39 months [95% CI: 7.35 to 13.43 months]; P = .008). Because of the high rate of overlap between the NPM1 and FLT3-ITD mutants, we wanted to analyze the relevance of NPM1 and FLT3-ITD mutations individually and together in the 4 defined groups (groups A to D). As shown in Figure 5B-C, if present alone, NPM1 mutations (group A) were associated with a significantly better OS and DFS compared with all other groups. Similar observations were made in patients younger than 60 years with normal karyotype (Figure 6A-B). This was associated with a significantly lower cumulative incidence of relapse (CIR) in patients with normal karyotype (CIR at 4 years for group A: 25%; group B: 57.2%; group C: 51.3%; group D: 32.7%; P = .004) (Figure 6C).
A multivariate analysis was performed to investigate whether NPM1 aberrations represent an independent prognostic factor. We included several known risk factors in the model (age, cytogenetics, WBCs, LDH, secondary AML [sAML], BM blasts, MLL-PTD, FLT3-TKD mutations) and NPM1 and FLT3 aberrations in the 4 defined groups. As shown in Tables 4, 5, NPM1 mutations alone represented an independent factor associated with a better OS in the entire cohort of patients and with a better OS and DFS in cases with normal karyotype.
Multivariate analysis for cytogenetics and clinical and biologic variables (all patients)
. | All patients within study . | . | . | . | Patients 60 y or younger . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | OS . | . | DFS . | . | OS . | . | DFS . | . | ||||||
. | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | ||||||
| Age, 60 y or younger vs older than 60 y | < .001 | 1.90 (1.65-2.15) | < .001 | 1.91 (1.50-2.42) | — | — | — | — | ||||||
| Cytogenetics*† | ||||||||||||||
| Low | < .001 | 0.48 (0.33-0.69) | .002 | 0.48 (0.30-0.77) | .005 | 0.55 (0.36-0.84) | .039 | 0.56 (0.32-0.97) | ||||||
| High | < .001 | 1.86 (1.57-2.2) | < .001 | 2.08 (1.51-2.87) | < .001 | 1.80 (1.42-2.80) | .001 | 1.98 (1.35-2.91) | ||||||
| WBC count† | ||||||||||||||
| Intermediate (> 9.2 ≤ 21.7 GPT/L) | < .005 | 1.30 (1.08-1.55) | .024 | 1.43 (1.05-1.96) | .082 | 1.26 (0.97-1.64) | NS | — | ||||||
| Highest, 21.7 or more GPT/L | < .001 | 1.48 (1.25-1.75) | < .001 | 1.88 (1.41-2.51) | .002 | 1.48 (1.16-1.89) | .005 | 1.69 (1.17-2.44) | ||||||
| De novo AML vs sAML | .001 | 0.73 (0.58-0.91) | .013 | 0.63 (0.44-0.91) | .002 | 0.65 (0.49-0.85) | .041 | 0.60 (0.36-0.98) | ||||||
| NPM1-FLT3 group | ||||||||||||||
| NPM1-mut/FLT3-ITDneg | .005 | 0.73 (0.58-0.91) | .086 | 0.75 (0.54-1.042) | .037 | 0.71 (0.51-0.98) | .082 | 0.67 (0.43-1.05) | ||||||
| NPM1-mut/FLT3-ITDpos | NS | — | NS | — | .089 | 1.33 (0.96-1.87) | NS | — | ||||||
| NPM1-wt/FLT3-ITDpos | NS | — | < .001 | 1.97 (1.38-2.82) | NS | — | .014 | 1.70 (1.12-2.59) | ||||||
. | All patients within study . | . | . | . | Patients 60 y or younger . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | OS . | . | DFS . | . | OS . | . | DFS . | . | ||||||
. | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | ||||||
| Age, 60 y or younger vs older than 60 y | < .001 | 1.90 (1.65-2.15) | < .001 | 1.91 (1.50-2.42) | — | — | — | — | ||||||
| Cytogenetics*† | ||||||||||||||
| Low | < .001 | 0.48 (0.33-0.69) | .002 | 0.48 (0.30-0.77) | .005 | 0.55 (0.36-0.84) | .039 | 0.56 (0.32-0.97) | ||||||
| High | < .001 | 1.86 (1.57-2.2) | < .001 | 2.08 (1.51-2.87) | < .001 | 1.80 (1.42-2.80) | .001 | 1.98 (1.35-2.91) | ||||||
| WBC count† | ||||||||||||||
| Intermediate (> 9.2 ≤ 21.7 GPT/L) | < .005 | 1.30 (1.08-1.55) | .024 | 1.43 (1.05-1.96) | .082 | 1.26 (0.97-1.64) | NS | — | ||||||
| Highest, 21.7 or more GPT/L | < .001 | 1.48 (1.25-1.75) | < .001 | 1.88 (1.41-2.51) | .002 | 1.48 (1.16-1.89) | .005 | 1.69 (1.17-2.44) | ||||||
| De novo AML vs sAML | .001 | 0.73 (0.58-0.91) | .013 | 0.63 (0.44-0.91) | .002 | 0.65 (0.49-0.85) | .041 | 0.60 (0.36-0.98) | ||||||
| NPM1-FLT3 group | ||||||||||||||
| NPM1-mut/FLT3-ITDneg | .005 | 0.73 (0.58-0.91) | .086 | 0.75 (0.54-1.042) | .037 | 0.71 (0.51-0.98) | .082 | 0.67 (0.43-1.05) | ||||||
| NPM1-mut/FLT3-ITDpos | NS | — | NS | — | .089 | 1.33 (0.96-1.87) | NS | — | ||||||
| NPM1-wt/FLT3-ITDpos | NS | — | < .001 | 1.97 (1.38-2.82) | NS | — | .014 | 1.70 (1.12-2.59) | ||||||
For all patients, n = 1328; for patients 60 years old or younger, n = 737.
Not significant (NS) were the following: platelets, LDH (log), sex, MLL-PTD, FLT3-TKD, BM blasts.
GPT/L indicates gigaparticles per liter; —, not applicable.
Cytogenetics were as defined by the Medical Research Council (MRC) study group; if more than two groups were built, a reference group was used.
For cytogenetics, the intermediate group was was set as the reference group; for WBC counts, lowest WBC count (9.2 or fewer GPT/L) was set as the reference group; and for de novo AML versus sAML NPM1-FLT3 group, NPM1-wt/FLT3-ITDneg was set as the reference group.
Multivariate analysis of outcome in patients with normal karyotype
. | All patients with normal karyotype . | . | . | . | Patients with normal karyotype, 60 y or younger . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | OS . | . | DFS . | . | OS . | . | DFS . | . | ||||||
. | P . | OR (95% Cl) . | P . | OR (95% Cl) . | P . | OR (95% Cl) . | P . | OR (95% Cl) . | ||||||
| Age, 60 y or younger vs older than 60 y | <.001 | 1.69 (1.38-2.07) | <.001 | 1.94 (1.42-2.66) | — | — | — | — | ||||||
| NPM1-FLT3 group* | ||||||||||||||
| NPM1-mut/FLT3-ITDneg | .043 | 0.76 (0.59-0.99) | .036 | 0.66 (0.45-0.97) | .019 | 0.63 (0.43-0.93) | .009 | 0.49 (0.29-0.84) | ||||||
| NPM1-mut/FLT3-ITDpos | NS | NS | NS | NS | ||||||||||
| NPM1-wt/FLT3-ITDpos | NS | .006 | 2.02 (1.22-3.34) | NS | NS | |||||||||
| WBC count* | ||||||||||||||
| Intermediate (>9.2 ≤ 21.7 GPT/L) | NS | NS | NS | NS | ||||||||||
| Highest, 21.7 or more GPT/L | .002 | 1.87 (1.25-2.79) | .001 | 2.25 (1.40-3.63) | .006 | 1.66 (1.15-2.34) | .019 | 1.86 (1.11-3.13) | ||||||
| De novo AML versus sAML | NS | .039 | 0.53 (0.29-0.97) | NS | NS | |||||||||
. | All patients with normal karyotype . | . | . | . | Patients with normal karyotype, 60 y or younger . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | OS . | . | DFS . | . | OS . | . | DFS . | . | ||||||
. | P . | OR (95% Cl) . | P . | OR (95% Cl) . | P . | OR (95% Cl) . | P . | OR (95% Cl) . | ||||||
| Age, 60 y or younger vs older than 60 y | <.001 | 1.69 (1.38-2.07) | <.001 | 1.94 (1.42-2.66) | — | — | — | — | ||||||
| NPM1-FLT3 group* | ||||||||||||||
| NPM1-mut/FLT3-ITDneg | .043 | 0.76 (0.59-0.99) | .036 | 0.66 (0.45-0.97) | .019 | 0.63 (0.43-0.93) | .009 | 0.49 (0.29-0.84) | ||||||
| NPM1-mut/FLT3-ITDpos | NS | NS | NS | NS | ||||||||||
| NPM1-wt/FLT3-ITDpos | NS | .006 | 2.02 (1.22-3.34) | NS | NS | |||||||||
| WBC count* | ||||||||||||||
| Intermediate (>9.2 ≤ 21.7 GPT/L) | NS | NS | NS | NS | ||||||||||
| Highest, 21.7 or more GPT/L | .002 | 1.87 (1.25-2.79) | .001 | 2.25 (1.40-3.63) | .006 | 1.66 (1.15-2.34) | .019 | 1.86 (1.11-3.13) | ||||||
| De novo AML versus sAML | NS | .039 | 0.53 (0.29-0.97) | NS | NS | |||||||||
For all patients with normal karyotype, n = 701; for those 60 years old or younger, n = 387.
Not significant (NS) were the following: platelets, LDH (log), sex, MLL-PTD, FLT3-TKD, BM blasts.
— indicates not applicable.
Reference group for NPM1-FLT3 was NPM1-wt/FLT3-ITDneg; for WBC count, those with 9.2 or fewer GPT/L.
OS, DFS, and CIR in AML patients (60 years or younger) with normal karyotype. (A) Comparison of the probability of OS according to the NPM1 and FLT3-ITD mutational status in the 4 defined groups. Group A (NPM1-mut alone) had a significantly higher actuarial probability of OS than group B (double mutants; P = .003) and D (all wt; P = .02), and trend was seen versus group C (FLT3-ITD alone; P = .11). (B) Probability of DFS in the 4 groups. Group A had a significantly higher probability of disease-free survival than group B (P = .02), group C (P < .001), and group D (P = .006). (C) CIR in the 4 groups. Analysis was done using the Gray k-sample algorithm in patients with normal karyotype showing a significantly reduced CIR for group A (CIR at 40 months, 25%) compared with the other groups (P = .004). Highest relapse rates were seen in group B (57%) and group C (51%); the CIR was 32.7% in group D.
OS, DFS, and CIR in AML patients (60 years or younger) with normal karyotype. (A) Comparison of the probability of OS according to the NPM1 and FLT3-ITD mutational status in the 4 defined groups. Group A (NPM1-mut alone) had a significantly higher actuarial probability of OS than group B (double mutants; P = .003) and D (all wt; P = .02), and trend was seen versus group C (FLT3-ITD alone; P = .11). (B) Probability of DFS in the 4 groups. Group A had a significantly higher probability of disease-free survival than group B (P = .02), group C (P < .001), and group D (P = .006). (C) CIR in the 4 groups. Analysis was done using the Gray k-sample algorithm in patients with normal karyotype showing a significantly reduced CIR for group A (CIR at 40 months, 25%) compared with the other groups (P = .004). Highest relapse rates were seen in group B (57%) and group C (51%); the CIR was 32.7% in group D.
Discussion
Using a fragment analysis procedure, we found NPM1 mutations in 27.5% of all analyzed AML patients. This number is lower than reported by Falini et al (35%)15 but in the same range as reported by 2 studies published during the preparation of this manuscript.35,38
The fragment analysis procedure used for the detection of NPM1 mutations is fast, easy, and more sensitive than direct sequencing. We have now optimized this method so that NPM1 and FLT3-ITD mutations can be screened in one multiplex PCR with subsequent fragment analysis, which provides the combined information to assess the prognostic impact of NPM1 mutations rapidly. Recently, 2 other groups38,39 published very similar Genescan-based approaches for the simultaneous detection of NPM1 and FLT3 mutations.
Using this assay we also found evidence that NPM1 mutations occurred before the FLT3-ITD mutations in most cases. Further support for this idea comes from the observation that in cases with several ITD mutations only one NPM1 mutation could be found. In all sequenced cases, we never found more than one NPM1 mutation, even in cases that had been cloned and sequenced. Taken together, these data indicate that NPM1 mutations are a primary event in most AML patients, preceding the acquisition of FLT3-ITD or other mutations. These data lend further support to the model proposed by Speck and Gilliland on the necessity of 2 events to induce AML.2
Sequence analysis of 229 cases confirmed the results by Falini et al15 that mutation A, the duplication of the 4 bases TCTG, is the most common change. Two other described changes (mutations B and D) were observed in 9.2% and 3.1%, respectively, but mutations C, E, and F could not be detected. Instead, we found 13 novel mutations, most of which also occurred at position 960 of the NPM1 coding sequence. Mutations DD-1, DD-6, and DD-7 were also described in the study by Suzuki et al.35 Interestingly, 97% of all mutations occurred at nucleotide position 960 and changed 2 of the tryptophan residues at amino acid positions 288 and 290, which are essential for the nuclear translocation of the protein.34 The other, less common mutations changed only the second tryptophan amino acid. In a study on pediatric patients, Cazzaniga et al described 3 of 7 (43%) of the NPM1 changes to be in the latter group.40 It remains to be clarified whether this is due to differences between children and adults and whether these changes are associated with different functional behavior as suggested by in vitro data.34
NPM1 mutations were found to be associated with specific clinical parameters. Like FLT3-ITD mutations, NPM1 mutations were associated with higher BM blasts and leukocyte counts, which were especially high when NPM1 mutations were present together with FLT3-ITD. Interestingly and in contrast to FLT3-ITD mutations, mutant NPM1 alone was associated with significantly higher platelet counts. Hsu and Yung showed that K562 cells transfected with a C-terminal NPM1 mutant have an increased ability for megakaryocytic differentiation.41 This might imply that blasts with NPM1 mutations retain a certain capacity for thrombocytic differentiation. A novel aspect was the highly significant increase of NPM1 mutations in female patients (33% versus 22%; P < .001). This is especially interesting because the incidence of AML in general is higher in males.1 Falini et al did not report an association with patient sex; however, their study was based mainly on immunohistochemistry and analyzed only about half of the patients.15 A similar association was not seen for FLT3-ITD mutations, which are also common in cases with normal karyotype (96 of 341 [28.2%] men versus 122 of 368 [33.1%] women; P = .167), and thus seems to be specific for NPM1. If confirmed by other groups, this finding might point to sex-specific differences in the mechanism of leukemia development between males and females.
Another important aspect of novel molecular abnormalities is the association with the clinical outcome. When all patients were analyzed, mutant NPM1 was found to be associated with a significantly better prognosis. Due to the very high prevalence of NPM1 mutations in AML with normal karyotype, we confined the analysis of the prognostic value to this patient population. However, we could not observe a major difference in the overall and disease-free survival in cases with normal karyotype between patients with and without NPM1 mutation. Because FLT3-ITD mutations can be found in more than 40% of the NPM1-mutated patients, we asked whether the presence or absence of FLT3-ITD has an impact on the prognosis in NPM1-mutant cases. As shown in Figures 5, 6 for patients with AML and normal karyotype, cases showing NPM1 mutations alone were found to have a significantly better OS and DFS as well as a lower cumulative incidence of relapse (Figure 6). Because autologous and allogeneic transplantation was performed in many of these patients, this might be a consequence of a higher rate of allogeneic stem cell transplantation. However, we did not find an increased number of patients with NPM1 mutations and allogeneic transplantation in first remission; most patients in this cohort received either chemotherapy or autologous stem cell transplantation (Table 6).
Comparison of SCT procedures performed in first CR in patients who had de novo AML, were 60 years old or younger and had normal karyotype (N = 154)
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 44 | 28 | 25 | 57 |
| Autologous, no. | 29 | 18 | 13 | 25 |
| Allogeneic, family, no. | 15 | 8 | 10 | 27 |
| Allogeneic, unrelated, no. | 0 | 2 | 2 | 5 |
. | NPM1-mut/FLT3-ITDneg . | NPM1-mut/FLT3-ITDpos . | NPM1-wt/FLT3-ITDpos . | NPM1-wt/FLT3-ITDneg . |
|---|---|---|---|---|
| No. | 44 | 28 | 25 | 57 |
| Autologous, no. | 29 | 18 | 13 | 25 |
| Allogeneic, family, no. | 15 | 8 | 10 | 27 |
| Allogeneic, unrelated, no. | 0 | 2 | 2 | 5 |
Why should patients with NPM1 mutations have a better prognosis than patients without this abnormality? NPM1 is involved in a complex way in stability, cellular distribution, and function of p53 and p19ARF.20,24-27 Li and coworkers recently described that wt-NPM1 protects hematologic cells from p53-induced apoptosis in conditions of cellular stress,42 an effect that appears to be regulated by the level of genotoxic stress.43 Thus it is tempting to speculate that failure of the mutant NPM1 to protect cells renders them more susceptible to high-level genotoxic stress induced by chemotherapy. In contrast, in patients who had acquired an additional FLT3-ITD mutation, the antiapoptotic and proproliferative pathways induced by FLT3-ITD, especially via STAT5, might dominate the leukemic phenotype.44 This interaction might explain why 2 other groups that did not separate NPM1 and FLT3-ITD mutations did not see differences in survival in NPM1-mutated cases.35,38
It is still unclear how the mutant NPM1 contributes to leukemogenesis. Very recently, Grisendi and coworkers showed that NPM1 knockout results in embryonic lethality between days E11.5 and E12.5 with developmental defects of forebrain and yolk sac hematopoiesis.45 This phenotype was associated with a hyperactive p53 and could be rescued in a p53 knockout background. Interestingly, haplodeficiency for NPM1 resulted in an MDS-like disease with abnormal platelet counts and dysplastic megakaryopoiesis in most mice investigated, which is in contrast to our clinical data, where NPM1 mutations were mostly seen in de novo AML cases. In vitro analyses indicated that cultured NPM1-haplodeficient cells have a reduced replication rate but after prolonged propagation overcame senescence and acquired an immortal phenotype.45 Taken together, these data strongly suggest that a deregulated p53 pathway is dominantly involved in the development in NPM1-mutated cases. It also points to the necessity of additional mutations. In support of this, a high rate of FLT3-TKD mutations was observed in NPM1-positive cases, supporting the classical 2-hit model.2 It will be interesting to see whether NPM1 mutations and FLT3-TKD mutations cooperate in leukemogenesis and increase the transforming potential of the otherwise less-transforming FLT3-TKD mutations.44,46
Taken together, mutations in NPM1 exon 12 can be found in many patients with AML and normal karyotype and appear to represent an independent subgroup of acute myeloid leukemias. The screening method used allows the simultaneous detection of NPM1 and FLT3-ITD mutations and therefore enables the rapid characterization of important prognostic parameters. In addition, patients with NPM1 mutations might be more susceptible to cytostatic treatment strategies. During the review of this work, 3 independent groups reported very similar results on smaller and more selected cohorts.47-49 Taken together, these data indicate that NPM1 mutations may be an important prognostic factor that may be used to stratify treatment of AML patients, especially patients with normal karyotype.
Appendix
We thank the following physicians of the German DSIL study group who entered their patients into the trial: D. Huhn, O. Knigge (Universitätsklinikum Charité, Campus Virchow, Berlin); E. Späth-Schwalbe, S. Hesse-Amojo (Krankenhaus Spandau, Berlin); O. Rick, W. Siegert (Charité Campus Mitte, Berlin); E. Thiel, L. Uharek (Universitätsklinikum Charité, Campus Benjamin-Franklin, Berlin); R. Kolloch, U. Krümpelmann (Krankenanstalten Gilead, Bielefeld); K.-H. Pflüger, T. Wolff (Evang. Diakonissenanstalt Bremen); H.-H. Heidtmann (St Joseph-Hospital, Bremerhaven); F. Marquard (Allgemeines Krankenhaus, Celle); M. Hähnel, F. Fiedler, R. Herbst (Krankenhaus Küchwald, Chemnitz); M. Gramatzki, G. Helm (Universitätsklinikum, Erlangen); J.-G. Saal (Malteser Krankenhaus, Flensburg); H.-G. Höffkes, M. Arland (Städtisches Klinikum, Fulda); E. Faβhauer (St Elisabeth-Krankenhaus, Halle); N. Schmitz, P. Dreger (Allgemeines Krankenhaus St Georg, Hamburg); H. Schmidt, K. Buhrmann (Kreiskrankenhaus, Hameln); H. Dürk (St Marien-Hospital, Hamm); M. Burk (Klinikum Stadt, Hanau); A.-D. Ho, U. Mahlknecht (Universitätsklinikum, Heidelberg); A. Bartholomäus (St Bernward Krankenhaus, Hildesheim); A. A. Fauser (Klinik f. Hämatologie/Onkologie und KMT, Idar-Oberstein); H. Link, F.-G. Hagmann (Westpfalzklinikum, Kaiserslautern); G. Köchling (Kreiskrankenhaus, Leer); K.-P. Schalk (St Vincent-Krankenhaus, Limburg/Lahn); S. Fetscher (Städtisches Krankenhaus Süd, Lübeck); T. Wagner (Universitätsklinikum, Lübeck); A. Neubauer (Universitätsklinikum, Marburg); H. Bodenstein, J. Tischler (Klinikum Minden, Minden); H. Pohlmann, N. Brack (Städtisches Krankenhaus München Harlaching, München); M. Wilhelm, H. Wandt, K. Schäfer-Eckart, (Städtisches Klinikum, Nürnberg); B. Seeber (Klinikum Offenbach, Offenbach); F. Hirsch (Kreiskrankenhaus, Offenburg); T. Geer, H. Heiβmeyer (Diakonie-Krankenhaus, Schwäbisch-Hall); J. Labenz (Ev. Jung-Stilling-Krankenhaus, Siegen); J. Kaesberger (Diakonissen-Krankenhaus, Stuttgart); W. E. Aulitzky, L. Leimer (Robert-Bosch-Krankenhaus, Stuttgart); M. R. Clemens, R. Mahlberg (Mutterhaus der Borromaerinnen, Trier); R. Schwerdtfeger (Deutsche Klinik für Diagnostik, Wiesbaden); R. Engberding, R. Winter (Stadtkrankenhaus, Wolfsburg); M. Sandmann (Klinikum St. Antonius, Wuppertal); H. Einsele; F. Weissinger, H. Rückle-Lanz (Universitätsklinikum, Würzburg).
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-08-3167.
A complete list of the participating institutions and members of the Deutsche Studieninitiative Leukämie (DSIL) appears in the “Appendix.”
Supported in part by grants from the Deutsche Krebshilfe (70-2210-Eh5) (G.E.) and by the Kompetenznetzwerk Akute und Chronische Leukämien sponsored by the Bundesministerium für Bildung und Forschung (BMBF).
C.T., S.K., E.C., and C.S. performed the experiments; T.I., M.S., and G.E. coordinated the clinical study; C.T. wrote the manuscript; and all authors checked the final version of the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank all participating centers of the German DSIL study group (formerly SHG) (see “Appendix”). S. Soucek and L. Lerche provided excellent help in statistical analyses. The skilful technical assistance by M. Karger, M. Hartwig, U. Löwel, J. Bornhäuser, and C. Busse is highly acknowledged.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal