Abstract
The involvement of the transcription factor c-Myb in promoting the proliferation and inhibition of erythroid cell differentiation has been established in leukemia cell models. The anemia phenotype observed in c-myb knockout and knockdown mice highlights a critical role for c-Myb in erythropoiesis. However, determining the reason for the failure of erythropoiesis in these mice and the precise function of c-Myb in erythroid progenitors remains elusive. We examined erythroid development under conditions of reduced c-Myb protein levels and report an unexpected role for c-Myb in the promotion of commitment to the erythroid lineage and progression to erythroblast stages. c-myb knockdown erythroid colony-forming unit (CFU-E) stage progenitors displayed an immature phenotype and aberrant expression of several hematopoietic regulators. To extend our findings, we analyzed the response of normal enriched erythroid progenitors to inducible disruption of a floxed c-myb allele. In agreement with the c-myb knockdown phenotype, we show that c-Myb is strictly required for expression of the c-Kit receptor in erythroid cells.
Introduction
The first cells committed to erythropoiesis, as defined by in vitro colony assays, are erythroid burst-forming units (BFU-Es), followed by erythroid colony-forming units (CFU-Es).1 The CFU-E undergoes a series of 3 to 6 cell divisions, giving rise to several stages of maturing erythroblasts that become hemoglobinized and extrude their nuclei to form reticulocytes and red blood cells. The development of red blood cells depends on extrinsic signals mediated by cytokines and microenvironmental factors. Thus, the interactions between the c-Kit receptor tyrosine kinase and stem cell factor (SCF) and between the erythropoietin receptor (EpoR) and erythropoietin (EPO) are crucial for survival, expansion, and differentiation of erythroid progenitors.2,3 c-Kit is expressed on immature cells up to the CFU-E stage,4 whereas EpoR is restricted to the erythroid lineage up to later erythroblast stages.5
Analysis of mice carrying targeted mutations of transcription factor genes has revealed the importance of the regulation of erythroid development at the level of transcription. For instance, SCL is essential for the commitment and differentiation of erythroid progenitors,6,7 whereas GATA-2 promotes the survival and proliferation of immature cells8 and GATA-1 is crucial for commitment, differentiation, and survival at later stages.9,10 In contrast, the Ets factor PU.1 inhibits erythroid development through antagonism with GATA-1, whereas it promotes the proliferation of erythroid progenitors.11,12
The transcription factor c-Myb is expressed at high levels in immature progenitors of all hematopoietic lineages and is involved in the regulation of proliferation, differentiation, and survival.13 During erythropoiesis, c-Myb expression is highest in CFU-Es and early erythroblasts.14 Extensive work on erythroleukemic cells, which are arrested close to the CFU-E stage, showed that c-Myb can act as an inhibitor of terminal erythroid differentiation.15 Embryos homozygous for a c-myb null allele (c-myb–) contain no definitive erythrocytes and die of anemia around embryonic day 15 (E15).16 Immature hematopoietic progenitors in the fetal liver of c-myb–/– embryos are unable to expand or develop into mature blood cells.17 Recently, a knockdown allele (c-mybKD) was generated in which a neomycin resistance cassette in intron 6 reduced expression to 5% to 10% of wild-type levels. Analysis of knockdown mice/embryos revealed requirements for c-Myb at key stages of hematopoiesis and suggested that erythroid development is sensitive to changes in c-Myb levels.18 Studies on mice carrying c-myb alleles with point mutations that compromise c-Myb function have confirmed the importance of this factor in erythropoiesis.19,20 However, major questions remain concerning the role of c-Myb in erythroid cells. How do reduced protein levels of c-Myb lead to anemia? Is the function of c-Myb in erythroid cells related to the control only of proliferation and not of differentiation? Furthermore, the molecular mechanism of c-Myb action is poorly understood in that characterized target genes13,21 cannot fully account for the phenotypic observations in these mouse models.
We have analyzed the phenotype of c-mybKD/KD fetal liver erythropoiesis in detail and provide evidence of a novel role for c-Myb in the promotion of commitment to and progression through erythropoiesis. In addition, our data suggest a differential requirement for c-Myb in the proliferation of early and late progenitors. To extend our findings, we established an approach for the inducible inactivation of a floxed c-myb allele in cultured erythroid progenitors and use this to confirm a strict requirement for c-Myb in the expression of the c-Kit receptor in erythroid cells.
Materials and methods
Mice
Mice were maintained on a C57/BL6 × 129Sv background. We have previously described the generation of the c-mybKD and c-mybF alleles.18 The c-myb– null allele16 was kindly provided by Michael Mucenski. The MxCre strain22 was obtained from the Jackson Laboratories. The c-myb alleles were genotyped as described.18 The Cre transgene was detected by polymerase chain reaction (PCR) using the primers TCGATGCAACGAGTGATGAG and TTCGGCTATACGTAACAGGG.
Blood counts
Adult mice were bled into ACD solution (6.8 mM citric acid, 11.2 mM trisodium citrate, 24 mM glucose), and blood counts were obtained using an ABX Pentra 60 (ABX Diagnostics, Montpellier, France) machine.
Flow cytometric analysis and sorting of fetal liver cells
Single-cell suspensions were generated from dissected fetal livers in PBS by passage through a 25-gauge needle. If required, red blood cells were lysed for 5 minutes in ACK buffer (0.15 M NH2Cl, 1 mM K2HCO3, Na2 EDTA, pH 7.3).
Cells were stained in PBS/5% FBS with combinations of the following antibodies: α-TER119–PE, α–c-Kit–Cy5, α–c-Kit–PE, α-CD45–biotin (followed by SA-APC), TER119-biotin (followed by SA-APC or SA-PE) (eBioscience, San Diego, CA), α-CD71 (Becton Dickinson [BD], San Jose, CA) (followed by α-IgG1–FITC) (Serotec, Raleigh, NC), α-CD34–biotin (BD) (followed by SA-PE), and α-CD41/61–PE (Emfret Analytics, Würzburg, Germany). α-CD16/32 (Fc-block; BD PharMingen) was used to block nonspecific binding. Cells were analyzed on a FACSCalibur flow cytometer using CellQuest software (BD). For cell sorting, red blood cells were lysed, and the stained samples were filtered through a 70-μm strainer. Fluorescence-activated cell sorter (FACS) analysis was performed on a MoFlo machine (Cytomation, Fort Collins, CO). For determination of the cell cycle distribution by DNA staining, cells were washed and resuspended in PBS, 5% FBS, 0.1% NP-40, and 25 μg/mL propidium iodide and were analyzed on a FACSCalibur.
Erythroid cell culture
Cells were plated under conditions supporting erythroid cell development, as described by Dolznig et al.23 SP34 medium contained serum-free medium (StemPro-34; Invitrogen, Carlsbad, CA) plus l-glutamine (2 mM), penicillin (50 U/mL), streptomycin (50 μg/mL), murine SCF (100 ng/mL; PeproTech, Rocky Hill, NJ), human erythropoietin (EPO-β, 2 U/mL; Roche, Indianapolis, IN), and 1 μM dexamethasone (Sigma, St Louis, MO). For long-term cultures, cells were maintained at 1.5 to 3 × 106/mL, and the media were partially refreshed daily.
For induction of terminal differentiation of CFU-E–enriched cultures (7-10 days in SP34), cells were washed twice and plated at 2 to 3 × 106/mL in StemPro-34 containing l-glutamine (2 mM), penicillin (50 U/mL), streptomycin (50 μg/mL), 3% FBS (AutogenBioclear), β-mercaptoethanol (0.1 mM), human holo-transferrin (250 μg/mL; Sigma), bovine insulin (10 μg/mL; Sigma), murine insulinlike growth factor-1 (IGF-1; 40 ng/mL; PeproTech), and human erythropoietin (10 U/mL; Roche).
CFU-E methylcellulose assay
CFU-Es were assayed in semisolid methylcellulose media containing IMDM, 15% FCS, 250 μg/mL holo-transferrin (Sigma), 10 μg/mL insulin (Sigma), 50 ng/mL SCF (PeproTech), and 8 U/mL EPO (Roche). Cultures were incubated at 37°C in a humidified 5% CO2 atmosphere. CFU-Es were scored on days 3 to 4.
IFN treatment
Fetal liver cells from E13 embryos were cultured for 7 to 8 days in SP34, at which point dead cells were removed by centrifugation over a layer of Ficoll-Paque (Amersham, Arlington Heights, IL). After cell culture for another 1 to 2 days, murine IFN-α–A (PBL Biomedical Laboratories, Piscataway, NJ) was added at 2000 U/mL. After 8 to 24 hours, cells were harvested for analysis or washed and plated in SP34 medium without IFN for a given period of time before they were subjected to analysis. The identical procedure was performed on untreated cells as controls.
RNA isolation, RT-PCR, real-time PCR
Cells were washed, and total RNA was extracted using Trizol Reagent (Invitrogen). RNA (0.1-5 μg) was reverse transcribed for 1 hour at 37°C with 400 U MoMuLV reverse transcriptase (Invitrogen) in the presence of 5 ng/μL oligodT, 0.2 mM dNTP, 1 U/μL RNase inhibitor, and 0.1 M DTT in first-strand buffer. For normalization, the cDNA samples were diluted appropriately to give an equal expression value in a real-time PCR assay for β-actin, as described later in this section. Normalization was confirmed by measurement of the relative expression of HPRT.
Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed using 1 μL diluted cDNA and ReddyMix PCR Master Mix (ABgene, Epsom, United Kingdom) in 50 μL containing the appropriate concentration of gene-specific oligonucleotides (Table 1). During cycling, four 10-μL samples were removed from each reaction in 2-cycle increments; the range of cycle numbers depended on the gene.
Oligonucleotide sequences of RT-PCR primers
Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| β-actin | CTTTGCAGCTCCTTCGTTGC | TCTGACCCATTCCCACCATCAC |
| HPRT | CCAGCGTCGTGATTAGCGATG | ATAGCCCCCCTTGAGCACACAGAG |
| βmajor-globin | GCACCTGACTGATGCTGAGAAGG | TGGGTCCAAGGGTAGACAACC |
| α-globin | TGCTCTCTGGGGAAGACAAAAG | CGTGGCTTACATCAAAGTGAGGG |
| Fli1 | AACGAACGGAGAGTCATTGTGC | TTCTGTGTTGTAGGCGGAGGTG |
| EpoR | TGACGCTGTCTCTCATTCTGGTCC | TCAAACTCGCTCTCTGGGCTTG |
| Runx1 | CAGGCAACGATGAAAACTAC | GCTCGGAAAAGGACAAAC |
| GATA1 | CCCTTGTGAGGCCAGAGA | ACCTGATGGAGCTTGAAATAG |
| GATA2 | GTCAGACGACAACCACCACCTTA | GGATTTGCTGGACATCTTCCGA |
| PU.1 | TGGAAGGGTTTTCCCTCACC | TGCTGTCCTTCATGTCGCCG |
| SCL | CTCCTATCACATTCTGCTATCAC | AATCTCTCACCCCTTCTCAC |
| c-fms | TAGGTGTGATGAAGCCCAG | TCAGTGATGACCAGGACAG |
| c-Kit | AAGGGGACACATTTACGGTGG | TCCAGAATCGTCAACTCTTGCC |
Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| β-actin | CTTTGCAGCTCCTTCGTTGC | TCTGACCCATTCCCACCATCAC |
| HPRT | CCAGCGTCGTGATTAGCGATG | ATAGCCCCCCTTGAGCACACAGAG |
| βmajor-globin | GCACCTGACTGATGCTGAGAAGG | TGGGTCCAAGGGTAGACAACC |
| α-globin | TGCTCTCTGGGGAAGACAAAAG | CGTGGCTTACATCAAAGTGAGGG |
| Fli1 | AACGAACGGAGAGTCATTGTGC | TTCTGTGTTGTAGGCGGAGGTG |
| EpoR | TGACGCTGTCTCTCATTCTGGTCC | TCAAACTCGCTCTCTGGGCTTG |
| Runx1 | CAGGCAACGATGAAAACTAC | GCTCGGAAAAGGACAAAC |
| GATA1 | CCCTTGTGAGGCCAGAGA | ACCTGATGGAGCTTGAAATAG |
| GATA2 | GTCAGACGACAACCACCACCTTA | GGATTTGCTGGACATCTTCCGA |
| PU.1 | TGGAAGGGTTTTCCCTCACC | TGCTGTCCTTCATGTCGCCG |
| SCL | CTCCTATCACATTCTGCTATCAC | AATCTCTCACCCCTTCTCAC |
| c-fms | TAGGTGTGATGAAGCCCAG | TCAGTGATGACCAGGACAG |
| c-Kit | AAGGGGACACATTTACGGTGG | TCCAGAATCGTCAACTCTTGCC |
Real-time PCR was performed using SYBR green with primers specific for β-actin, HPRT, α-globin, βmajor-globin, GATA2, Fli1, and c-Kit (Table 1). cDNA dilutions (0.6-2 μL) were used in a 15-μL reaction with ABsolute QPCR SYBR Green Mix (ABgene) and 70 to 300 nM each primer. Reactions were performed and analyzed on a RotorGene 3000 system (Corbett Research, Sydney, Australia). After an initial denaturation step (95°C for 15 minutes), cycling was performed at 95°C for 15 seconds, 59 to 64°C for 20 seconds, and 72°C for 20 to 30 seconds (40 cycles). Amplification efficiency and the take-off point (Ct value) for each reaction were calculated with the use of RotorGene 6.0 software (Corbett Research) based on the second derivative method. For calculation of the β-actin–normalized relative expression values for each sample a mathematical model24 that takes gene-dependent differences in the amplification efficiency into account. At least 2 separate reactions with 3 replicates per sample were run. For significance testing, the normalized relative expression values were log transformed to fulfil the normality criterion. A 2 × 2 ANOVA was performed for each time point, and the P value for the interaction, termed “cell type × treatment,” was calculated.
Southern blotting
Genomic DNA was prepared using standard methods and was digested with HindIII. After agarose electrophoresis, the DNA was blotted onto a Hybond-N+ membrane (Amersham, Arlington Heights, IL). Probe P (intron 2 of c-myb; Figure 5A), which was obtained by PCR amplification of genomic DNA with oligonucleotide primers AACGTAGGGAAACTCTGTAG and AAGATAAGACCATGGATGAG, was radiolabeled using PrimeIt II Random Prime Labeling Kit (Stratagene, La Jolla, CA) and α-[32P]dCTP (Amersham). After hybridization, a phosphor screen (Molecular Dynamics, Eugene, OR) was exposed for 1 to 5 days and scanned on a Typhoon 9200 PhosphorImager (Amersham). The intensities of the bands corresponding to the c-mybΔ (2911 bp) and the c-myb– (1607 bp) alleles were obtained, and the ratio was calculated as a representation of the recombination rate.
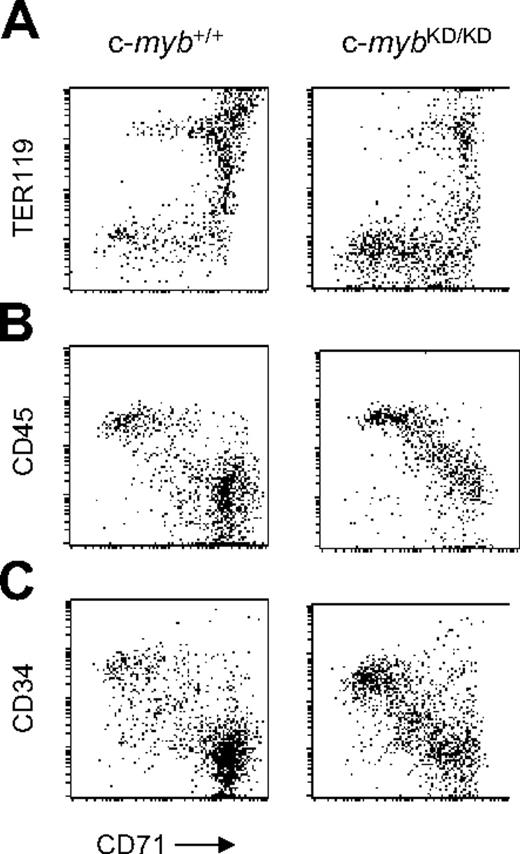
Accumulation of immature progenitor cells in the fetal liver of c-mybKD/KD embryos. Presented are fluorescence values (log-scale) from flow cytometric analyses of fetal liver single-cell suspensions from E13 c-myb+/+ and c-mybKD/KD embryos stained with monoclonal antibodies. Gated live cells are shown. (A) Cells were stained with α-CD71(FITC) and α-TER119(APC). (B-C) Cells were stained with α-CD71(FITC), α-c-Kit(PE-Cy5), and either α-CD45(APC) (B) or α-CD34(PE) (C). Gated c-Kit+ cells are shown.
Accumulation of immature progenitor cells in the fetal liver of c-mybKD/KD embryos. Presented are fluorescence values (log-scale) from flow cytometric analyses of fetal liver single-cell suspensions from E13 c-myb+/+ and c-mybKD/KD embryos stained with monoclonal antibodies. Gated live cells are shown. (A) Cells were stained with α-CD71(FITC) and α-TER119(APC). (B-C) Cells were stained with α-CD71(FITC), α-c-Kit(PE-Cy5), and either α-CD45(APC) (B) or α-CD34(PE) (C). Gated c-Kit+ cells are shown.
Hemoglobin staining
Cytospins were treated for 10 to 15 seconds in methanol, 5 minutes in 1% ortho-dianisidine (3,3′-dimethoxy-benzidine; Sigma) in methanol, and 2.5 minutes in 2.5% hydrogen peroxide/70% ethanol and were then washed for 2.5 minutes in distilled water. Counterstaining was performed with Harris hematoxylin (1.5 minutes) and a final wash in running water for 8 minutes. Four fields containing 250 cells on average were counted.
Results
Reduced levels of c-Myb result in retarded commitment and progression of erythroid progenitors
To characterize fetal liver erythropoiesis, we combined staining with CD71 (transferrin receptor) and TER119 to distinguish the CFU-E stage (CD71+TER119–) from earlier or nonerythroid cells (CD71–TER119–) and later erythroblast stages (TER119+).25 We confirmed an increased proportion of immature CD71low/+TER119– and nonerythroid CD71–TER119– cells in c-mybKD/KD E13 fetal liver (Figure 1A). We also stained simultaneously for CD71, c-Kit, and CD45 and gated for c-Kit+ cells, representing hematopoietic progenitors (Figure 1B). CD45 is expressed on leukocytes and hematopoietic progenitors but is down-regulated during erythropoiesis after the BFU-E stage.26 c-mybKD/KD fetal liver displayed a higher abundance of CD71–/lowCD45+. Staining for CD71, c-Kit, and CD34, which is also down-regulated early in erythropoiesis,26 revealed a similar pattern (Figure 1C). We conclude that reduced levels of c-Myb lead to an accumulation of nonerythroid progenitor cells and cells committing to erythropoiesis.
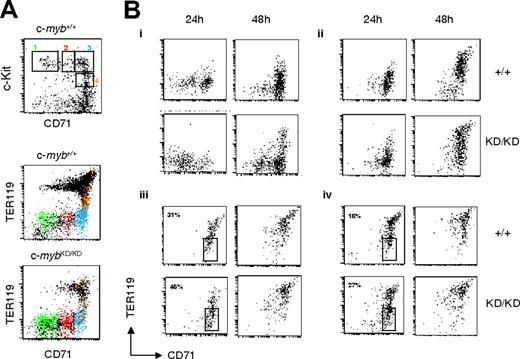
To test whether the accumulation of immature cells resulted from a partial developmental block, we sorted progenitor populations from wild-type and c-mybKD/KD E14 fetal liver and examined their development in culture. Cells were sorted on the basis of their expression of CD71 and c-Kit (Figure 2A, upper panel; lower panels show the corresponding expression of CD71 vs TER119). CD41– cells were selected to exclude cells with megakaryocytic potential. Sorted cells were plated in serum-free medium (SP34) containing SCF/EPO/dexamethasone, which favors the development of erythroid cells while restricting differentiation toward other lineages,23 cultured for 24 or 48 hours and analyzed for CD71 and TER119 expression (Figure 2B). Wild-type CD71– c-Kit+ cells, corresponding to the most immature progenitors, committed to erythropoiesis (CD71+TER119–) by 24 hours and began to up-regulate TER119 expression by 48 hours (Figure 2B, R1). c-mybKD/KD cells were able to partially reach these stages, but their progression was slowed. The progression of CD71lowc-Kit+ c-mybKD/KD cells past the CFU-E stage was similarly affected (Figure 2B, R2). Sorted c-myb+/+ cells at the CFU-E stage (R3) or late CFU-E (CD71+c-KitlowTER119low, R4) reached erythroblast stages (TER119high) within 48 hours. Similarly, their c-mybKD/KD counterparts reached these stages, but with a lag of 24 hours, as emphasized by the percentages of cells maintaining their CD71+TER119– phenotype after 24 hours, which was 31% and 16% for wild-type R3 and R4 sorted cells respectively, compared with 46% and 27% for the corresponding c-mybKD/KD sorted populations (Figure 2B).
Reduced levels of c-Myb are sufficient for proliferation of early pre–CFU-E progenitors
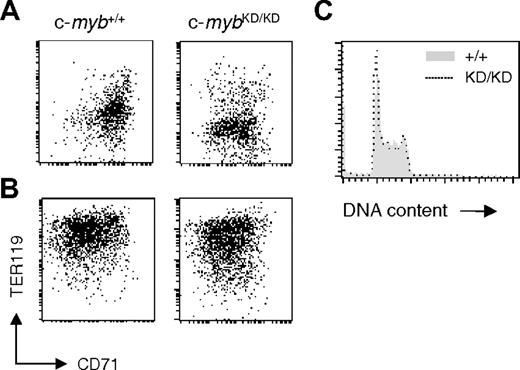
c-Myb has been shown to be required for cell proliferation in hematopoietic cell lines.13 We tested whether the retardation of erythropoiesis associated with reduced c-Myb levels is a consequence of compromised proliferation. c-myb+/+ and c-mybKD/KD E14 fetal liver cells were plated in SP34 medium containing SCF/EPO/dexamethasone, as previously described by Dolznig et al.23 c-myb+/+ cultures reached a steady state of growth after 5 to 7 days, having been enriched for cells representing erythroid progenitors at around the CFU-E stage. At day 9, c-myb+/+ cells were predominantly CD71+TER119low (Figure 3A) c-Kit+ and had a blastlike morphology (data not shown). When these cells were transferred to serum-containing medium with high levels of EPO and insulin and no SCF or dexamethasone, they underwent terminal erythroid differentiation and reached late erythroblast and reticulocyte stages at 72 hours (CD71–/lowTER119high; Figure 3B and data not shown). In agreement with the observations made on sorted cells, c-mybKD/KD day 9 cultures were enriched for CD71low/+TER119– cells (Figure 3A), representing a more immature stage compared with wild-type cultures. Nevertheless, these cells had erythroid potential comparable with c-myb+/+ cells, displaying a similar ability to differentiate in response to high EPO/insulin (Figure 3B). No difference was observed in either the cell cycle or the extent of apoptosis of c-myb+/+ and c-mybKD/KD cells at steady state after 9 days in SP34 (Figure 3C and data not shown). These results suggest that early pre–CFU-E progenitors do not require high levels of c-Myb protein for intense ex vivo proliferation and that the effects of c-Myb on the proliferation and development of early erythroid progenitors can probably be uncoupled.
Progenitors from c-mybKD/KD fetal liver display retarded commitment and progression through erythropoiesis. Fetal liver single-cell suspensions from E14 c-myb+/+ and c-mybKD/KD embryos were stained with α-CD71(FITC), α-CD41(PE), α-c-Kit(PE-Cy5), and α-TER119(APC). CD41– live cells were sorted based on their CD71 and c-Kit staining. (A) Cells in sorting regions 1-4 (top panel) are displayed in the corresponding color in the middle and bottom panels to indicate their TER119 expression pattern. Note that a minor shift in the TER119 signal of c-mybKD/KD cells is attributed to overcompensation caused by reduced c-Kit levels (see panel B). (B) Sorted cells were cultured in SP34 medium containing SCF (100 μg/mL), EPO (2 U/mL), and dexamethasone (1 μM). At the indicated time points, cells were stained with α-CD71 (FITC) and α-TER119 (APC) and analyzed by flow cytometry. Panels Bi to Biv show the development of cells from CD71/c-Kit–sorted regions 1 to 4 (A), respectively. Percentages in Biii and Biv represent proportions of cells at the CFU-E stage. Dead cells were excluded by propidium iodide staining. All plots are in log-scale.
Progenitors from c-mybKD/KD fetal liver display retarded commitment and progression through erythropoiesis. Fetal liver single-cell suspensions from E14 c-myb+/+ and c-mybKD/KD embryos were stained with α-CD71(FITC), α-CD41(PE), α-c-Kit(PE-Cy5), and α-TER119(APC). CD41– live cells were sorted based on their CD71 and c-Kit staining. (A) Cells in sorting regions 1-4 (top panel) are displayed in the corresponding color in the middle and bottom panels to indicate their TER119 expression pattern. Note that a minor shift in the TER119 signal of c-mybKD/KD cells is attributed to overcompensation caused by reduced c-Kit levels (see panel B). (B) Sorted cells were cultured in SP34 medium containing SCF (100 μg/mL), EPO (2 U/mL), and dexamethasone (1 μM). At the indicated time points, cells were stained with α-CD71 (FITC) and α-TER119 (APC) and analyzed by flow cytometry. Panels Bi to Biv show the development of cells from CD71/c-Kit–sorted regions 1 to 4 (A), respectively. Percentages in Biii and Biv represent proportions of cells at the CFU-E stage. Dead cells were excluded by propidium iodide staining. All plots are in log-scale.
c-myb knockdown CFU-E–stage progenitors display an immature phenotype
To investigate molecular mechanisms underlying the effects of lower levels of c-Myb, we characterized gene expression in c-mybKD/KD CFU-E–stage progenitors. CD71+c-Kit+CD41– cells were sorted from E14 fetal livers, and RNA was extracted. RT-PCR was performed for β-actin cDNA for normalization and for several genes relevant to hematopoiesis (Figure 4A). To assess whether the erythroid gene expression program was induced in c-mybKD/KD cells, we performed RT-PCR for the transcription factor genes SCL and GATA1 and for the erythropoietin receptor (EpoR) gene. These genes play critical roles in the commitment and execution of erythroid differentiation.3,6,9 No prominent difference could be detected between wild-type and c-myb knockdown samples (Figure 4A).
Next, we examined expression of the transcription factor GATA-2, which is required for the maintenance of immature hematopoietic progenitors but is down regulated during erythroid differentiation.8 c-mybKD/KD cells expressed 5.1-fold higher levels of GATA-2 mRNA than wild-type cells, suggesting that they have a more immature phenotype (Figure 4B). Finally, we examined expression of the transcription factor genes Runx1, PU.1, and Fli1, which play critical roles in regulating the development of immature hematopoietic progenitors29-31 and are normally down-regulated during erythropoiesis.30,31 Strikingly, RNA levels of all 3 genes were increased in c-mybKD/KD relative to wild-type CFU-E stage progenitors (Figure 4A), correlating with the immature character of these cells. A 3.4-fold up-regulation of Fli1 RNA was confirmed by real-time PCR (Figure 4B). To exclude the possibility of contaminating nonerythroid cells as the cause for this result, we performed RT-PCR for the myelomonocytic marker gene c-fms and found a decrease rather than an increase in c-mybKD/KD cells (Figure 4A).
Reduced levels of c-Myb are sufficient for intense proliferation of immature erythroid progenitors. Fetal liver cells from E14 c-myb+/+ and c-mybKD/KD embryos were plated in SP34 medium containing SCF (100 μg/mL), EPO (2 U/mL), and dexamethasone (1 μM). (A) At day 9 of culture, cells were stained with α-CD71 (FITC) and α-TER119 (APC) and analyzed by flow cytometry. (B) At day 9 of culture, cells were transferred to media favoring terminal erythroid differentiation and cultured for 3 additional days before being analyzed by flow cytometry for CD71 and TER119 expression. (C) At day 9 of culture in SP34, cells were permeabilized with 0.1% NP-40 and stained with 25 μg/mL propidium iodide for flow cytometric analysis of DNA content. Histograms represent linear fluorescence intensities.
Reduced levels of c-Myb are sufficient for intense proliferation of immature erythroid progenitors. Fetal liver cells from E14 c-myb+/+ and c-mybKD/KD embryos were plated in SP34 medium containing SCF (100 μg/mL), EPO (2 U/mL), and dexamethasone (1 μM). (A) At day 9 of culture, cells were stained with α-CD71 (FITC) and α-TER119 (APC) and analyzed by flow cytometry. (B) At day 9 of culture, cells were transferred to media favoring terminal erythroid differentiation and cultured for 3 additional days before being analyzed by flow cytometry for CD71 and TER119 expression. (C) At day 9 of culture in SP34, cells were permeabilized with 0.1% NP-40 and stained with 25 μg/mL propidium iodide for flow cytometric analysis of DNA content. Histograms represent linear fluorescence intensities.
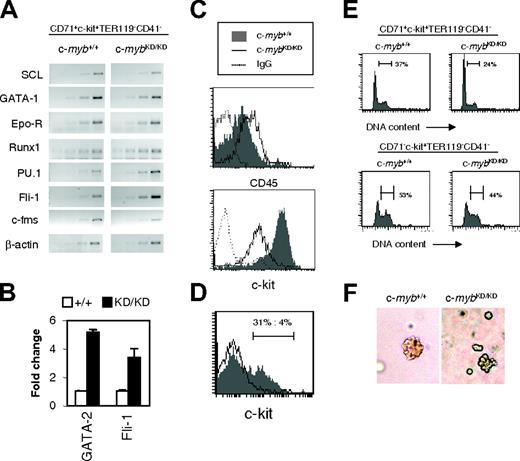
c-mybKD/KD CFU-E stage progenitors display aberrant expression of hematopoietic regulators and abnormal differentiation ex vivo. (A) CD71+c-kit+TER119–CD41– progenitors (Figure 2Biii) corresponding to the CFU-E stage were sorted by FACS from the fetal liver of c-myb+/+ and c-mybKD/KD E14 embryos. RT-PCR analysis was performed on RNA extracted from sorted cells. Ethidium bromide–stained agarose gels of PCR products from 2-cycle increments are shown. Normalization by β-actin was confirmed by real time PCR analysis (not shown). (B) Real-time PCR was performed on the cDNA samples described for panel A using SYBR green, primers specific for the indicated genes, and β-actin (for normalization). Reactions were repeated at least 6 times. Average fold change–normalized expression values are shown. Error bars represent the SEM. (C) Fetal liver cells were stained with α-CD71 (FITC), α-CD45 (APC) and α-TER119 (APC) (top panel) or α-CD71 (FITC), α-TER119 (PE), and α-c-Kit (PE-Cy5) (bottom panel). Histograms represent fluorescence intensities (log-scale) of cells gated as CD71+TER119– (CFU-E stage). (D) Fetal liver cells were stained and CFU-E stage cells were sorted as for panel A and were cultured for 48 hours, as described in Figure 2. Cells were stained with α-c-Kit (PE) and were analyzed by flow cytometry. Log-scale of PE-fluorescence intensity is shown. (E) CD71+c-Kit+TER119–CD41– (Figure 2Biii) CFU-E stage progenitors (top panels) and CD71– c-Kit+TER119–CD41– (Figure 2Bi) less mature progenitors (bottom panels) from E14 fetal liver were sorted by FACS and cultured in SP34 for 2 days. Cells were permeabilized with 0.1% NP-40 and were stained with 25 μg/mL propidium iodide for flow cytometric analysis of DNA content. Histograms represent linear fluorescence intensities. (F) Lower c-Myb expression limits the size of CFU-E colonies. Fetal liver cells were stained and CFU-E stage cells were sorted as for panel A and then plated in methylcellulose containing SCF and EPO for 3 days. Colonies were observed using an Olympus CKX41 microscope (Olympus, London, United Kingdom) and a 20 ×/0.40 numeric aperture Php objective under phase contrast. Images were acquired using an Olympus Camedia C3030 camera and were processed using Adobe Photoshop version 4.0 (Adobe Systems, San Jose, CA).
c-mybKD/KD CFU-E stage progenitors display aberrant expression of hematopoietic regulators and abnormal differentiation ex vivo. (A) CD71+c-kit+TER119–CD41– progenitors (Figure 2Biii) corresponding to the CFU-E stage were sorted by FACS from the fetal liver of c-myb+/+ and c-mybKD/KD E14 embryos. RT-PCR analysis was performed on RNA extracted from sorted cells. Ethidium bromide–stained agarose gels of PCR products from 2-cycle increments are shown. Normalization by β-actin was confirmed by real time PCR analysis (not shown). (B) Real-time PCR was performed on the cDNA samples described for panel A using SYBR green, primers specific for the indicated genes, and β-actin (for normalization). Reactions were repeated at least 6 times. Average fold change–normalized expression values are shown. Error bars represent the SEM. (C) Fetal liver cells were stained with α-CD71 (FITC), α-CD45 (APC) and α-TER119 (APC) (top panel) or α-CD71 (FITC), α-TER119 (PE), and α-c-Kit (PE-Cy5) (bottom panel). Histograms represent fluorescence intensities (log-scale) of cells gated as CD71+TER119– (CFU-E stage). (D) Fetal liver cells were stained and CFU-E stage cells were sorted as for panel A and were cultured for 48 hours, as described in Figure 2. Cells were stained with α-c-Kit (PE) and were analyzed by flow cytometry. Log-scale of PE-fluorescence intensity is shown. (E) CD71+c-Kit+TER119–CD41– (Figure 2Biii) CFU-E stage progenitors (top panels) and CD71– c-Kit+TER119–CD41– (Figure 2Bi) less mature progenitors (bottom panels) from E14 fetal liver were sorted by FACS and cultured in SP34 for 2 days. Cells were permeabilized with 0.1% NP-40 and were stained with 25 μg/mL propidium iodide for flow cytometric analysis of DNA content. Histograms represent linear fluorescence intensities. (F) Lower c-Myb expression limits the size of CFU-E colonies. Fetal liver cells were stained and CFU-E stage cells were sorted as for panel A and then plated in methylcellulose containing SCF and EPO for 3 days. Colonies were observed using an Olympus CKX41 microscope (Olympus, London, United Kingdom) and a 20 ×/0.40 numeric aperture Php objective under phase contrast. Images were acquired using an Olympus Camedia C3030 camera and were processed using Adobe Photoshop version 4.0 (Adobe Systems, San Jose, CA).
Given that CD45 down-regulation is a hallmark of commitment to erythropoiesis, we tested its surface expression on CD71+TER119– CFU-E stage cells. As shown in Figure 4C, c-mybKD/KD cells expressed increased levels of CD45 (2-fold), confirming their immature character. Given the ability of CD45 to interfere with EpoR signaling, this represents another possible mechanism of inhibition of erythropoiesis in c-myb knockdown progenitors.32 We also examined the surface expression of c-Kit on CD71+TER119–CD41– cells and found a marked down-regulation on c-mybKD/KD cells (7-fold; Figure 4C). To ensure that this was not caused by the selection of an irrelevant population by FACS, we determined expression levels on cells cultured for 9 days in SP34 medium, representing early erythroid progenitors, and found a similar difference (data not shown). The development of early erythroid progenitors, including the CFU-E stage, strongly depends on SCF/c-Kit signaling.2,35 Therefore, reduced levels of c-Kit surface expression could provide a good explanation for the impaired progression and response of c-myb knockdown cells to erythropoietic stimuli.
Reduced levels of c-Myb result in accelerated growth arrest of CFU-E stage cells
Careful examination of Figure 2B reveals a subtle difference in the effect of reduced c-Myb expression on differentiation progression comparing pre–CFU-E stage cells (R1, CD71– c-Kit+TER119–CD41– or R2, CD71lowc-Kit+TER119–CD41–) and CFU-E stage cells and proerythroblasts (R3, CD71+c-Kit+TER119–CD41–; or R4, CD71+c-kit+TER119lowCD41–). Hence, the former exhibited clearly slowed progression at 24 hours and 48 hours when derived from c-mybKD/KD fetal liver (Figure 2B, R1 and R2), whereas c-mybKD/KD CFU-E stage cells and proerythroblasts exhibited slowed progression at 24 hours but appeared equally or even more differentiated at 48 hours compared with the wild type based on TER119 expression (Figure 2B, R3 and R4). We explored this further by examining additional features of the growth and differentiation of CD71+c-Kit+TER119–CD41– c-mybKD/KD fetal liver CFU-E stage progenitors after plating them in SP34 medium. Interestingly, although a considerable fraction (31%) of c-myb+/+ cells still expressed c-Kit on the surface at 48 hours (Figure 4D), few (4%) c-mybKD/KD cells remained c-Kit+. Analysis of the cell cycle distribution of these cells revealed a decreased proportion of cells in S and G2/M phases in c-mybKD/KD cultures (24% as opposed to 37% in wild type) (Figure 4E, upper panels). These results suggest that, despite their immature character in vivo and their inhibited progression, c-mybKD/KD CFU-E stage cells undergo accelerated growth arrest when they are induced to differentiate ex vivo. Similar analysis of CD71– c-Kit+TER119–CD41– cells indicated that, compared with the effect on CFU-E stage cells, reduced levels of c-Myb have less influence on the proliferation of more immature cells (Figure 4E, lower panels). In support of the conclusion that CD71+c-Kit+TER119–CD41– progenitors are significantly affected in terms of their proliferative capacity, we observed that CFU-E–like colonies that developed from CD71+c-kit+TER119–CD41––sorted cells plated in semisolid media were smaller and had abnormal structures when the cells were derived from c-mybKD/KD fetal liver (Figure 4F).
We suggest that, in contrast to immature pre–CFU-E progenitors, CFU-E stage progenitors are more dependent on high levels of c-Myb for performing their terminal cell divisions and, on reaching approximately the CFU-E/proerythroblast stage, c-mybKD/KD cells undergo uncontrolled or uncoordinated differentiation with more rapid cell-cycle exit.
Inducible Cre-mediated inactivation of c-myb in erythroid progenitors
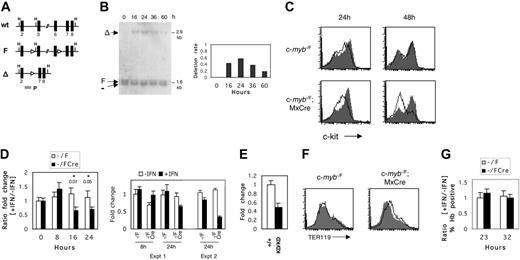
A possible drawback of studying c-mybKD/KD cells is the inability to distinguish between direct and indirect effects of c-Myb. Therefore, we established an inducible ex vivo system for the inactivation of c-myb in normal erythroid progenitors. To achieve this, we made use of a targeted c-myb allele (c-mybF) containing loxP sites flanking exons 3 to 6, which encode the DNA-binding domain (Figure 5A),18 in conjunction with the MxCre transgene, the expression of which can be induced by IFN.22 c-myb–/F/Cre E13 embryos were used as a source of erythroid progenitors, and c-myb–/F embryos were used as controls. Fetal liver cells were cultured for 8 days in SP34 medium to enrich for CFU-E progenitors (Figure 3A-B). At this point, cultures were treated for 16 to 24 hours with 2000 U/mL recombinant murine IFN-α–A or were left untreated. To assess the efficiency of recombination, we harvested genomic DNA at 16 to 60 hours after the addition of IFN and performed Southern blot analysis of the relative abundance of the c-mybΔ allele (Figure 5B). Recombination could be detected as early as 16 hours after the addition of IFN (40%) and reached a maximum at 24 hours (varying between 60% and 90% across numerous experiments), indicating that c-myb–/Δ cells have a lower growth rate at later time points than wild-type cells.
c-Myb is essential for c-Kit expression in erythroid progenitors
CFU-E progenitors in the fetal liver of c-mybKD/KD embryos expressed reduced levels of c-Kit (Figure 4C). To determine whether c-Myb regulates c-Kit expression in normal erythroid progenitors, we measured surface c-Kit at 24 and 48 hours after induced c-myb inactivation (Figure 5C). Treatment with IFN resulted in a clear reduction of c-Kit levels on c-myb–/F/Cre cells at 24 hours, and by 48 hours most were c-Kit–. At 24 hours, most cells declined in c-Kit expression (peak fluorescence intensity, approximately 75), whereas a second minority population of c-Kit– cells (peak fluorescence intensity, approximately 20) was approximately equivalent to that seen in the IFN-treated control cells. At 48 hours, when the overall extent of deletion of c-myb in the population had declined (Figure 5B), all cells were c-Kit– or c-Kit+ (peak fluorescence intensity, approximately 150), the latter presumably because of the selective outgrowth of undeleted cells. To exclude the possibility of selective ablation of c-Kit+ cells in c-myb–/F/Cre cultures, we measured the rate of apoptosis at 24 and 40 hours and found no significant changes (data not shown). To test whether c-Myb regulates c-Kit RNA levels, we examined the kinetics of RNA expression in response to c-myb inactivation. Real-time PCR (Figure 5D) revealed a 1.5-fold reduction of c-Kit RNA levels in c-myb–/F/Cre cells by 16 hours after the addition of IFN (P = .01 for cell type × treatment in 2 × 2 ANOVA). The down-regulation persisted at 24 hours (P = .05). The kinetics of c-Kit down-regulation correlated well with the kinetics of disruption of the c-mybF allele (Figure 5B). Because the average deletion efficiency of the c-mybF allele was 70%, the 1.5- to 2-fold measured reduction of c-Kit mRNA represents an underestimate. That the change observed in c-Kit RNA expression is not an artifact of a nonspecific effect of IFN can be seen from the absolute levels of c-Kit RNA in the absence or presence of IFN in the control (c-myb–/F) and experimental (c-myb–/F/Cre) samples throughout the time course (Figure 5D, right panel, experiment 1 (Expt 1), the corresponding ratios for which are depicted in the left panel). Hence, at both 8 hours (the first time point at which any nonspecific effect of IFN can be assessed) and 24 hours, the control cells showed no substantial difference in c-Kit RNA expression as a result of the presence of IFN, whereas at 24 hours an IFN-dependent decrease in c-Kit RNA expression was seen in the experimental cells. An independent time-course experiment yielded a similar result in terms of no significant nonspecific effect of IFN but a decrease in c-Kit RNA levels in c-myb–/F/Cre cells, consistent with the extent of c-myb deletion (Figure 5D, right panel, Expt 2). In support of the conditional c-myb deletion result, we quantified c-Kit RNA levels in c-mybKD/KD CFU-E stage progenitors, which express reduced levels of c-Myb protein. Quantitative PCR analysis revealed a 2-fold reduction of c-Kit RNA in this cell type (Figure 5E).
Inducible inactivation of c-myb reveals its requirement for maintenance of c-Kit expression in erythroid progenitors. (A) Schematic representation of c-myb alleles: wild type (wt), targeted floxed (F), and recombined by Cre-recombinase (Δ). Exons are shown as black boxes. Arrowheads represent loxP sites. The probe (P) used in hybridization to Southern blots is indicated by the gray box. H indicates HindIII. (B) Detection of Cre-mediated recombination of the c-mybF allele in cultured erythroid progenitors. Fetal liver cells from E13 c-myb–/F/Cre embryos were cultured as described in Figure 3 for 8 days to enrich for erythroid progenitors. IFN–α-A (2000 U/mL) was then added to the cultures and washed off at 24 hours. Genomic DNA was harvested at the indicated time points and digested with HindIII. Southern blot analysis was performed with probe P, and signals were quantified by phosphorimaging. The deletion rate of the c-mybF allele is represented as the ratio of intensities of the c-mybΔ signal to the constant c-myb– signal (right panel). (C) Flow cytometric analysis of c-Kit surface expression on cultured c-myb–/F/Cre and c-myb–/F cells in response to IFN treatment. Day 8 fetal liver cultures were treated with IFN–α-A (2000 U/mL) for 24 hours (open histogram) or were left untreated (filled histogram). At 24 or 48 hours, cells were stained with α-c-Kit-PE and analyzed by flow cytometry. (D) Real-time RT-PCR analysis of c-Kit mRNA expression in response to c-myb inactivation. Cells were treated with IFN–α-A, as described, and RNA was harvested at the indicated time points and reverse transcribed. Real-time PCR was performed as described in “Materials and methods.” (Left) Ratio of normalized relative expression levels from samples with IFN to samples without IFN was calculated for c-myb–/F/Cre and c-myb–/F separately. The average of at least 6 replicates is shown. ANOVA (2 × 2) was performed, and P values are given for the interaction term (cell type × treatment). (Right) Absolute expression (normalized to 8-hour control IFN) of c-Kit RNA determined by real time RT-PCR is shown for the c-myb–/F and c-myb–/F/Cre samples with or without IFN at 8 and 24 hours. Data for the 24-hour time point from a second independent experiment are illustrated on the right. (E) Real-time RT-PCR analysis of c-Kit mRNA expression in c-myb+/+ and c-mybKD/KD CFU-E stage cells sorted from E14 fetal liver. Normalized relative expression levels are shown. Error bars represent SEM. (F) c-myb inactivation does not lead to a general induction of differentiation. Enriched erythroid progenitors from c-myb–/F/Cre and c-myb–/F fetal livers were treated with IFN–α-A (2000 U/mL) for 24 hours (open histogram) or were left untreated (filled histogram) and subsequently were cultured for another 24 hours. Cells were stained with α-TER119 (PE) and analyzed by flow cytometry. (G) Cells were treated with IFN–α-A, as described in panel F, and were collected on cytospins at the indicated time points. After o-dianisidine/hematoxylin staining, 4 fields were counted for the proportion of hemoglobin-positive cells. The ratio of the frequencies of Hb+ cells in samples with IFN to samples without IFN is represented. Error bars represent the SEM.
Inducible inactivation of c-myb reveals its requirement for maintenance of c-Kit expression in erythroid progenitors. (A) Schematic representation of c-myb alleles: wild type (wt), targeted floxed (F), and recombined by Cre-recombinase (Δ). Exons are shown as black boxes. Arrowheads represent loxP sites. The probe (P) used in hybridization to Southern blots is indicated by the gray box. H indicates HindIII. (B) Detection of Cre-mediated recombination of the c-mybF allele in cultured erythroid progenitors. Fetal liver cells from E13 c-myb–/F/Cre embryos were cultured as described in Figure 3 for 8 days to enrich for erythroid progenitors. IFN–α-A (2000 U/mL) was then added to the cultures and washed off at 24 hours. Genomic DNA was harvested at the indicated time points and digested with HindIII. Southern blot analysis was performed with probe P, and signals were quantified by phosphorimaging. The deletion rate of the c-mybF allele is represented as the ratio of intensities of the c-mybΔ signal to the constant c-myb– signal (right panel). (C) Flow cytometric analysis of c-Kit surface expression on cultured c-myb–/F/Cre and c-myb–/F cells in response to IFN treatment. Day 8 fetal liver cultures were treated with IFN–α-A (2000 U/mL) for 24 hours (open histogram) or were left untreated (filled histogram). At 24 or 48 hours, cells were stained with α-c-Kit-PE and analyzed by flow cytometry. (D) Real-time RT-PCR analysis of c-Kit mRNA expression in response to c-myb inactivation. Cells were treated with IFN–α-A, as described, and RNA was harvested at the indicated time points and reverse transcribed. Real-time PCR was performed as described in “Materials and methods.” (Left) Ratio of normalized relative expression levels from samples with IFN to samples without IFN was calculated for c-myb–/F/Cre and c-myb–/F separately. The average of at least 6 replicates is shown. ANOVA (2 × 2) was performed, and P values are given for the interaction term (cell type × treatment). (Right) Absolute expression (normalized to 8-hour control IFN) of c-Kit RNA determined by real time RT-PCR is shown for the c-myb–/F and c-myb–/F/Cre samples with or without IFN at 8 and 24 hours. Data for the 24-hour time point from a second independent experiment are illustrated on the right. (E) Real-time RT-PCR analysis of c-Kit mRNA expression in c-myb+/+ and c-mybKD/KD CFU-E stage cells sorted from E14 fetal liver. Normalized relative expression levels are shown. Error bars represent SEM. (F) c-myb inactivation does not lead to a general induction of differentiation. Enriched erythroid progenitors from c-myb–/F/Cre and c-myb–/F fetal livers were treated with IFN–α-A (2000 U/mL) for 24 hours (open histogram) or were left untreated (filled histogram) and subsequently were cultured for another 24 hours. Cells were stained with α-TER119 (PE) and analyzed by flow cytometry. (G) Cells were treated with IFN–α-A, as described in panel F, and were collected on cytospins at the indicated time points. After o-dianisidine/hematoxylin staining, 4 fields were counted for the proportion of hemoglobin-positive cells. The ratio of the frequencies of Hb+ cells in samples with IFN to samples without IFN is represented. Error bars represent the SEM.
Taken together, we demonstrated a strong correlation between c-Myb and c-Kit levels in normal erythroid progenitors, both in vivo and ex vivo. We conclude that c-Myb is required for c-Kit expression.
Because our observation of c-Kit down regulation could be associated with a general spontaneous differentiation, we measured TER119 levels on c-myb–/F/Cre cells treated with IFN–α-A for 24 hours and cultured for another 24 hours (Figure 5F). IFN caused only a minor increase in TER119 levels, whereas no change could be detected in c-myb–/F cells. We also determined the effect of IFN treatment on the frequency of spontaneously hemoglobinized cells in c-myb–/F and c-myb–/F/Cre. No difference could be detected in the fraction of o-dianisidine–positive cells between the 2 genotypes at 23 or 32 hours after the addition of IFN (Figure 5G). These results suggest that general spontaneous differentiation is not inhibited by c-Myb in erythroid progenitors. Moreover, the down-regulation of c-Kit expression in response to c-myb inactivation seemed to be a specific effect and not a result of uncontrolled differentiation.
Discussion
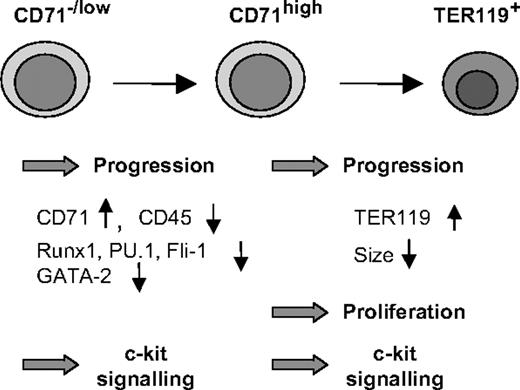
By sorting defined progenitor populations of fetal liver cells followed by ex vivo culture, we showed that the transition of uncommitted progenitors to erythropoiesis is inhibited by reduced levels of c-Myb (Figure 6). This observation explains the accumulation of progenitor cells coexpressing low levels of CD71 and the markers CD45 and CD34 in the fetal liver of c-mybKD/KD embryos. Strikingly, we also found that the progression of early committed erythroid progenitors was slowed as a consequence of reduced c-Myb levels, whereas later stages of erythropoiesis required normal levels of c-Myb to complete differentiation correctly. These findings implicate c-Myb in processes other than growth control, suggesting that it is involved in the response of uncommitted and committed erythroid cells to erythropoietic stimuli and the execution of an erythroid program.
Coordination of erythropoiesis by c-Myb. c-Myb has a dual role as a regulator of erythroid development. Both in uncommitted/early stages (CD71–/low) and in later stages (CFU-E/erythroblast—CD71+TER119–/low), high levels of c-Myb are required for an efficient response of progenitors to erythropoietic stimuli and progression to later stages. c-Myb is indirectly involved in the down-regulation of GATA-2, Runx1, PU.1, Fli-1, and CD45, a process that is a prerequisite for terminal differentiation. At later stages of erythropoiesis, high levels of c-Myb allow the cells to undergo terminal cell divisions. Finally, c-Myb maintains high levels of c-Kit expression on erythroid progenitors, thereby rendering them responsive to external signals regulating proliferation and differentiation.
Coordination of erythropoiesis by c-Myb. c-Myb has a dual role as a regulator of erythroid development. Both in uncommitted/early stages (CD71–/low) and in later stages (CFU-E/erythroblast—CD71+TER119–/low), high levels of c-Myb are required for an efficient response of progenitors to erythropoietic stimuli and progression to later stages. c-Myb is indirectly involved in the down-regulation of GATA-2, Runx1, PU.1, Fli-1, and CD45, a process that is a prerequisite for terminal differentiation. At later stages of erythropoiesis, high levels of c-Myb allow the cells to undergo terminal cell divisions. Finally, c-Myb maintains high levels of c-Kit expression on erythroid progenitors, thereby rendering them responsive to external signals regulating proliferation and differentiation.
That c-Myb might influence commitment processes related to the erythroid and megakaryocytic lineages was suggested by recent studies on the c-mybKD allele18 and other compromised c-myb alleles,19,20 which indicate that c-Myb inhibits megakaryocytic commitment and differentiation while simultaneously being required for normal erythroid development. In agreement with this, we measured an increased frequency of CFU-E stage cells expressing the megakaryocytic marker CD41 in the fetal liver of c-mybKD/KD embryos (data not shown). Given the currently restricted knowledge on the regulation of erythroid versus megakaryocytic commitment, it is an intriguing possibility that c-Myb positively regulates molecular pathways that simultaneously promote the erythroid program and suppress the megakaryocytic program.
Which c-Myb–dependent molecular pathways promote erythroid development?
The growth characteristics of pre–CFU-E progenitors expressing reduced levels of c-Myb indicate that cell cycle slowing can be excluded as a cause of the compromised erythroid development. Our results indicate rather that c-Myb influences a network of regulator molecules. It is noteworthy that we did not detect the down-regulation of any erythroid-specific mRNA in c-mybKD/KD CFU-E stage progenitors, implying that instruction of the erythroid expression program is not affected by reduced c-Myb. However, we found increased expression of the transcription factors Runx1, PU.1, and Fli-1, which are normally down-regulated during erythroid differentiation.33,34 It is plausible that elevated expression of Fli-1 and PU.1 could interfere with the erythroid program and result in slowed progression. Hence, it has been shown that Fli-1 inhibits the differentiation of erythroid progenitors31,36 and can antagonize the function of the erythroid transcription factor EKLF.37 Similarly, PU.1 suppresses erythroid commitment and differentiation through its functional antagonism with GATA-1.11 We also found increased expression of GATA-2 in c-mybKD/KD CFU-E stage cells. Although GATA-2 has been shown to participate in the priming of erythroid-specific gene transcription, it also promotes proliferation and inhibits differentiation,8,38 and it is down-regulated during differentiation.39 We tested whether c-Myb directly inhibits Runx1, PU.1, Fli-1, and GATA-2 expression by measuring RNA levels in response to inducible c-myb disruption in enriched erythroid progenitors. No significant change was observed (data not shown), suggesting that these genes are unlikely to be direct targets of c-Myb in erythroid progenitors and that the observed elevated expression in c-mybKD/KD cells might be related to their developmental retardation.
In contrast to the changes in transcription factor RNAs, the strong correlation between the manipulated expression levels of c-Myb and the levels of c-Kit, both at the protein and the RNA levels, firmly supports the hypothesis that the c-Kit gene is regulated by c-Myb. Previous studies have provided evidence for this hypothesis using c-myb antisense oligonucleotides,40 whereas a truncated c-Myb protein was shown to be required for c-Kit expression in transformed myelomonocytic cells,41 and both the human and the murine c-Kit promoter have been shown to contain c-Myb binding sites that are able to mediate transactivation by c-Myb.41,42 However, our study is the first to demonstrate that c-Myb is strictly required in vivo for c-Kit expression in normal erythroid progenitors.
In conclusion, c-Kit represents a good candidate for mediating the function of c-Myb in promoting the progression of erythroid progenitors. Further insight will be gained from experiments, currently under way, that restore high levels of c-Kit in the c-myb knockdown or knockout background.
Differential requirement for c-Myb in early and late erythroid cells
Pre–CFU-E progenitors expressing reduced levels of c-Myb proliferated like wild-type cells in response to EPO, SCF, and dexamethasone in short-term and steady state cultures. This suggests that, at least at this stage, any effect of c-Myb on the cell cycle is unrelated to its effect on erythroid development. On the other hand, CFU-E stage progenitors from the fetal liver of c-mybKD/KD embryos displayed accelerated cell cycle exit, which was associated with a more rapid down-regulation of c-Kit surface expression. It seems that despite the immature character of these cells in vivo, once committed to terminal differentiation, they require normal levels of c-Myb to undergo the maturation-associated terminal cell divisions characteristic of CFU-E development. The more rapid down-regulation of c-Kit compared with wild type, which does not take place in less mature cells, could account for the accelerated cell cycle exit of these progenitors. The ability of SCF to promote proliferation and to inhibit terminal differentiation in CFU-E stage cells supports this hypothesis.43 When we analyzed the response of erythroid progenitors to the inducible inactivation of c-myb, we observed an absence of general erythroid differentiation, in agreement with the knockdown phenotype and the rapid cell cycle arrest paralleling c-Kit down-regulation (data not shown). We propose that different c-Myb–regulated pathways control proliferation and erythropoietic development.
Until now, the inhibition of erythroid differentiation has been considered a key function of c-Myb. However, this latter conclusion was established from studies on murine erythroleukemic cells through the overexpression of c-Myb27,44 or the use of a dominant-negative form of Myb, which also acts on B-Myb targets.28 Furthermore, in agreement with our observations, Kamano et al45 found that normal levels of c-Myb expression were required for efficient hemoglobin synthesis during hemin-induced differentiation of K562 cells.
Taken together, we have demonstrated that in contrast to earlier stages, high levels of c-Myb are more important for the terminal proliferation of CFU-E stage cells. This finding provides, along with the role of c-Myb in promoting commitment and progression through erythropoiesis, an additional explanation for the decreased abundance of erythroblast stages under conditions of reduced c-Myb levels (Figure 6).
In this study we have highlighted an unexpected role for c-Myb as a factor promoting commitment to erythropoiesis and progression from early to late stages of differentiation (Figure 6). We have shown that this function of c-Myb is probably not related to the cell cycle but rather to the control of a network of hematopoietic regulators. The expression of c-Kit in erythroid progenitors was tightly dependent on c-Myb levels. Finally, we demonstrated that c-Myb acts as a coordinator at the CFU-E stage by promoting further progression while supporting terminal cell divisions. The challenge now will be to determine which molecular pathways downstream of c-Myb are responsible for its different functions.
Prepublished online as Blood First Edition Paper, February 16, 2006; DOI 10.1182/blood-2005-07-2968.
Supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the staff of the Biomedical Services Unit for animal care and Roger Byrd for cell sorting. We also thank all members of the Frampton group for their support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal