Abstract
We analyzed minimal residual disease (MRD) by consensus polymerase chain reaction (PCR), quantitative PCR (qPCR), and flow cytometry in 40 patients with chronic lymphocytic leukemia (CLL) who underwent stem cell transplantation; 97.4%, 89%, and 100% of the patients could be studied by consensus PCR, qPCR, and flow cytometry, respectively. Overall, 164 of 248 samples were negative for MRD by consensus PCR. Among those, CLL cells were detected by qPCR and by flow cytometry in 77 (47%) and 39 (23%) of the 164 samples, respectively. All 84 samples positive on PCR had detectable CLL cells by qPCR and flow cytometry. A good correlation was seen between MRD levels by flow cytometry and by qPCR (n = 254; r = 0.826; P < .001). Fifteen of 25 patients receiving autografts suffered a relapse, with increasing levels of MRD being observed before relapse in all of them. MRD detection within the first 6 months after autologous transplantation identified patients with a high relapse risk. In contrast, in allografted patients (n = 15) MRD did not correlate with outcome. In conclusion, quantitative methods to assess MRD (flow cytometry and qPCR) are more accurate than consensus PCR to predict clinical evolution. These results might be useful to investigate treatments aimed at preventing relapse in patients with CLL who have received an autograft.
Introduction
Classically, treatment of chronic lymphocytic leukemia (CLL) was based on rather ineffective treatments such as alkylating agents, resulting in a modest complete response (CR) rate and a slight improvement, if any, in survival. The past 2 decades, however, have witnessed a revolution in CLL therapy. Thanks to the combined use of purine analogs and other cytotoxic agents or monoclonal antibodies, CRs are being obtained in a large proportion of patients.1-3 Furthermore, stem cell transplants are being increasingly used in patients with CLL, also resulting in a high CR rate.4,5 In some of these responses, no evidence of minimal residual disease (MRD) can be demonstrated, and, importantly, there is some indication that MRD– status might translate into longer survival.6-8
A number of methods to detect residual tumor cells have been developed.9,10 In CLL, MRD has been estimated mainly by flow cytometry.8,11-13 Alternatively, polymerase chain reaction (PCR)–based techniques, using either consensus14,15 or specific allele complementarity determining region 3 (CDR3) primers,16,17 have also been used. However, there is limited information on the applicability of each of these methods, their relative merits, and clinical significance.18
Against this background, we have undertaken a study with 2 main objectives: (1) to compare flow cytometry, PCR, and quantitative PCR (qPCR) in the assessment of MRD and (2) to determine the clinical impact of MRD monitoring in patients with CLL who have received a stem cell transplant.
Patients, materials, and methods
Patients
Forty patients, 24 men and 16 women, with a median age of 47.5 years (range, 29-61 years) with CLL who underwent stem cell transplantation (SCT) between 1991 and 2004 at the Hospital Clinic of Barcelona were included in the study. Diagnosis of CLL was made according to National Cancer Institute–Working Group (NCI-WG) criteria.19 Fifteen patients received an allogeneic transplant (from a HLA-identical sibling donor in 13 cases, and from an unrelated donor in 2 cases) and 25 patients received an autologous transplant. The main characteristics of the patients are shown in Table 1. Clinical results from 32 of 40 patients included in this report have been previously reported elsewhere.12 The median follow-up after the procedure was 47 months (range, 14-107 months) in the autograft group and 71 months (range, 5.5-163 months) in allograft patients.
Main pretransplantation characteristics of patients
. | Autotransplant . | Allotransplant . |
|---|---|---|
| No. patients | 25 | 15 |
| Median age, y (range) | 48 (29-61) | 46 (29-55) |
| No. men | 17 | 7 |
| Median time from diagnosis to transplantation, mo (range) | 29 (11-183) | 30 (6-100) |
| Median no. lines of therapy (range) | 2 (1-5) | 2 (1-6) |
| Rai stage status at transplantation | ||
| 0 | 19 | 10 |
| I-II | 4 | 3 |
| III-IV | 2 | 2 |
| Bone marrow infiltration greater than 30% | 5/24 | 8/10 |
| IGVH mutational status* | ||
| Mutated | 4 | 7 |
| Unmutated | 18 | 7 |
| Conditioning regimen, no. patients | ||
| Myeloablative regimen | ||
| Cy/TBI | 21 | 13 |
| BEAM | 4 | — |
| Nonmyeloablative regimen | ||
| Flu/Melph | — | 1 |
| Flu/TBI | — | 1 |
. | Autotransplant . | Allotransplant . |
|---|---|---|
| No. patients | 25 | 15 |
| Median age, y (range) | 48 (29-61) | 46 (29-55) |
| No. men | 17 | 7 |
| Median time from diagnosis to transplantation, mo (range) | 29 (11-183) | 30 (6-100) |
| Median no. lines of therapy (range) | 2 (1-5) | 2 (1-6) |
| Rai stage status at transplantation | ||
| 0 | 19 | 10 |
| I-II | 4 | 3 |
| III-IV | 2 | 2 |
| Bone marrow infiltration greater than 30% | 5/24 | 8/10 |
| IGVH mutational status* | ||
| Mutated | 4 | 7 |
| Unmutated | 18 | 7 |
| Conditioning regimen, no. patients | ||
| Myeloablative regimen | ||
| Cy/TBI | 21 | 13 |
| BEAM | 4 | — |
| Nonmyeloablative regimen | ||
| Flu/Melph | — | 1 |
| Flu/TBI | — | 1 |
Cy/TBI indicates cyclophosphamide 120 mg/kg and total body irradiation (13 Gy); BEAM, bischloroethylnitrosourea (BCNU; 300 mg/m2, ×1 day), etoposide (200 mg/m2, ×4 days), cytarabine (400 mg/m2, ×4 days), melphalan (140 mg/m2, ×1 day); Flu/Melph, fludarabine (150 mg/m2) and melphalan (140 mg/m2); Flu/TBI, fludarabine 90 mg/m2 and total body irradiation (2 Gy); and —, not applicable.
Mutational status is available in 22 patients receiving an autologous stem cell transplant and in 14 receiving an allogeneic stem cell transplant.
This study was approved by the Ethical Committee of the University of Barcelona Hospital Clinic. Informed consent was provided according to the Declaration of Helsinki.
Response criteria
Response was assessed according to NCI-WG on CLL guidelines19 3 months after SCT. In patients achieving either CR or partial response (PR), the presence of MRD was assessed by means of molecular and cytofluorometry techniques.
MRD was analyzed in samples from peripheral blood or bone marrow or both obtained before transplantation and, as a rule, every 3 months during the first year after transplantation and every 12 months thereafter. MRD was also studied when clinically indicated (eg, disease progression). Clinical relapse was defined as an increase of at least 50% in the size of the lymph nodes, spleen, or liver if they were previously enlarged or the detection of enlargement; an increase of at least 50% in the number of circulating lymphocytes; or a transformation to a more aggressive histology, following the NCI-WG criteria.
Consensus PCR for the IGVH
gDNA was isolated from mononuclear cells or whole blood using standard procedures. Amplification of DNA was performed with a consensus primer from the framework 3 (FR3) region of the variable region and a primer from the 3′ portion of the joining region (JH) of the immunoglobulin heavy chain gene (IgH).14 PCR clonal products observed between 80 and 120 bp in length were either identified under UV light after staining of an acrylamide gel with ethidium bromide or by GeneScan using a FAM-labeled JH primer. In posttransplantation samples, the result was considered monoclonal if the clonal product had the same size of that observed at diagnosis.
Amplification and sequencing of the IGVH
To obtain the variable (VH), diversity (DH), and joining (JH) segments of the IgH, 200 ng DNA was amplified in 6 PCRs with primers from the framework 1 (FR1) from each of the VH families combined with one JH consensus primer following the BIOMED-2 protocol.10 Clonal FR1-IgH PCR products were purified using a rapid PCR purification system (Marligen Biosciences, Ijamsville, MD) and directly sequenced, in both directions to avoid misreading errors, using the BigDye Terminator Cycle Sequencing kit (Big Dye 3.1; Applied Biosystems, Foster City, CA) in an automated DNA sequencer (Abi Prism 3100; Applied Biosystems).
The sequences were aligned with the published VH,DH, and JH germline sequences using IgBlast20 and the International ImMunoGeneTics database.21 For each patient, allele-specific oligonucleotides (ASOs) corresponding to the CDR3 region, the most hypervariable region of the IgH, were designed using the Primer Express software (Applied Biosystems).
ASO real-time quantitative PCR (qPCR)
qPCR was carried out in an Abi Prism 7700 thermal cycler in a total volume of 25 μL containing 500 ng gDNA, 200 nM JH consensus TaqMan probe, and 900 nM of the individual ASO and the appropriate consensus JH primers.22 Reaction conditions were as follows: 50°C 2 minutes, 95°C 10 minutes, and 40 cycles of denaturation at 95°C for 15 seconds followed by a combined annealing-extension step at 60° to 66°C for 30 seconds depending on each ASO (Table 2). To test the quantity and integrity of the DNA, 50 ng was additionally amplified using 100 nM of probe and 200 nM of primers specific for β-actin or albumin. All PCRs were tested in duplicate. However, when the threshold cycle (Ct) differed in more than one cycle, samples were amplified in triplicate.
CDR3 junctional sequences and design of ASOs for each patient
Case . | 5′-VH . | N-DH-N . | JH-3′ . | DH . | JH-3′ . | Taa . |
|---|---|---|---|---|---|---|
| 1 | TGTGCGAGAGA | tcctGATATTGTAGTAGTACCAGCTGCTATgaattactactactacggt | ATGGACGTC | D2-2 | JH6 | 60 |
| 2 | GCGAGAGA | gcaggctatcGGGTATAGCAGgcc | CTTTGACTAC | D6-25 | JH4 | 60 |
| 3 | GCGAGAG | tcgccggaacgcgagaagTACGATTTTTGGAGTGGTTATactcaaagggcgcgt | ACTACTTTGACTAC | D3-3 | JH4 | 60 |
| 4 | GCGAGAGG | ctggctaggatattgtagtagtaccagctgcgcagtggggacGTATTACGATTTTTGGAGTGGTTATTcctcaact | TTTGACTAC | D3-3 | JH4 | 60 |
| 5 | TGTGCACGGATAC | caGATTTTTGGAGTGCTTATTccaccgtgtaa | GTACTTCGATCTC | D3-3 | JH2 | 60 |
| 6 | TGTGCGAGA | cgAGTGCGAAGT | ACAACTGGTTCGACCCC | D1-26 | JH5 | 60 |
| 7 | GCG | ctcccctcgGGATATAGTGCCTACAATT | ACTACTTTGACTAC | D5-12 | JH4 | 60 |
| 8 | GCGTTAG | gtccattaggGTATTACGATTTTTGGAGTGGTTATCAACCCaaatactttgg | CTACGGTATGGACGTC | D3-3 | JH6 | 60 |
| 9 | GCG | agaggatgggGCAGCACTTGGGAagtacgaaga | TACTACTACTACGGTATGGACGTC | D6-13 | JH5 | 60 |
| 10 | TGTGCGAGAGA | tcagTGGCTGCCTATTAat | TACTTTGACTAC | D3-9 | JH4 | 60 |
| 11 | GCGAGAG | ggACGGTTCGGGGAGTTATTgggttatacttggactactacggtatg | GACGTC | D3-10 | JH6 | 60 |
| 12 | TGTGCGAGAG | cTAGTGGGAG | CTATGACTAC | D1-26 | JH4 | 60 |
| 13 | GCAAGAGA | ccgGTATTACGATTTTTGGAGTGGTTATTATgtac | TTGACTAC | D3-3 | JH4 | 65 |
| 14 | GCGAGA | cagcagttcgccccaTATTACGATTTTTGGAGTGGTTcctcttttcgatt | CTACTACTACGGTA | D3-3 | JH6 | 60 |
| 15 | GCGAGAG | tcgcgggagtcGCATATTGTGGTGGTGACTGCTATTggagagagt | ACTACTTTGACTAC | D2-21 | JH4 | 60 |
| 16 | GCGAGA | gatccccGCCATAGCAGCAGCTGGTACccctactactactacggtatg | GACGCT | D6-13 | JH6 | 60 |
| 17 | GCGAACG | gTGTCAGGGGTGACTACgtgtatccg | TCCGGTATGGACGTC | D4-17 | JH6 | 60 |
| 18 | GCGAGACA | tcgactAGGATATTGTAGTAGTACCAGCTGC | TACTACTACTACTACGGTA | D2-2 | JH6 | 60 |
| 19 | GCGAAAGA | ttcggggtatcaGTATTACGATTTTTGGAGTGGTTATTATAacgata | ATTACTACTACTACTACGGTA | D3-3 | JH6 | 60 |
| 20 | GCGAG | gaaggagtcTTACGATTTTTGGAGTGGTTATTtgtcccgt | TACTACTACTACGGTA | D3-3 | JH6 | 60 |
| 21 | GCGAGA | actttgagaattccGTATTACTATGATAGTAGTGGTTATTATTggggct | ACAAC | D3-22 | JH5 | 60 |
| 22 | GCG | agacggggGGGTATTGTAGTGGTGGTAGCTGCCATT | TGGGACTAC | D2-15 | JH4 | 60 |
| 23 | GCG | agatggtctttcggtTATTGTGCTGGTGGCATCTGttacggtggaaactaccaatactacgctatg | GACGTC | D2-15 | JH6 | 60 |
| 24 | GCGAG | ccgcGATATTGTAGTGGTGGTAGCTGCTACCCCgg | AATACTACTACTACATGGACGTC | D2-15 | JH6 | 66 |
| 25 | TGTATTACTGTGC | cgtaTGTGGTGGTTATTACTATGC | TGACTAC | D2-21 | JH4 | 60 |
| 26 | CTGTGCGAAAGA | tagggatgACTATGGTaattacgt | CTTTGACTAC | D3-10 | JH4 | 60 |
| 27 | GTGAGAGA | tacctccaGTATAGTAGGAGGTGGTCCccttgaccgt | TGG | D6-13 | JH5 | 60 |
| 28 | GCAAGA | gatgcttggcggcccgctcgccccgcGTATTATGATTACGTTTGGGGGAGTTATCGCCAcggccccca | TGATGCTTTTGATATC | D3-16 | JH3 | 60 |
| 29 | ACCACAG | cccgtcggtgcttgcgccccCGATATTTTGACTGGTTATTGGACCcgggggta | TGCTACTACGGTATGGACGTC | D3-9 | JH6 | 60 |
| 30 | GCGAGA | gattcccacttGTATTACGATATTTTGACTGGTTATTCTActgaaatggag | GACTAC | D3-9 | JH4 | 60 |
| 31 | GCGAGAGA | tcgccccggGGGTATTGTAGTAGTACCAGCTGCTATGtgctactacggta | TGGACGTC | D2-2 | JH6 | 60 |
| 32 | GCGGCCG | cTGCAGATTTTTGGAGTGGTTCTa | AAGAC | D3-3 | JH4 | 60 |
| 33 | TGCGAGTG | gTTACTATGATTCGGGGAc | CTACTACGGTGTGGACGTC | D3-10 | JH6 | 60 |
| 34 | GCAAAAGAT | tctgttcagatgAGCAACCCCTGGTACgaaggaggggg | CTTCTGGTTCGACCCC | D6-13 | JH5 | 60 |
| 35 | GCGAGAAA | accaagtTATTACTATGATAGTAGTGGTTATTACTACtgga | ACTACTACTACTACGGTATGGACGTC | D3-22 | JH6 | NF |
| 36 | GCGAAAG | gtgctagTTACTTTGGTTCGGGGAGTTTTTATAACgagcc | CTCTGACCACTGG | D3-10 | JH4 | NF |
Case . | 5′-VH . | N-DH-N . | JH-3′ . | DH . | JH-3′ . | Taa . |
|---|---|---|---|---|---|---|
| 1 | TGTGCGAGAGA | tcctGATATTGTAGTAGTACCAGCTGCTATgaattactactactacggt | ATGGACGTC | D2-2 | JH6 | 60 |
| 2 | GCGAGAGA | gcaggctatcGGGTATAGCAGgcc | CTTTGACTAC | D6-25 | JH4 | 60 |
| 3 | GCGAGAG | tcgccggaacgcgagaagTACGATTTTTGGAGTGGTTATactcaaagggcgcgt | ACTACTTTGACTAC | D3-3 | JH4 | 60 |
| 4 | GCGAGAGG | ctggctaggatattgtagtagtaccagctgcgcagtggggacGTATTACGATTTTTGGAGTGGTTATTcctcaact | TTTGACTAC | D3-3 | JH4 | 60 |
| 5 | TGTGCACGGATAC | caGATTTTTGGAGTGCTTATTccaccgtgtaa | GTACTTCGATCTC | D3-3 | JH2 | 60 |
| 6 | TGTGCGAGA | cgAGTGCGAAGT | ACAACTGGTTCGACCCC | D1-26 | JH5 | 60 |
| 7 | GCG | ctcccctcgGGATATAGTGCCTACAATT | ACTACTTTGACTAC | D5-12 | JH4 | 60 |
| 8 | GCGTTAG | gtccattaggGTATTACGATTTTTGGAGTGGTTATCAACCCaaatactttgg | CTACGGTATGGACGTC | D3-3 | JH6 | 60 |
| 9 | GCG | agaggatgggGCAGCACTTGGGAagtacgaaga | TACTACTACTACGGTATGGACGTC | D6-13 | JH5 | 60 |
| 10 | TGTGCGAGAGA | tcagTGGCTGCCTATTAat | TACTTTGACTAC | D3-9 | JH4 | 60 |
| 11 | GCGAGAG | ggACGGTTCGGGGAGTTATTgggttatacttggactactacggtatg | GACGTC | D3-10 | JH6 | 60 |
| 12 | TGTGCGAGAG | cTAGTGGGAG | CTATGACTAC | D1-26 | JH4 | 60 |
| 13 | GCAAGAGA | ccgGTATTACGATTTTTGGAGTGGTTATTATgtac | TTGACTAC | D3-3 | JH4 | 65 |
| 14 | GCGAGA | cagcagttcgccccaTATTACGATTTTTGGAGTGGTTcctcttttcgatt | CTACTACTACGGTA | D3-3 | JH6 | 60 |
| 15 | GCGAGAG | tcgcgggagtcGCATATTGTGGTGGTGACTGCTATTggagagagt | ACTACTTTGACTAC | D2-21 | JH4 | 60 |
| 16 | GCGAGA | gatccccGCCATAGCAGCAGCTGGTACccctactactactacggtatg | GACGCT | D6-13 | JH6 | 60 |
| 17 | GCGAACG | gTGTCAGGGGTGACTACgtgtatccg | TCCGGTATGGACGTC | D4-17 | JH6 | 60 |
| 18 | GCGAGACA | tcgactAGGATATTGTAGTAGTACCAGCTGC | TACTACTACTACTACGGTA | D2-2 | JH6 | 60 |
| 19 | GCGAAAGA | ttcggggtatcaGTATTACGATTTTTGGAGTGGTTATTATAacgata | ATTACTACTACTACTACGGTA | D3-3 | JH6 | 60 |
| 20 | GCGAG | gaaggagtcTTACGATTTTTGGAGTGGTTATTtgtcccgt | TACTACTACTACGGTA | D3-3 | JH6 | 60 |
| 21 | GCGAGA | actttgagaattccGTATTACTATGATAGTAGTGGTTATTATTggggct | ACAAC | D3-22 | JH5 | 60 |
| 22 | GCG | agacggggGGGTATTGTAGTGGTGGTAGCTGCCATT | TGGGACTAC | D2-15 | JH4 | 60 |
| 23 | GCG | agatggtctttcggtTATTGTGCTGGTGGCATCTGttacggtggaaactaccaatactacgctatg | GACGTC | D2-15 | JH6 | 60 |
| 24 | GCGAG | ccgcGATATTGTAGTGGTGGTAGCTGCTACCCCgg | AATACTACTACTACATGGACGTC | D2-15 | JH6 | 66 |
| 25 | TGTATTACTGTGC | cgtaTGTGGTGGTTATTACTATGC | TGACTAC | D2-21 | JH4 | 60 |
| 26 | CTGTGCGAAAGA | tagggatgACTATGGTaattacgt | CTTTGACTAC | D3-10 | JH4 | 60 |
| 27 | GTGAGAGA | tacctccaGTATAGTAGGAGGTGGTCCccttgaccgt | TGG | D6-13 | JH5 | 60 |
| 28 | GCAAGA | gatgcttggcggcccgctcgccccgcGTATTATGATTACGTTTGGGGGAGTTATCGCCAcggccccca | TGATGCTTTTGATATC | D3-16 | JH3 | 60 |
| 29 | ACCACAG | cccgtcggtgcttgcgccccCGATATTTTGACTGGTTATTGGACCcgggggta | TGCTACTACGGTATGGACGTC | D3-9 | JH6 | 60 |
| 30 | GCGAGA | gattcccacttGTATTACGATATTTTGACTGGTTATTCTActgaaatggag | GACTAC | D3-9 | JH4 | 60 |
| 31 | GCGAGAGA | tcgccccggGGGTATTGTAGTAGTACCAGCTGCTATGtgctactacggta | TGGACGTC | D2-2 | JH6 | 60 |
| 32 | GCGGCCG | cTGCAGATTTTTGGAGTGGTTCTa | AAGAC | D3-3 | JH4 | 60 |
| 33 | TGCGAGTG | gTTACTATGATTCGGGGAc | CTACTACGGTGTGGACGTC | D3-10 | JH6 | 60 |
| 34 | GCAAAAGAT | tctgttcagatgAGCAACCCCTGGTACgaaggaggggg | CTTCTGGTTCGACCCC | D6-13 | JH5 | 60 |
| 35 | GCGAGAAA | accaagtTATTACTATGATAGTAGTGGTTATTACTACtgga | ACTACTACTACTACGGTATGGACGTC | D3-22 | JH6 | NF |
| 36 | GCGAAAG | gtgctagTTACTTTGGTTCGGGGAGTTTTTATAACgagcc | CTCTGACCACTGG | D3-10 | JH4 | NF |
Underlined sequences represent allele-specific oligonucleotides. The lower-case letters indicate the inserted nucleotides.
VH indicates variable segment of the immunoglobulin heavy chain gene; N, nucleotides; DH, diversity segment; JH, joining segment; Taa, annealing temperature; and NF, not found.
The amount of residual CLL cells in follow-up samples was estimated by extrapolating the results obtained by qPCR in a standard curve generated for each patient. These curves were obtained from DNA samples at diagnosis that were diluted into water to reach final dilutions of 100, 10–1, 10–2, 10–3, and 10–4. Standard curves for albumin and β-actin were also generated using commercial gDNA (Applied Biosystems). All standard curves obtained were considered optimal for quantification if the coefficient of correlation was higher than 0.95 and the slope was between 3.0 and 3.9. The results were expressed as a ratio between clonal IgVH and the β-actin or albumin genes.
The specificity of the qPCR for each ASO primer was assessed by the absence of amplification in 3 DNA samples from healthy individuals. In cases with nonspecific amplification, the annealing temperature was increased. To test the sensitivity of the qPCR, serial dilutions of CLL cells (100 to 10–6) into normal blood samples were performed in some patients. The sensitivity achieved was of 10–5 in 6 of the 7 performed dilutional experiments. In the remaining patient, qPCR detected one CLL cell in 1000 cells, this patient being excluded from the qPCR assessment. As a result, MRD negativity was defined as the detection of fewer than 10–5 CLL cells.
Phenotypic detection of MRD
Up until 2001, MRD was studied by using triple antigenic combinations (κ-FITC/λ-PE/CD19-PE-Cy5, CD20-FITC/CD5-PE/CD19-PE-Cy5, CD22-FITC/CD23-PE/CD19-PE-Cy5).12 Afterward, quadruple antigenic combinations were used (CD20-FITC/CD79b-PE/CD19-PerCP Cy5.5/CD5-APC, CD22-FITC/CD23-PE/CD19-PerCP Cy5.5/CD5-APC, and κ-FITC/λ-PE/CD19-PerCP Cy5.5/CD5-APC). In bone marrow samples an additional combination (CD20-FITC/CD38-PE/CD19PerCP Cy5.5/CD5APC) was added to better discriminate CLL cells from precursor B cells.13 Stained samples were acquired in a single step and the signal of 300 000 cells was recorded.23 CLL cells were identified according to their specific phenotype, the proportion of atypical cells being recorded for each of the combinations studied. The sensitivity achieved by flow cytometry was of 10–4.12,13 The correlation of MRD values of CD20-based with CD22-based panels was excellent (correlation of Pearson r = 0.999, P < .001). The MRD value in each experiment was calculated as the mean value obtained with the different antigenic combinations, excluding κ/λ antigens.
Statistical analysis
The correlation coefficient of Pearson was used to compare MRD values detected by qPCR and flow cytometry and between peripheral blood and bone marrow samples. Actuarial curves for relapse risk and overall survival (OS) were obtained according to the Kaplan-Meier method.24 OS was considered from the date of transplantation until the last follow-up. Relapse risk was calculated from the date of transplantation until relapse. Patients dying due to a nonrelated cause were censored at the date of the event. The comparison between MRD subgroups was determined by using the log-rank test.25 P values below .05 were considered statistically significant. The analysis was performed by using the SPSS 11.0 statistical software (Chicago, IL).
Results
Analysis of MRD by consensus PCR, qPCR, and flow cytometry
Forty patients were studied by consensus PCR. In 37 of 38 patients with an available sample at diagnosis, a clonal band was obtained. Two patients were excluded from the analysis due to the lack of DNA sample before transplantation. As a result, consensus PCR was applicable to 97.4% of patients.
By qPCR, 37 patients with an available pretransplantation sample were studied. In 36 cases, a clonal band was successfully sequenced and specific primers designed (Table 2). An efficient, specific, and sensitive qPCR could be obtained in 33 patients, whose follow-up samples were analyzed for the presence of MRD. Therefore, qPCR was applicable to 33 (89%) of 37 patients. Flow cytometry by using triple or quadruple combinations could be applied to all patients.
Detection of MRD by consensus PCR in comparison with qPCR and flow cytometry
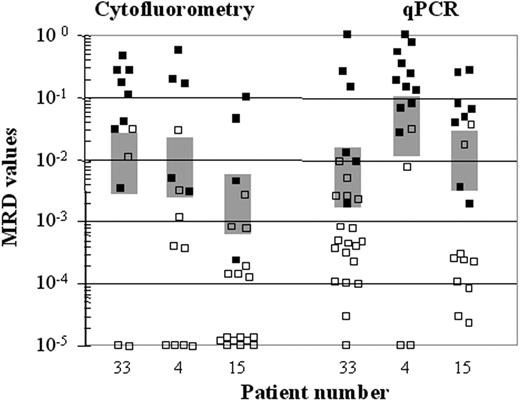
A total of 248 samples from 28 patients were analyzed by the 3 techniques. By consensus PCR, of the 248 samples, 84 were MRD+ and 164 were MRD–. All MRD+ samples by consensus PCR were also found to be MRD+ by qPCR and flow cytometry, whereas among MRD– samples by consensus PCR, CLL cells were detected by qPCR and flow cytometry in 77 (47%) and 39 (23%) of the 164 samples, respectively. MRD levels ranging from 10–2 to 10–3 as detected by quantitative methods were not necessarily detected by consensus PCR (Figure 1).
Comparison of MRD assessment by consensus PCR with flow cytometry and quantitative PCR. MRD assessment by these techniques in samples from 3 representative patients (cases 1, 2, 3); ▪, MRD+ samples by consensus PCR; □, MRD– samples. MRD levels (y-axis) quantified by flow cytometry and quantitative PCR are shown in logarithmic scale. The majority of samples with MRD levels higher than 10–2 were positive by consensus PCR, whereas those with levels lower than 10–3 resulted in MRD– by consensus PCR. There was a zone represented by a light gray box, in which samples were either positive or negative independently of the MRD level.
Comparison of MRD assessment by consensus PCR with flow cytometry and quantitative PCR. MRD assessment by these techniques in samples from 3 representative patients (cases 1, 2, 3); ▪, MRD+ samples by consensus PCR; □, MRD– samples. MRD levels (y-axis) quantified by flow cytometry and quantitative PCR are shown in logarithmic scale. The majority of samples with MRD levels higher than 10–2 were positive by consensus PCR, whereas those with levels lower than 10–3 resulted in MRD– by consensus PCR. There was a zone represented by a light gray box, in which samples were either positive or negative independently of the MRD level.
Correlation of MRD detected by qPCR and flow cytometry
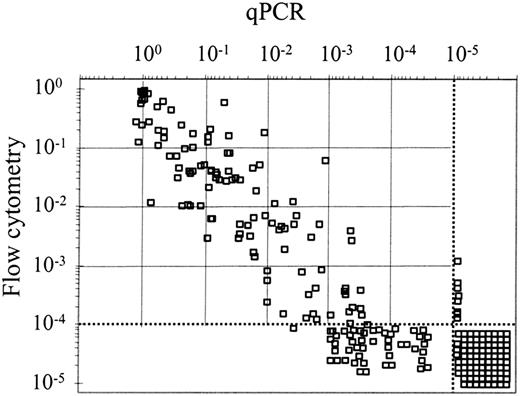
In 254 samples from 30 patients MRD was simultaneously evaluated by qPCR and flow cytometry, with a good correlation (r = 0.826, P < .001) being observed (Figure 2). In 49 samples corresponding to 15 patients, MRD could be detected by qPCR but not by flow cytometry. Thirty-three of these samples had MRD levels higher than 10–4 by qPCR. In contrast, only 8 samples from 3 patients were positive by flow cytometry and negative by qPCR. Moreover, there was a good correlation between MRD values as quantified by qPCR or flow cytometry in 54 samples from 6 dilutional experiments (r = 0.84, P < .001; data not shown).
Levels of MRD according to sample source
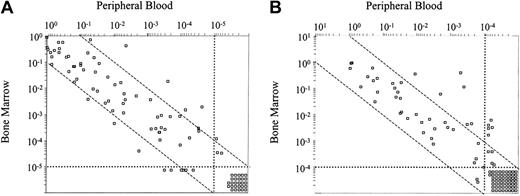
In 103 paired peripheral blood and bone marrow samples from 28 patients analyzed by qPCR, a concordance between results in both sample sites was observed (r = 0.786, P < .001; Figure 3A). However, in 13 samples (12.6%) from 8 cases, MRD values differed in more than one logarithm between both sample sites (6 higher in bone marrow, 7 higher in peripheral blood).
By flow cytometry, 109 paired peripheral blood and bone marrow samples from 32 patients were studied (Figure 3B). Similarly to qPCR, a good correlation was observed in MRD values detected in peripheral blood and bone marrow (r = 0.856, P < .001). Eleven samples from 8 patients presented a difference of more than a logarithm, MRD being higher in bone marrow than in peripheral blood.
In 3 patients, differences of more than one log in MRD levels between peripheral blood and bone marrow were observed by qPCR or flow cytometry or both in the majority of samples analyzed during the study period.
Clinical impact of MRD in autologous transplantation
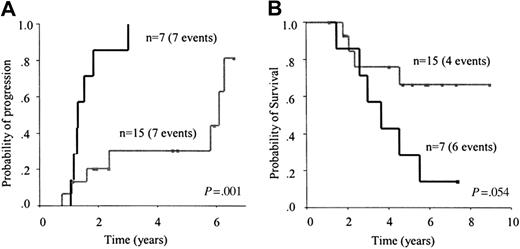
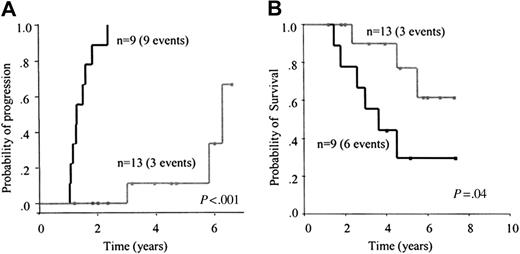
Clinical value of MRD detected by consensus PCR. Twenty-two patients with a monoclonal pattern were evaluated within 3 to 6 months after autologous transplantation. At that time, 7 of them had a PCR monoclonal pattern and 15 a PCR polyclonal pattern. All patients with a monoclonal pattern eventually relapsed (5-year risk, 100%) compared with 7 of 15 patients with a polyclonal pattern (5-year risk, 30%; 95% CI, 4%-56%; P = .001; Figure 4A). The median time to progression for patients with a monoclonal pattern was 16 months versus 73 months for patients with a polyclonal pattern. Although no significant differences were observed, a trend for a longer survival was observed in patients with a polyclonal pattern 3 to 6 months (early) after transplantation (P = .054; Figure 4B).
Over the course of follow-up, 7 patients switched from a polyclonal to a monoclonal pattern (median, 23 months; range, 11-48 months). The detection of a monoclonal pattern at any time after autografting anticipated clinical relapse, the median time from conversion to a monoclonal pattern to clinical relapse being 14 months (range, 6-76 months).
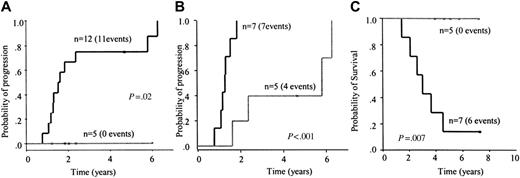
Clinical value of MRD detected by qPCR. At 3 to 6 months after autologous transplantation, MRD values ranged from less than 10–5 to 7.1 × 10–1, with 5 of 17 patients having MRD levels lower than 10–5. Patients with MRD 10–5 or greater had a higher relapse risk than those with MRD less than 10–5 (5-year risk, 75%; 95% CI, 50%-99% versus 0%, P = .02; Figure 5A), with a median time to progression of 19 months versus not reached, respectively.
MRD+ patients could be separated into 2 different risk groups based on the amount of residual disease (≥ 10–3 and < 10–3 to ≥ 10–5; Figure 5B). Patients with MRD greater than 10–3 progressed faster than patients with MRD levels between 10–3 and 10–5 (15 months versus 70 months, respectively; P = .001). In these 2 groups, significant differences in OS were also observed (36 months versus not reached, P = .001; Figure 5C).
During follow-up, one of the 5 MRD– patients became MRD+ at 60 months after transplantation; 18 months later, this patient continues to be in CR. The detection of MRD by qPCR at any time after autologous transplantation anticipated clinical relapse (median time to relapse, 24 months; range, 6-82 months).
Clinical value of MRD detected by flow cytometry. At 3 to 6 months after autologous transplantation, MRD values in 22 evaluable patients ranged from less than 10–4 to 4.7 × 10–2. Thirteen patients showed no MRD and 9 presented MRD levels ≥ 10–4. The relapse risk was significantly higher in patients with MRD of 104 or greater than in those with undetectable MRD (5-year risk, 100% versus 11%; 95% CI, 0%-31%, respectively; P < .001; Figure 6A). The median times to progression were 16 months and 75 months, respectively.
Patients with MRD levels less than 10–4 had a significantly longer survival than those with MRD levels of 10–4 or more (not reached versus 43 months, respectively; P = .04; Figure 6B).
Correlation of MRD levels assessed by quantitative PCR and flow cytometry. In 254 samples from 30 patients, quantification of CLL cells (expressed in logarithmic scale) measured by quantitative PCR and flow cytometry correlated well (r = 0.826, P < .001). The sensitivity achieved by each of these techniques are labeled with dotted lines. MRD+ samples (n = 49) by qPCR and negative by flow cytometry and those MRD– samples (n = 8) by qPCR but positive by flow cytometry are depicted out of the dotted lines. MRD– samples (n = 86) by the 2 techniques are shown below and to the right of the dotted lines.
Correlation of MRD levels assessed by quantitative PCR and flow cytometry. In 254 samples from 30 patients, quantification of CLL cells (expressed in logarithmic scale) measured by quantitative PCR and flow cytometry correlated well (r = 0.826, P < .001). The sensitivity achieved by each of these techniques are labeled with dotted lines. MRD+ samples (n = 49) by qPCR and negative by flow cytometry and those MRD– samples (n = 8) by qPCR but positive by flow cytometry are depicted out of the dotted lines. MRD– samples (n = 86) by the 2 techniques are shown below and to the right of the dotted lines.
Relationship between the number of CLL cells detected in peripheral blood and the degree of bone marrow infiltration in patients with CLL who received stem cell transplants. Paired peripheral blood and bone marrow were analyzed in 103 samples by qPCR (A) and in 109 samples by flow cytometry (B). A good correlation was observed between peripheral blood and bone marrow levels assessed by qPCR and flow cytometry, with correlation coefficients of 0.786 and 0.856, respectively (P < .001). Samples with an MRD amount differing more than one logarithm between both sites are depicted out of the dashed lines.
Relationship between the number of CLL cells detected in peripheral blood and the degree of bone marrow infiltration in patients with CLL who received stem cell transplants. Paired peripheral blood and bone marrow were analyzed in 103 samples by qPCR (A) and in 109 samples by flow cytometry (B). A good correlation was observed between peripheral blood and bone marrow levels assessed by qPCR and flow cytometry, with correlation coefficients of 0.786 and 0.856, respectively (P < .001). Samples with an MRD amount differing more than one logarithm between both sites are depicted out of the dashed lines.
During follow-up, 8 of 14 patients became MRD+. The detection of MRD by flow cytometry anticipated clinical relapse (median time to relapse, 22 months; range, 8-65 months).
Clinical impact of MRD assessment in allogeneic transplantation
MRD status prior to transplantation was available in 13 of 15 patients, of whom only one was MRD–. MRD positivity levels ranged from 7.8 × 10–4 to 1.0. Early (3-6 months) after transplantation, a monoclonal PCR was found in 2 of 13 patients, a positive qPCR in 10 of 13 patients, and a positive flow cytometry study in 5 of 11 patients. MRD detection at this early time point did not predict clinical relapse.
Over time, decreasing MRD levels as assessed by qPCR or flow cytometry or both were seen in 6 patients. In 5 of them, a switch from MRD+ to MRD– status was observed within the first year after transplantation. In the remaining patient, intermittent MRD levels (3.65 × 10–4 to < 10–5) were detected, although at the last follow-up (+60 months) MRD was negative. On the contrary, none of the 3 MRD– patients has become MRD+.
Among MRD persistently positive patients (n = 6), 2 different MRD kinetics were seen. In 4 cases, MRD levels remained stable by qPCR or flow cytometry or both; none of these patients has had a relapse at 6, 48, 72, and 84 months after transplantation. In 2 patients, increasing MRD levels were observed; one of these patients had a relapse at 53 months after transplantation and the remaining patient continues in clinical CR at the last follow-up (+60 months). In addition, another patient presented a late relapse (+120 months) at an extranodal site without detection of residual disease in peripheral blood and bone marrow.
Discussion
Over the past 2 decades, important progress has been made in the treatment of CLL. As a result, not only do a larger proportion of patients achieve CR, but in some of these responders there is no evidence of persistent leukemic cells. This is important because achieving MRD– status has been correlated with a longer survival,1,6,8,11-14,26 although the possibility that patients achieving MRD– status constitute a population with an intrinsically better prognosis cannot be excluded.27
Progression and OS. Risk of progression (A) and OS (B) according to the result of early assessment of MRD, at 3 to 6 months, after autologous transplantation by consensus PCR. The subset of patients with a negative MRD result (gray line) showed a significantly lower relapse risk and a trend to a longer survival compared with patients with disease positive for MRD (black line).
Progression and OS. Risk of progression (A) and OS (B) according to the result of early assessment of MRD, at 3 to 6 months, after autologous transplantation by consensus PCR. The subset of patients with a negative MRD result (gray line) showed a significantly lower relapse risk and a trend to a longer survival compared with patients with disease positive for MRD (black line).
To determine MRD, different techniques are used. Consensus PCR is a qualitative method with variable sensitivity (1 cell in 102-104 normal cells),14,17 whereas qPCR allows for an accurate quantification of MRD with a reported sensitivity of 1 in 104 to 105 cells,16,17 although the technique is complex, time consuming, and expensive to prepare. On the other hand, multiparameter flow cytometry is considered to have a sensitivity of 1 in 104 cells and is available to most centers on a regular basis.12,13,17
The relative merits of each of the techniques to assess MRD in patients with CLL who have received a transplant have only been analyzed in one study,17 which concluded that qPCR and flow cytometry are equally useful to measure MRD. In that study, however, the clinical usefulness of these techniques could not be demonstrated, most likely due to the short follow-up and the small number of cases in which clinical correlations could be established. Apart from this study, there are no other reports comparing the different techniques to assess MRD in the same group of patients with CLL and, in particular, correlating such studies with clinical outcome.
We have compared MRD assessment by consensus PCR, qPCR, and flow cytometry in 40 patients with CLL consecutively submitted to stem cell transplantation in our center. Also, such results were correlated with the clinical outcome to ascertain which technique is most convenient and reliable from the clinical standpoint.
Flow cytometry could be applied in 100% of the patients and PCR in 97.4%. That small difference (1 case) is due to the fact that PCR depends on the acquisition of a clonal product in samples at diagnosis. Nevertheless, the proportion of patients in our study that could be investigated by PCR was higher than the 73% reported in another study,14 It is worth noting that a high proportion of patients from our series (70%) had unmutated IGVH genes. IGVH mutations along with the length of the amplified fragment have recently been reported to be possible pitfalls for consensus PCR.17 In addition, 4 patients (11%) in our study could not be followed by qPCR, due to the impossibility of obtaining the CDR3 sequence or of developing an efficient and sensitive qPCR assay.
Clinical outcome after autologous transplantation according to early assessment of MRD, at 3 to 6 months, by quantitative PCR. Patients with undetectable MRD (gray line) had a significantly lower risk of relapse compared with patients with detectable MRD (black line). Among MRD+ patients, 2 different groups of risk were observed according to MRD level. Thus, those patients with MRD 10–3 or greater (black line) had a significantly higher relapse risk (B) and shorter survival (C) than patients with MRD level between 10–3 and 10–5 (gray line).
Clinical outcome after autologous transplantation according to early assessment of MRD, at 3 to 6 months, by quantitative PCR. Patients with undetectable MRD (gray line) had a significantly lower risk of relapse compared with patients with detectable MRD (black line). Among MRD+ patients, 2 different groups of risk were observed according to MRD level. Thus, those patients with MRD 10–3 or greater (black line) had a significantly higher relapse risk (B) and shorter survival (C) than patients with MRD level between 10–3 and 10–5 (gray line).
Our study demonstrates that MRD levels assessed by qPCR and flow cytometry yield concordant results in the majority of samples and patients. Moreover, the proportion of CLL cells detected in paired peripheral blood and bone marrow samples from 28 patients assessed by qPCR and 32 patients assessed by flow cytometry was very similar. In only 3 patients were differences of more than 1 log in MRD levels between peripheral blood and bone marrow consistently observed (2 higher in bone marrow, 1 higher in peripheral blood). Therefore, as a rule, there is a very good correlation between results obtained in peripheral blood and bone marrow. Because of this, and for patients' convenience, peripheral blood should be the first site investigated when MRD is assessed, with bone marrow giving additional information in some cases. This may be particularly true in those patients treated with monoclonal antibodies, because a study has recently described how bone marrow yields more reliable results than peripheral blood during or after treatment with alemtuzumab.28 None of our patients had been treated with monoclonal antibodies.
With respect to the prognostic value, a correlation between MRD detection and clinical relapse in autografted patients has already been demonstrated by us and others in a number of studies,7,12,13,15 with such a correlation being confirmed in this larger series. Although it could be argued that patients achieving MRD– status have an intrinsically less aggressive disease and, hence, with a better prognosis,8 it is important to note that patients included in this series had, by definition and according to the treatment criteria, poor prognosis.
Clinical outcome after autologous transplantation according to early assessment of MRD, at 3 to 6 months, by flow cytometry. Patients with undetectable MRD (gray line) had a significantly lower risk of relapse and longer survival compared with patients with detectable MRD (black line).
Clinical outcome after autologous transplantation according to early assessment of MRD, at 3 to 6 months, by flow cytometry. Patients with undetectable MRD (gray line) had a significantly lower risk of relapse and longer survival compared with patients with detectable MRD (black line).
In the present study, we have also shown that despite moderate differences in their sensitivity, PCR, qPCR, and flow cytometry are all able to identify patients likely to progress after autotransplantation. Thus, MRD+ patients had a significantly higher relapse risk than MRD– patients. In fact, MRD detection as early as 3 to 6 months after transplantation predicted relapse. Of note, the higher the MRD level as determined by qPCR, the more rapid the clinical progression. Thus, patients with a higher MRD had a higher relapse risk and, more importantly, a shorter survival than those with lower MRD levels. Interestingly, similar observations have recently been reported at the Annual Meeting of the American Society of Hematology.18
None of the 5 patients with undetectable MRD after autologous transplantation has had a relapse at the time of this report, with the follow-up after transplantation ranging from 15 to 72 months. This observation, although based on a small group of patients, underlines the importance of achieving MRD– status in patients with CLL.6,8
In contrast to what is observed in autografts, allogeneic transplantation results in long-term disease control in some patients, suggesting that they can be clinically cured.29-32 There is now robust evidence that the graft-versus-leukemia effect makes possible the coexistence of some residual leukemic cells with a prolonged clinical CR.12,33,34
To summarize, this study shows that MRD correlates with time to progression and survival in patients with CLL submitted to autologous transplantation. We have also demonstrated that MRD detection as early as 3 to 6 months after autotransplantation allows the identification of patients who will progress. More importantly, time to progression and survival in patients receiving autografts depend on the MRD levels, quantitative methods (ie, qPCR, flow cytometry) identifying patients with different risks. This information provides background for designing clinical trials (eg, maintenance therapy with monoclonal antibodies) aimed at preventing relapse. In contrast, in allografted patients MRD detection has limited value, and no treatment interventions in these patients seem justified based only on MRD status. Finally, our results indicate that quantitative techniques are superior to qualitative methods in the assessment of MRD in CLL, and that in most cases, flow cytometry applied to peripheral blood provides useful information on MRD status.
Prepublished online as Blood First Edition Paper, January 31, 2006; DOI 10.1182/blood-2005-09-3634.
Supported by grants 01/1581, 04/1051, Red de Genómica de Cáncer 03/10 and Red Estudio de Neoplasias linfoides 03/179, Instituto Nacional de Salud Carlos III-Ministerio de Sanidad from Fondo de Investigaciones Sanitarias (Spain), and the José Carreras International Leukemia Foundation (EM-04; CR-04). C.M. holds a contract from the Ministerio de Sanidad (CM-04/00 187).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal