Abstract
Delta-like 4 (Dll4), a membrane-bound ligand for Notch1 and Notch4, is selectively expressed in the developing endothelium and in some tumor endothelium, and it is induced by vascular endothelial growth factor (VEGF)-A and hypoxia. Gene targeting studies have shown that Dll4 is required for normal embryonic vascular remodeling, but the mechanisms underlying Dll4 regulatory functions are currently not defined. In this study, we generated primary human endothelial cells that overexpress Dll4 protein to study Dll4 function and mechanism of action. Human umbilical vein endothelial cells retrovirally transduced with Dll4 displayed reduced proliferative and migratory responses selectively to VEGF-A. Expression of VEGF receptor-2, the principal signaling receptor for VEGF-A in endothelial cells, and coreceptor neuropilin-1 was significantly decreased in Dll4-transduced endothelial cells. Consistent with Dll4 signaling through Notch, expression of HEY2, one of the transcription factors that mediates Notch function, was significantly induced in Dll4-overexpressing endothelial cells. The γ-secretase inhibitor L-685458 significantly reconstituted endothelial cell proliferation inhibited by immobilized extracellular Dll4 and reconstituted VEGFR2 expression in Dll4-overerexpressing endothelial cells. These results identify the Notch ligand Dll4 as a selective inhibitor of VEGF-A biologic activities down-regulating 2 VEGF receptors expressed on endothelial cells and raise the possibility that Dll4 may be exploited therapeutically to modulate angiogenesis.
Introduction
Notch signaling plays a crucial role in cell fate determinations of a variety of cell types during development and postnatally.1-5 Four Notch receptors have been identified in mammals, Notch1,6 Notch2,7 Notch3,8 and Notch4,9 and 5 ligands, Jagged110 and Jagged211 belonging to the Serrate family, and Delta1,12 Delta3,13 and Delta-like 4 (Dll4)14-16 belonging to the Delta family. Ligand binding to Notch receptors triggers the proteolytic release of Notch intracellular domain which translocates into the nucleus to form a nuclear complex with the transcription factor RBP-J (also named CSL and CBF1/Su(H)/Lag-1) and activates transcription of downstream target genes.17 In mammals, primary target genes of the Notch-intracellular domain/RBP-J complex include the HES (Hairy/Enhancer of Split)18,19 and HEY (HES-related with YRPW motif, also named HERP, HES-related repressor protein)20-23 family of genes, which act as transcription factors.
A number of observations indicate that the Notch signaling pathway plays a critical role in vascular development and homeostasis.24-26 In particular, the Notch1 and Notch4 genes are expressed in endothelial cells within the embryonic vasculature,9,27-29 and mice with targeted deletions of Notch1 alone or Notch1 plus Notch4 display severe defects in embryonic vascular remodeling with the mutant embryos dying at approximately gestational day E9.5 (embryonic day 9.5).30 Expression of activated Notch4 in the developing mouse vasculature also caused abnormal vessel structure and patterning, resulting in embryonic death at approximately day E10.5.31 In addition, expression of active Notch4 in human dermal microvascular endothelial cells inhibited endothelial cell sprouting on collagen.32
Dll4 is the most recently identified Notch ligand14-16,33 and was found to interact with Notch1 and Notch4.14,34 In situ hybridization and immunocytochemistry studies showed that the predominant site of Dll4 expression is the vasculature, particularly the arteries, arterioles, and capillaries during development,14,16,35 and small arteries, microvessels, and tumor vessels in adult mice.35 This selectivity is unique among Notch ligands.30 Recently, mice with targeted deletions of the Dll4 gene were generated, revealing characteristic vascular remodeling defects similar to those previously observed in the Notch1 mutant and the Notch1 and Notch4 double mutant mice.30,35-37 Strikingly, mice with heterozygous deletions of the Dll4 gene also failed to remodel the primary vascular plexus in the yolk sack and died at the embryo stage, providing evidence for the critical importance of Dll4 expression levels in vascular development.30,35-37 The VEGF gene is the only other known example of inactivation of a single allele resulting in marked vascular defects and embryonic lethality in mice.38,39 In vitro, hypoxia can induce the expression of both vascular endothelial growth factor (VEGF) and Dll4 in endothelial cells.16,35,40,41
Taking advantage of retroviral transduction, we have expressed Dll4 in primary human endothelial cells to delineate Dll4 function in these cells. We show that Dll4 inhibits expression of VEGF receptor-2 (VEGFR2) and neuropilin-1 (NRP1) coreceptor and by this mechanism likely modulates VEGF-A-induced endothelial cell function.
Materials and methods
Constructs
The cDNA of human full-length Dll4 (GenBank no. AF253468) was cloned from placental cDNA using as primers hDL4B sense, 5′-GGATCCCATATGGCGGCAGCGTCCCGTAGCGCCTC-3′, and hDL4E antisense, 5′-GAATTCTTATACCTCCGTGGCAATGACACATTCATTC-3′, followed by TA cloning into pGEM-T easy vector (Promega, Southampton, United Kingdom). Cloning accuracy was verified by DNA sequencing. Full-length Dll4 was cut from the pGEM-T easy vector using BamHI and EcoRI restriction enzymes and ligated into the retroviral plasmid LZRSpBMN-linker-IRES-EGFP. Ligation sites were sequenced, and accuracy of insertion was verified.
Cells, cell culture, and reagents
Human umbilical vein endothelial cells (HUVECs)42 were used at passages 3 to 7. The Phoenix amphotropic viral packaging cell line (Orbigen, San Diego, CA) was grown in DMEM medium (Gibco-Invitrogen, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Biofluids Rockville, MD) and 1.6 mM l-glutamine (Sigma Chemical, St Louis, MO). Recombinant human Dll4 was from R&D Systems (Minneapolis, MN). The γ-secretase inhibitor L-685458 (Sigma Chemical or Peptide Institute, Osaka, Japan), dissolved in dimethyl sulfoxide (DMSO; Sigma Chemical), was used at final concentrations of 0.1 to 7.5 μM.
Virus packaging and infection
Phoenix cells grown on 10-cm dishes to 50% confluency were transfected with retroviral constructs using FuGENE6 (Roche Molecular Biochemicals, Indianapolis, IN) or Lipofectamine2000 (Invitrogen, Carlsbad, CA) for 24 hours. Fresh HUVEC culture medium with 2 μg/mL Puromycin (Sigma Chemical) was added. After reaching confluency (24-48 hours), cells were washed and cultured in Opti-MEM I (Gibco, Invitrogen) for 36 to 48 hours. The virus-containing culture supernatant was filtered (0.4 μm) and added with Polybrene (4 ng/mL; Sigma Chemical) to 50% confluent HUVECs at passage 2.After 3 hours at 37°C, cells were incubated in HUVEC culture medium for an additional 24 hours, at which time cells were washed. Subsequently, the cells were maintained under standard conditions.
RNA preparation and quantitative RT-PCR
Total RNA was extracted (Absolutely RNA Microprep Kit; Stratagene, La Jolla, CA). Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) was performed (One Step RT-PCR kit; Qiagen, Valencia, CA) with SYBR Green PCR master mix (Applied Biosystem, Foster City, CA), as described.43 The reaction was carried out in an Abi Prism 7900HT sequence detection system (Applied Biosystems). Primers used included GAPDH sense (5′-GCCACCCAGAAGACTGTGGATGGC) and antisense (5′-CATGATGGCCATGAGGTCCACCAC), Dll4 sense (5′-GACCACTTCGGCCACTATGT) and antisense (5′-CCTGTCCACTTTCTTCTCGC), HEY1 sense (5′-AACTGTTGGTGGCCTGAATC) and antisense (5′-AATTCTTTGTGTTGCTGGGG), HEY2 sense (5′-TTCAAGGCAGCTCGGTAACT) and antisense (5′-GGGCATTTTACTTCCCCAAT), ephrinB2 sense (5′-GAAAATACCCCTCTCCTCAACT) and antisense (5′-CTTCGGAACCGAGGATGTTGTTC), Neuropilin-1 sense (5′-CAAGGCGAAGTCTTTTGAGG) and antisense (5′-CACCTGTGAGCTGGAAGTCA), VEGFR1 sense (5′-GCACCTTGGTTGTGGCTGAC) and antisense (5′-CGTGCTGCTGCTTCCTGGTCC), VEGFR2 sense (5′-GGAAATCATTATTCTAGTAGGCACGACG) and antisense (5′-CCTGTGGATACACTTTCGCGATG), FGFR1 sense (5′-GGAGGATCGAGCTCACTGTGG) and antisense (5′-CGGAGAAGTAGGTGGTGTCAC), VEGF-A sense (5′-CCTTGCTGCTCTACCTCCAC) and antisense (5′-ATGTTGGACTCCTCAGTGGG), Notch1 sense (5′-GCAACAGCTCCTTCCACTTC) and antisense (5′-GCCTCAGACACTTTGAAGCC), Notch2 sense (5′-CCCAATGGGCAAGAAGTCTA) and antisense (5′-CACAATGTGGTGGTGGGATA), Notch3 sense (5′-TCTTGCTGCTGGTCATTCTC-3′) and antisense (5′-TGCCTCATCCTCTTCAGTTG-3′), and Notch4 sense (5′-CACTGAGCCAAGGCATAGAC) and antisense (5′-ATCTCCACCTCACACCACTG). RT-PCR reactions were performed at 50°C for 30 minutes, 94°C for 15 minutes, 35 cycles of 94°C for 1 minute, 60°C (58°C for Notch3 and 55°C for Notch1, Notch2, and Notch4) for 1 minute, and 72°C for 1 minute, and 72°C for 10 minutes, followed by a dissociation step of 15 minutes at 95°C, 15 minutes at 60°C, and 15 minutes at 95°C.
Cell-cycle analysis and flow cytometry
Cell-cycle analysis was carried out in HUVECs synchronized by 24 hours in “starvation medium” consisting of M199 (Gibco-Invitrogen) with 2.5% FBS (Biofluids), 25 μg/mL porcine heparin (Sigma Chemical) or in exponentially growing HUVECs. Cells were incubated in starvation medium supplemented with 50 ng/mL VEGF-A (R&D Systems). Where noted, cells were pulsed with 10 μM 5′-Bromo-2′-deoxyuridine (BrdU; Sigma Chemical) by incubation for 1 hour prior to harvest. At intervals (0-72 hours), cells were detached with 0.05% Trypsin/ethylenediaminetetraacetic acid (EDTA) (Gibco, Invitrogen), washed, suspended in 0.5 to 1 mL cold 70% ethanol in PBS. After washing, incubation with 2 M HCl/Triton X-100 for 30 minutes at room temperature, neutralization and suspension in PBS containing 0.5% Tween-20 and 1% BSA, cells were incubated with FITC-labeled mouse monoclonal anti-BrdU antibody (Becton Dickinson Immunocytometry Systems, San Jose, CA). For propidium iodide (PI) incorporation, cells were suspended in PBS with 50 μg/mL PI (Sigma Chemical) and 100 U/mL Ribonuclease A (RNase; Sigma Chemical), incubated at 37°C for 30 minutes, and rinsed with PBS. At least 10 000 events were acquired, and results were analyzed with CellQuest (Becton Dickinson, Franklin Lakes, NJ) and MODFIT LT software (Topsham, ME), as described.44 For flow cytometric analysis of enhanced green fluorescent protein (EGFP) expression, HUVECs were suspended in 0.5 mL PBS and examined.43 Flow cytometric analysis of surface antigens was performed as described.42 For VEGFR2 staining, we used mouse monoclonal anti-VEGFR2 (ab9530; Abcam, Cambridge, MA) and Alexa594-labeled goat antimouse antibody (Molecular Probes, Eugene, OR). Control staining was with mouse IgG1 and Alexa594-labeled goat antimouse antibody. For neuropilin-1 staining, we used PE-labeled mouse monoclonal anti-BDCA-4 (human neuropilin-1) antibody (AD5-17F6 clone; Miltenyi Biotec GmbK, Bergisch Gladbach, Germany) and for CD31 staining a PE-labeled mouse monoclonal anti-human CD31 antibody (Pharmingen, BD Biosciences, San Diego, CA). Data were collected using a fluorescence-activated cell sorting (FACS) Calibur flow cytometer (Becton Dickinson) and were analyzed using CELLQuest software (Becton Dickinson).
Immunocytochemistry
A rabbit immune serum against human Dll4 (named R3) was produced by immunization with recombinant intracellular domain of Dll4 produced in Escherichia coli. Human Dll4 intracellular domain was cloned into the expression vector pET-28C-D4ICD, using primers: sense 5′-GGATCCCATATGCGTCAG-CTGCGTCTTCGTCGTCCGG-3′ and hDl4E antisense (see “Constructs”). PCR product was cloned into pGEM-T easy vector to produce the pGEM-T-D4ICD plasmid. The plasmid was then digested with NdeI and EcoRI restriction enzymes, and the fragment was inserted into pET-28C to produce the pET-28C-D4ICD expression vector. The vector was transformed into BL21 Codon+ competent cells for expression of recombinant protein, (MX3H6 × 10.D4ICD). Dll4 intracellular domain was purified from inclusion bodies and used for rabbit immunization. Cytospin preparations of HUVECs (10-20 000 cells/slide, 38.2 g for 6 minutes) were fixed with acetone for 10 minutes at room temperature, washed with PBS over 20 minutes, blocked with 10% fetal bovine serum in PBS, incubated with R3 rabbit antibody (1:300 dilution in PBS containing 3% FBS, 1 hour at 37°C), washed in PBS over 20 minutes, and then incubated with a goat antirabbit antibody labeled with horseradish-peroxidase (DAKO, Carpinteria, CA) for 30 minutes at 37°C. After washing, slides were incubated 30 minutes at room temperature in 3-amino-9-ethylcarbazol (AEC) substrate (21 mL 0.1 M Na acetate 79 mL plus 0.1 M acetic acid plus 6 mL AEC, 100 μL 30% H2O2) and counterstained with Meyer hematoxylin for 5 to 10 minutes.
Western blotting and VEGF ELISA
Immunoblotting was performed as described.45 For Dll4 recognition, we used rabbit R3 anti-human Dll4 antibody (dilution 1:250) with an affinity-purified, peroxidase-linked, donkey IgG antirabbit antibody (Amersham Biosciences, Piscataway, NJ); a mouse monoclonal anti-human/mouse Dll4 (MAB1389, 1:500 dilution; R&D Systems) with a peroxidase-linked donkey IgG antimouse antibody; or an affinity-purified goat anti-Dll4 peptide antibody (C-20, 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) with a peroxidase-linked donkey anti-goat IgG antibody (Santa Cruz Biotechnology). For identification of β-actin, we used a goat IgG anti-actin antibody (C11, 1:1000 dilution; Santa Cruz Biotechnology) with a donkey anti-goat IgG HRP-conjugated (Santa Cruz Biotechnology). Antibody detection was by chemiluminescence using enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech, Piscataway, NJ). VEGF-A was measured by enzyme-linked immunosorbent assay (ELISA; R&D Systems; lower limit of detection approximately 3 pg/mL).
Cell proliferation assay
HUVEC proliferation was measured as described.42 HUVECs were seeded in triplicate in 96-well plates (2-4 × 103 cells/well in 0.2 mL RPMI 1640 culture medium supplemented with 18% heat-inactivated FBS [Biosource, Camarillo, CA]) and 18 U/mL porcine heparin, with or without human VEGF-A165 (3-24 ng/mL; R&D Systems) or human bFGF (3-28 ng/mL; R&D Systems) and incubated for 3 days. HUVECs were also cultured onto microtiter 96-well plates that were preincubated for 18 hours at 4°C with 50 μL recombinant human Dll4 (1 μg/mL in PBS or gelatin; R&D Systems). When L-685458 was used, medium was replenished every 24 hours with inhibitor or DMSO control. Proliferation was measured by 3H-thymidine deoxyribose uptake (0.5 μCi [0.0185 MBq]/well, 25 Ci/mmol [925 GBq/mmol]; New England Nuclear, Boston, MA) during the last 18 to 22 hours of culture. Results are expressed as mean (± SEM) cpm/culture.
Matrigel cord formation assay
The in vitro matrigel assay was performed essentially as described.45 HUVECs were plated (40 000-75 000 cells) onto 24-well tissue culture plates coated with 200 to 300 μL solidified matrigel (an extract of the Englebreth-Holm-Swarm tumor, Collaborative; BD Pharmingen, San Diego, CA). After 16 to 18 hours of incubation, cells were photographed (Retiga 1300 digital camera; Qimaging, Burnaby, BC, Canada) under phase-contrast microscopy (Olympus 1 × 51 with a 10 × 0.25 NA PhL lens; Olympus Optical, Melville, NY), and images were obtained with IPLab for Windows software (Scanalytics, Fairfax, VA) imported into Adobe Photoshop (Adobe Systems, San Jose, CA). Network formation was measured by counting the number of cord angles/field (each field is defined as the area visualized by a 4 × magnification lens). Each experimental condition was tested in 8 separate experiments.
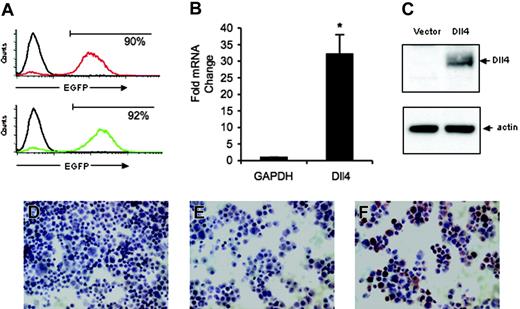
Dll4 expression in retrovirally transduced primary human endothelial cells. (A) Flow cytometric measurement of EGFP expression in HUVECs infected 3 days earlier with control retrovirus or Dll4 retrovirus. Representative images from 8 independent infections. (B) Levels of Dll4 expression in HUVECs 3 to 15 days after transduction with control vector or Dll4 retrovirus were evaluated by quantitative RT-PCR relative to GAPDH expression. The results reflect the mean (± SEM) from 14 separate determinations from 8 infections. *P = .012. (C) Dll4 and actin expression in HUVECs transduced with control vector or Dll4 retrovirus evaluated by immunoblotting with R3 rabbit anti-Dll4 antibody and reblotting with goat anti-β-actin antibody. (D-F) Immunocytochemical analysis of Dll4 expression in HUVECs transduced with Dll4 (F) or vector only (D) using rabbit anti-Dll4 antibodies; control staining of Dll4-transduced HUVECs with preimmune rabbit serum (E).
Dll4 expression in retrovirally transduced primary human endothelial cells. (A) Flow cytometric measurement of EGFP expression in HUVECs infected 3 days earlier with control retrovirus or Dll4 retrovirus. Representative images from 8 independent infections. (B) Levels of Dll4 expression in HUVECs 3 to 15 days after transduction with control vector or Dll4 retrovirus were evaluated by quantitative RT-PCR relative to GAPDH expression. The results reflect the mean (± SEM) from 14 separate determinations from 8 infections. *P = .012. (C) Dll4 and actin expression in HUVECs transduced with control vector or Dll4 retrovirus evaluated by immunoblotting with R3 rabbit anti-Dll4 antibody and reblotting with goat anti-β-actin antibody. (D-F) Immunocytochemical analysis of Dll4 expression in HUVECs transduced with Dll4 (F) or vector only (D) using rabbit anti-Dll4 antibodies; control staining of Dll4-transduced HUVECs with preimmune rabbit serum (E).
Migration assay
Endothelial migration assays were performed using 0.2% gelatin-coated polycarbonate filters (pore size 8 μm) of transwells; Costar, Cambridge, MA). HUVECs transduced with empty vector or Dll4 (5 × 105/well) were placed in the upper chamber in migration medium (RPMI 1640 containing 0.5% BSA and 10 mM HEPES). The lower chamber contained migration medium with or without 100 ng/mL VEGF-A or 100 ng/mL bFGF. After 16 to 20 hours of incubation at 37°C, viable cells in the lower chamber were counted.
Statistical analysis
Group differences were evaluated by Student t test; P values less than .05 were considered significant.
Results
Up-regulation of Dll4 in primary human endothelial cells
We selected retrovirus-mediated transduction to overexpress Dll4 in primary human endothelial cells derived from the umbilical vein (HUVECs) to achieve moderate levels of Dll4 expression. In culture, HUVECs constitutively express low-level Dll4 mRNA detected by RT-PCR.42 We found that approximately 90% of HUVECs expressed enhanced green fluorescent protein (EGFP) on day 3 after infection with the control retrovirus (Figure 1A). A similar proportion of HUVECs (92%) expressed EGFP on day 3 after infection with Dll4 retrovirus (Figure 1A). When infection efficiency fell below 60%, cells were FACS sorted for 100% EGFP expression. The proportion of EGFP-positive cells decreased on the average by approximately 15% over 7 to 10 days after infection. All results were obtained from 8 separate populations of cells in which greater than 50% of cells were positive for EGFP.
By quantitative real-time PCR analysis, we found that Dll4 transcript levels were significantly higher in Dll4 cells than in control cells. In 14 determinations from 8 separate infections, levels of Dll4 expression normalized to GAPDH were 32- ± 5.9-fold (mean ± SEM) higher (P < .001) in Dll4 cells than in the control cells (Figure 1B).
By immunoblotting, we found that antibody R3 we raised against the intracellular domain of Dll4 identified a band migrating at approximately 70 kDa in cell extracts from Dll4-transduced cells but not in cell extracts from the control cells (Figure 1C). The predicted molecular weight of full-length Dll4 is calculated to be 75 kDa. This difference could not be attributed to uneven loading as demonstrated by membrane reblotting with antibodies directed against actin (Figure 1C). Similar results showing Dll4 protein expression in Dll4 cells were derived from membrane reblotting with a purified rat monoclonal antibody raised against recombinant Dll4 (R&D Systems) and with an affinity-purified goat polyclonal antibody directed against a C-terminal peptide of Dll4 (Santa Cruz Biotechnology) (not shown). We conclude that Dll4-transduced HUVECs express Dll4 protein, whereas control HUVECs express insufficient amounts of Dll4 protein to be detected by Western blotting.
Up-regulation of Dll4 protein in transduced HUVECs was confirmed by immunocytochemical staining of cytospin preparations. HUVECs infected with the Dll4 retrovirus were Dll4-positive (brown staining) when immunostained by the R3 antibody (Figure 1F). By contrast, HUVECs infected with the wild-type vector (Figure 1D) were essentially negative. In addition, a preimmune rabbit serum (from the same rabbit producing R3) did not immunostain HUVECs transduced with Dll4 (Figure 1E). Thus, these results demonstrate that primary human endothelial cells can be transduced to overexpress Dll4.
Effects of Dll4 overexpression on endothelial cell proliferation and cell-cycle progression
We examined the effects of Dll4 overexpression on HUVECs growth in response to the proangiogenic factors vascular endothelial growth factor (VEGF)-A and basic fibroblast growth factor (bFGF). To best evaluate the effects of Dll4 overexpression rather than clonal variation, we selected for functional experiments HUVEC cultures in which similar proportions of cells were EGFP-positive after infection with control and Dll4 retrovirus, reflecting effective and comparable transduction levels. When stimulated for 3 days in the presence of VEGF-A (25 ng/mL), Dll4-transduced HUVECs consistently proliferated to a significantly lower degree (P < .001) than the control cells (Figure 2A).
This difference was observed over a wide range of VEGF-A concentrations (Figure 2B; representative experiment). Consistent with this reduced proliferative capacity of Dll4-overexpressing HUVECs, there were significantly fewer cells in VEGF-A (25 ng/mL)-supplemented cultures of Dll4-transduced HUVECs compared with the control HUVECs (not shown). In a representative experiment (of 4 performed), there were 115 × 103 Dll4-transduced viable HUVECs as opposed to 216 × 103 control HUVECs after 3-day incubation. In contrast to their reduced proliferative response to VEGF-A, Dll4-transduced and control HUVECs proliferated similarly in response to bFGF at varying concentrations (Figure 2C; representative experiment). Thus, Dll4 overexpression in primary human endothelial cells is associated with reduced proliferation in response to VEGF-A.
Effects of Dll4 expression on endothelial cell proliferation. (A) HUVECs transduced 3 to 10 days earlier with either vector only or Dll4 (70%-90% of cells expressing EGFP) were cultured for 72 hours in medium alone or medium supplemented with VEGF-A (25 ng/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SEMs) from 5 separate experiments, each performed in triplicate cultures. *P < .001. (B) Dose dependency of VEGF-A (3-24 ng/mL) induced HUVEC proliferation 3 to 10 days after transduction with vector only or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate cultures (representative of 4 determinations). (C) Analysis of HUVEC proliferation in response to bFGF (3-28 ng/mL) 3 to 10 days after transduction with vector alone or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate determinations (representative of 4 determinations).
Effects of Dll4 expression on endothelial cell proliferation. (A) HUVECs transduced 3 to 10 days earlier with either vector only or Dll4 (70%-90% of cells expressing EGFP) were cultured for 72 hours in medium alone or medium supplemented with VEGF-A (25 ng/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SEMs) from 5 separate experiments, each performed in triplicate cultures. *P < .001. (B) Dose dependency of VEGF-A (3-24 ng/mL) induced HUVEC proliferation 3 to 10 days after transduction with vector only or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate cultures (representative of 4 determinations). (C) Analysis of HUVEC proliferation in response to bFGF (3-28 ng/mL) 3 to 10 days after transduction with vector alone or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate determinations (representative of 4 determinations).
To characterize this reduced proliferative capacity of Dll4-overexpressing HUVECs in response to VEGF-A, we examined cell-cycle distribution during VEGF-A stimulation. First, the cells were synchronized by culture for 24 hours under starvation conditions (serum concentration in the culture medium was reduced 2.5%; ascorbate and ECGS omitted) and then stimulated with VEGF-A (50 ng/mL) in starvation medium. Initially (0 time point), the cell distribution in the different phases of the cell cycle was similar in control and Dll4 HUVECs, reflecting a similar proliferative capacity of these cells in the presence of endothelial cell growth supplement, which contains bFGF and acidic FGF, used for culture maintenance (Figure 3A, representative experiment of 4 performed). However, at subsequent time points (24 and 48 hours), a clear difference in cell-cycle progression between control cells and Dll4-overexpressing HUVECs was noted. In the control cultures (vector), the percentage of cells in the G0/G1 phase of cell cycle fell from 84% (time 0) to 53% after 48-hour starvation in the presence of VEGF-A, whereas the percentage of cells in the S phase of the cell cycle correspondingly increased from 6.7% to 36%. Under the same culture conditions and testing, the percentage of Dll4-transduced HUVECs (Dll4) in the G0/G1 phase of cell cycle fluctuated between 83% and 90%, and the percentage of cells in S phase fluctuated only between 5% and 6% (Figure 3A). These experiments provided evidence that, after stimulation with VEGF-A, HUVECs overexpressing Dll4 have a reduced capacity to enter S phase compared with control cells.
Analysis of the effects of Dll4 overexpression on cell-cycle distribution. (A) HUVECs transduced with Dll4 or vector only (60%-90% of cells expressing EGFP) were synchronized by 24-hour incubation in starvation medium supplemented with 2.5% serum and then stimulated with VEGF-A (50 ng/mL) for 24 or 48 hours. Cell-cycle distribution was evaluated by flow cytometric analysis of relative DNA content, after cell fixation in cold 70% ethanol and incorporation of propidium iodide (PI). The results, analyzed by MODFIT LT software, reflect the percentage of cells found in the G0/G1, S, and G2/M phases of the cell cycle at time 0 (end of synchronization), 24 and 48 hours after culture with VEGF-A. Representative experiment of 4 performed. (B) Exponentially growing HUVECs transduced with vector only or Dll4 (60%-90% of cells expressing EGFP) were cultured in serum-reduced (2.5%) medium containing VEGF-A (50 ng/mL) for 24 to 72 hours. Cells were pulsed with BrdU (10 μM) over 1 hour prior to harvest. After fixation in 70% ethanol and Triton-X 100 permeabilization, the cells were stained with FITC-labeled mouse monoclonal anti-BrdU antibodies and subsequently allowed to incorporate PI. Cell-cycle distribution was evaluated by flow cytometry. The results reflect the percentage of cells in the S phase of cell cycle measured at time 0 (1 hour after culture in serum-reduced medium), 24, 48, and 72 hours after incubation in VEGF-A-supplemented serum-reduced culture medium.
Analysis of the effects of Dll4 overexpression on cell-cycle distribution. (A) HUVECs transduced with Dll4 or vector only (60%-90% of cells expressing EGFP) were synchronized by 24-hour incubation in starvation medium supplemented with 2.5% serum and then stimulated with VEGF-A (50 ng/mL) for 24 or 48 hours. Cell-cycle distribution was evaluated by flow cytometric analysis of relative DNA content, after cell fixation in cold 70% ethanol and incorporation of propidium iodide (PI). The results, analyzed by MODFIT LT software, reflect the percentage of cells found in the G0/G1, S, and G2/M phases of the cell cycle at time 0 (end of synchronization), 24 and 48 hours after culture with VEGF-A. Representative experiment of 4 performed. (B) Exponentially growing HUVECs transduced with vector only or Dll4 (60%-90% of cells expressing EGFP) were cultured in serum-reduced (2.5%) medium containing VEGF-A (50 ng/mL) for 24 to 72 hours. Cells were pulsed with BrdU (10 μM) over 1 hour prior to harvest. After fixation in 70% ethanol and Triton-X 100 permeabilization, the cells were stained with FITC-labeled mouse monoclonal anti-BrdU antibodies and subsequently allowed to incorporate PI. Cell-cycle distribution was evaluated by flow cytometry. The results reflect the percentage of cells in the S phase of cell cycle measured at time 0 (1 hour after culture in serum-reduced medium), 24, 48, and 72 hours after incubation in VEGF-A-supplemented serum-reduced culture medium.
Additionally, we confirmed this observation by measuring the fraction of cells in the S phase of the cell cycle based on BrdU and PI incorporation. Exponentially growing, unsynchronized HUVECs were incubated for 72 hours in culture medium containing 2.5% serum and 50 ng/mL VEGF. Under these restrictive culture conditions, the proportion of BrdU-positive cells in the S phase of the cell cycle decreased over 72 hours in both control and Dll4-transduced HUVECs, despite the presence of VEGF-A, but the reduction was greater in Dll4-overexpressing cells as compared with control cells (Figure 3B).
Effects of Dll4 overexpression on endothelial cell migration and matrix-dependent cord formation
These results provided evidence that Dll4-overexpressing endothelial cells are defective in their ability to replicate in response to VEGF-A. We examined whether this defect was limited to VEGF-A-induced proliferation or extended to other nonmitogenic functions of VEGF-A. In migration assays, VEGF-A (100 ng/mL) and bFGF (100 ng/mL) promoted a significant response from control HUVECs (Figure 4A). However, VEGF-A induced only modest migration from Dll4-overexpressing HUVECs, which was significantly (P = .021) reduced compared with the control HUVECs. This difference was not associated with a more global defect in cell migration, because Dll4-overexpressing HUVECs and control HUVECs migrated to a similar degree (P = .74) in response to bFGF (Figure 4A).
In additional analyses of nonmitogenic activities of VEGF-A, we examined the ability of endothelial cells to undergo a morphogenic change into cordlike structures. When incubated on extracellular matrices, such as collagen, fibrin, or matrigel (a mixture of the extracellular matrix proteins laminin, collagen IV, heparan sulfate proteoglycans, and entactin-nidogen), primary endothelial cells can form a characteristic network of cordlike structures.43,45 With HUVECs, matrigel-induced cord formation takes place over 6 to 16 hours of incubation, does not require cell division, and is critically dependent on endogenous VEGF-A and other factors.45-49 As shown in a representative experiment (Figure 4B), the control HUVECs formed the characteristic network of orderly branching cordlike structures, Dll4-overexpressing HUVECs formed many fewer such structures so that the network was often incomplete. Quantitative analysis of network formation in 8 independent experiments by measuring cord branching angles revealed that Dll4-overexpressing HUVECs were significantly (P = .031) defective in their ability to form networks compared with control cells (Figure 4C).
Effects of Dll4 overexpression on endothelial cell migration and extracellular matrix-dependent cord formation. (A) VEGF-A (100 ng/mL)- and bFGF (100 ng/mL)-induced transwell migration of control and Dll4-overexpressing HUVECs (5 × 105 cells/well) over 16 to 20 hours of incubation. The results reflect the mean fold increase (± SEM) in cell migration compared with medium alone in 3 experiments performed in triplicate. (B) Representative images reflecting cord formation by control and Dll4-overexpressing HUVECs (4-7.5 × 104 cells/well) plated in complete culture medium on matrigel-coated wells (24-well plate) and incubated 16 to 18 hours. Phase-contrast microscopy (original magnification, × 10). (C) Quantitative analysis of matrigel-induced cord formation in control and Dll4-overexpressing HUVECs. Cord formation was measured as a function of the number of cord angles per visual field (phase-contrast microscopy, 4 × magnification). The results represent the mean (± SD) from 8 independent experiments. *P = .001.
Effects of Dll4 overexpression on endothelial cell migration and extracellular matrix-dependent cord formation. (A) VEGF-A (100 ng/mL)- and bFGF (100 ng/mL)-induced transwell migration of control and Dll4-overexpressing HUVECs (5 × 105 cells/well) over 16 to 20 hours of incubation. The results reflect the mean fold increase (± SEM) in cell migration compared with medium alone in 3 experiments performed in triplicate. (B) Representative images reflecting cord formation by control and Dll4-overexpressing HUVECs (4-7.5 × 104 cells/well) plated in complete culture medium on matrigel-coated wells (24-well plate) and incubated 16 to 18 hours. Phase-contrast microscopy (original magnification, × 10). (C) Quantitative analysis of matrigel-induced cord formation in control and Dll4-overexpressing HUVECs. Cord formation was measured as a function of the number of cord angles per visual field (phase-contrast microscopy, 4 × magnification). The results represent the mean (± SD) from 8 independent experiments. *P = .001.
VEGFR2 and ephrinB2 modulation in endothelial cells that overexpress Dll4
We examined whether the reduced capacity to assemble into cordlike structures by Dll4-overexpressing cells might be attributable to reduced expression of endogenous VEGF, which is required for this morphogenic process. By quantitative RT-PCR analysis, we found that levels of VEGF mRNA were similar (P = .82) in the control- and Dll4-transduced HUVECs (Figure 5A). In addition, culture supernatants from the control cells and Dll4-overexpressing cells contained similar levels of VEGF (52 and 61 pg/mL, respectively) after 20 hours of incubation on matrigel. This provides evidence that Dll4 does not regulate VEGF expression in HUVECs and suggests that reduced VEGF-A expression is not the reason underlying defective matrigel-dependent network formation by Dll4-overexpressing HUVECs.
Regulation of VEGFR2 and NRP1 expression in Dll4-transduced endothelial cells. (A) Quantitative RT-PCR analysis of VEGF-A, VEGFR1, VEGFR2, FGFR1, and NRP1 expression in HUVECs transduced with vector only or Dll4. The results reflect relative mRNA levels (normalized to GAPDH) in Dll4-transduced and control HUVECs and are expressed as the fold mRNA change in Dll4 versus control HUVECs. The results represent the mean (± SEM) from 4 to 9 independent determinations. *P < .05. (B) Surface levels of CD31, VEGFR2, and NRP1 expression in vector-only (Vector) and Dll4-transduced (Dll4) HUVECs measured by flow cytometry. Control reflects background surface fluorescence staining with appropriate controls.
Regulation of VEGFR2 and NRP1 expression in Dll4-transduced endothelial cells. (A) Quantitative RT-PCR analysis of VEGF-A, VEGFR1, VEGFR2, FGFR1, and NRP1 expression in HUVECs transduced with vector only or Dll4. The results reflect relative mRNA levels (normalized to GAPDH) in Dll4-transduced and control HUVECs and are expressed as the fold mRNA change in Dll4 versus control HUVECs. The results represent the mean (± SEM) from 4 to 9 independent determinations. *P < .05. (B) Surface levels of CD31, VEGFR2, and NRP1 expression in vector-only (Vector) and Dll4-transduced (Dll4) HUVECs measured by flow cytometry. Control reflects background surface fluorescence staining with appropriate controls.
On the basis of the observation that Dll4 overexpression is associated with selectively reduced endothelial cell responses to VEGF-A, we examined VEGFR2 expression in Dll4-overexpressing HUVECs. It is known that VEGFR2 serves as the principal signaling receptor for endothelial cell proliferation, migration, survival, and angiogenesis induced by VEGF-A.50 By quantitative RT-PCR analysis, we consistently (5 determinations) found that VEGFR2 mRNA levels were significantly (P = .035) reduced in Dll4-overexpressing HUVECs in comparison with control HUVECs, whereas levels of VEGFR1 and FGFR1 mRNAs were similar (P > .1; Figure 5A). Additionally, we consistently (7 determinations) found that RNA levels of NRP1, a coreceptor for VEGF-A that enhances VEGF-A binding to VEGFR2,51 were significantly (P = .003) reduced in Dll4-transduced cells compared with control cells (Figure 5A). By flow cytometry, levels of surface VEGFR2 and NRP1 expression were markedly reduced in Dll4-overexpressing HUVECs compared with control HUVECs, whereas levels of surface CD31 were similar (Figure 5B).
Notch expression in HUVECs and contribution of Notch engagement to the phenotype of Dll4-transduced HUVECs
We looked for evidence of Notch expression in the HUVECs. By RT-PCR, we determined that the vector-transduced HUVECs used in the current experiment, express mRNA for the Notch receptors Notch1, Notch2, and Notch4 but only low-level Notch3 mRNA (Figure 6A). By quantitative RT-PCR analysis, we found that HEY2 mRNA levels were consistently and significantly (P = .013) increased in Dll4-transduced HUVECs compared with controls, whereas HEY1 mRNA levels were similar (P = .2; Figure 6B). Also, we found that ephrinB2 mRNA levels were significantly (P = .019) higher in Dll4-overexpressing HUVECs than in the control HUVECs, consistent with previous observations that HEY1/HEY2-deficient mice do not express ephrinB2.52 Thus, overexpression of Dll4 in HUVECs was associated with a significantly enhanced expression of HEY2 and ephrinB2, consistent with enhanced Notch signaling in these cells.
To more directly assess the contribution of Notch signaling to the phenotype of Dll4-transduced HUVECs and distinguish it from the potential contribution of reverse signaling by the intracellular domain of Dll4, we used recombinant human Dll4 (rhDLL4), which lacks the intracellular and transmembrane domains. We found that rhDLL4 immobilized onto culture wells reduced HUVEC proliferation in response to VEGF-A (Figure 6C). In addition, we found that HUVECs cultured on plates coated with rhDLL4 showed decreased surface VEGFR2 expression compared with HUVECs cultured on diluent-coated plates (Figure 6D). These results demonstrate that the extracellular domain of Dll4 is sufficient to inhibit endothelial cell proliferation in response to VEGF-A and to reduce surface VEGFR2 expression. These effects are similar to those induced by full-length Dll4, suggesting that the intracellular and transmembrane domains of Dll4 are not required for activity in the current system, and that Dll4 likely acts through Notch rather than Dll4 signaling.
Proteolytic processing of Notch by γ-secretase is an essential step after receptor activation.17 As a consequence, γ-secretase inhibitors block activation of the Notch pathway. To further establish that extracellular Dll4 inhibits VEGF-A-induced HUVEC proliferation acting through Notch, we used the γ-secretase inhibitor L-685458.53 As shown in Figure 6E, L-685458 specifically and dose dependently reconstituted VEGF-A-induced HUVEC proliferation reduced by rhDLL4. These results are consistent with Dll4 signaling through Notch receptors.
We also tested the effects of the γ-secretase inhibitor L-685458 on VEGFR2 expression in control and Dll4-overexpressing HUVECs (Figure 6F). When levels of VEGFR2 mRNA were reduced by at least 5-fold in Dll4-transduced HUVECs compared with vector-transduced HUVECs, L-685458 (2 μM, 72-hour incubation) significantly (P = .041) reconstituted VEGFR2 mRNA levels in Dll4-overexpressing HUVECs compared with vector-transduced HUVECs. Reconstitution of VEGFR2 expression was more dependent on defined experimental conditions in Dll4-overexpressing HUVECs compared with HUVECs induced by rhDLL4, likely reflecting the complexities of the retroviral expression system. Together, these results provide evidence that Notch signaling contributes to reduced expression of VEGFR2 and reduced proliferation to VEGF-A in Dll4-transduced primary endothelial cells.
Expression of Notch receptors in endothelial cells and Notch function in Dll4-transduced cells. (A) mRNAs for Notch receptors 1 to 4 detected by RT-PCR in HUVECs transduced with vector. (B) Quantitative RT-PCR analysis of HEY1, HEY2, ephrinB2, and EphB2 expression in control HUVECs transduced with vector only or Dll4-overexpressing HUVECs. The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4 versus control HUVECs from 2 to 11 separate determinations. *P < .05. (C) Effects of immobilized soluble recombinant human Dll4 (rhDLL4) on the proliferation of HUVECs. The endothelial cells were cultured for 72 hours in the presence of VEGF-A (25 ng/mL) on plates coated with rhDLL4 (1 μg/mL) or diluent (0.1% BSA); proliferation was measured by 3H thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .05. (D) Surface levels of VEGFR2 expression in HUVECs cultured for 48 hours onto rhDLL4-coated plates (1 μg/mL) (rhDLL4) or diluent-coated plates (gelatin) measured by flow cytometry. Control reflects background surface fluorescence staining with appropriate controls. Representative of 2 performed. (E) Effects of the γ-secretase inhibitor L-685458 (0.1-7.5 μM) on the proliferation of HUVECs cultured with rhDLL4-coated plates (1 μg/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .01 (rhDLL4 with 4 μM L-685458 inhibitor versus VEGF alone). (F) Quantitative RT-PCR analysis of GAPDH and VEGFR2 expression in vector- and Dll4-transduced HUVECs after 72-hour culture with L-685458 (2 μM) or diluent only (0.02% DMSO). The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4-transduced versus control HUVECs from 4 separate experiments, in which levels of VEGFR2 expression in the absence of inhibitor were reduced by at least 5-fold in Dll4-transduced compared with control HUVECs (*P < .05).
Expression of Notch receptors in endothelial cells and Notch function in Dll4-transduced cells. (A) mRNAs for Notch receptors 1 to 4 detected by RT-PCR in HUVECs transduced with vector. (B) Quantitative RT-PCR analysis of HEY1, HEY2, ephrinB2, and EphB2 expression in control HUVECs transduced with vector only or Dll4-overexpressing HUVECs. The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4 versus control HUVECs from 2 to 11 separate determinations. *P < .05. (C) Effects of immobilized soluble recombinant human Dll4 (rhDLL4) on the proliferation of HUVECs. The endothelial cells were cultured for 72 hours in the presence of VEGF-A (25 ng/mL) on plates coated with rhDLL4 (1 μg/mL) or diluent (0.1% BSA); proliferation was measured by 3H thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .05. (D) Surface levels of VEGFR2 expression in HUVECs cultured for 48 hours onto rhDLL4-coated plates (1 μg/mL) (rhDLL4) or diluent-coated plates (gelatin) measured by flow cytometry. Control reflects background surface fluorescence staining with appropriate controls. Representative of 2 performed. (E) Effects of the γ-secretase inhibitor L-685458 (0.1-7.5 μM) on the proliferation of HUVECs cultured with rhDLL4-coated plates (1 μg/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .01 (rhDLL4 with 4 μM L-685458 inhibitor versus VEGF alone). (F) Quantitative RT-PCR analysis of GAPDH and VEGFR2 expression in vector- and Dll4-transduced HUVECs after 72-hour culture with L-685458 (2 μM) or diluent only (0.02% DMSO). The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4-transduced versus control HUVECs from 4 separate experiments, in which levels of VEGFR2 expression in the absence of inhibitor were reduced by at least 5-fold in Dll4-transduced compared with control HUVECs (*P < .05).
Discussion
In this study, we have expressed Dll4 in primary human endothelial cells by use of retroviral gene transduction and have identified Dll4 as a negative regulator of endothelial cell responses to VEGF-A. We found that Dll4 down-regulates endothelial cell expression of VEGFR2, which is the major mediator of the mitogenic, survival, migration-promoting, and angiogenic activities of VEGF-A.50 We also found that Dll4 down-regulates endothelial cell expression of NRP1, which functions as a coreceptor for members of the VEGF-A family, enhancing VEGFR2 activity.51 We and others have shown that Dll4 RNA is induced by hypoxia or VEGF-A in endothelial cells.16,35,41 Our current work extends these observations to demonstrate that Dll4 serves to dampen endothelial cell responses to VEGF-A by reducing the expression of its principal signaling receptor and coreceptor.
Previous studies have generally concluded that Notch activation in endothelial cells serves to reduce angiogenic responses. Constitutively active Notch4 inhibited the sprouting of primary human dermal microvascular endothelial cells and reduced VEGF-induced angiogenesis in the chick chorioallantoic membrane in vivo, in part because of conformational changes in β1-integrins.32 Overexpression of active Notch1 or Notch4 inhibited the proliferation of HUVECs, in part through repression of p21Cip1 expression.54 Constitutively active Notch1 inhibited the migration of mouse embryonic endothelial cells.55 In addition, constitutively active Notch1 or Notch4 reduced HUVEC proliferation in response to VEGF but not bFGF.56 Consistent with Dll4 being a Notch1 and Notch4 ligand,14,34 we have shown that the phenotype of endothelial cells in which Dll4 is overexpressed bears similarity to the phenotype of endothelial cells with active Notch1 or Notch4. Recently, overexpression in HUVECs of Jagged1, another Notch ligand expressed by endothelial cells, induced cell-cycle arrest and reduced cell proliferation, similar to the effect of active Notch1 or Notch4 under the same conditions.54 These observations would suggest that in endothelial cells there is at least some functional redundancy not only of Notch1 and Notch4 but also of Jagged1 and Dll4.
Overexpression of Dll4 in endothelial cells was accompanied by a significant increase in the expression of the transcription factor HEY2 with little or no change in the expression of HEY1. Previously, active Notch1 and Notch4 were shown to up-regulate expression of HEY1 in endothelial cells,56 indicating activation of different transcription factors by the ligand Dll4 and the active Notch1 or Notch4 receptors. Because Dll4 has been reported to also bind Notch2,15 which is expressed on HUVECs and other endothelial cells,54 it is possible that active Notch2 may up-regulate HEY2. If Notch receptors can activate different transcription factors depending on the nature of the ligand and perhaps even the degree and precise conditions of activation, this would provide a basis for explanation of the complexities of the Notch system. It is possible that the Dll4 intracellular domain may itself exert specific signaling functions, consistent with what was previously hypothesized for Dll1.57,58 However, we have demonstrated that down-regulation of VEGFR2 expression and diminished proliferative response to VEGF-A can be achieved through use of the extracellular domain of Dll4 alone. This suggests that the intracellular domain of Dll4 is not necessary for these effects. Furthermore, reduced VEGF-A-induced proliferation caused by extracellular Dll4 can be overcome by the addition of the γ-secretase inhibitor L-685458, providing strong evidence that Dll4 is signaling through Notch receptors. In addition, an important contribution of Notch signaling to the phenotype of Dll4-overexpressing cells is supported by the results presented here, showing that the inhibitor to Notch signaling L-685458 significantly reconstituted VEGFR2 expression in Dll4-transduced endothelial cells.
We discovered here that overexpression of Dll4 is associated with reduced expression of VEGFR2 and NRP1 in endothelial cells, and this reduction is likely responsible for the abnormally low proliferative and migratory responses to VEGF-A in Dll4-overexpressing endothelial cells. Previously, constitutively active Notch1 and Notch4 in endothelial cells were reported to down-regulate VEGFR2 expression probably by decreasing VEGFR2 promoter activity,56 and overexpression of HEY1 in these cells reduced levels of VEGFR2 mRNA.49 Our results confirm this link between Notch signaling and down-regulation of VEGFR2 and extend this observation in discovering that Dll4 also reduces NRP1 expression. The possibility that NRP1 expression may be regulated by Notch signaling was raised by recent experiments showing that the large arteries, which developed in mice with targeted deletions of Notch1 or HEY1/HEY2, did not express NRP1.52
The role of NRP1 as a receptor for class-3 semaphorins, which mediate neuronal guidance,59,60 and for several isoforms of VEGF-A, enhancing VEGF-A activity,51 is well established. Recently, we and others found that NRP1 regulates endothelial cell adhesion to extracellular matrix and to other cells,43,61 indicating that NRP1 is important in angiogenesis both as a modulator of VEGF-A activity and a mediator of endothelial cell adhesion. This conclusion is consistent with the marked vascular abnormalities developing in mice with endothelial cell-targeted deletions of NRP162 and with embryonic death associated with defects in the heart, vasculature, and nervous system occurring in NRP1-deficient mice.63,64 By down-regulating both VEGFR2 and NRP1 expression, Dll4 is positioned to function as an endogenous inhibitor of angiogenesis.
The preferential expression of Dll4 in the developing arteries, arterioles, and capillaries as opposed to veins,14,16 the arterial-specific expression of HEY2,65 the loss of the CD44 and ephrinB2 arterial endothelial markers in the large arteries of Notch1 or HEY1/HEY2-deficient mice,52 and the HEY2-induced expression of many arterial endothelial cell markers in HUVECs65 have suggested that the Dll4-Notch1/4-HEY1/2 pathway specifies arterial endothelial fate during development.30 The observation herein that HUVECs overexpressing Dll4 display increased levels of the arterial marker ephrinB2 mRNA is consistent with this concept. Dll4 expression in the tumor vascular endothelium and angiogenic endothelium of the ovaries and its induction by exposure to hypoxia and VEGF-A16,40,41 further suggest that Dll4 may also play a role in postnatal endothelial cell differentiation processes during angiogenesis.
Our identification of Dll4 as a repressor of endothelial cell responses to VEGF-A, a principal proangiogenic factor, assigns a previously unknown function to Dll4 and extends current concepts of angiogenesis regulation. The challenge of future experiments will be to evaluate the potential usefulness of Dll4 as an inhibitor of angiogenesis for the treatment of cancer and other disorders characterized by excessive angiogenesis.
Prepublished online as Blood First Edition Paper, October 11, 2005; DOI 10.1182/blood-2005-03-1000.
Supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, and in part by “the 6th Framework Programme of the European Union [Angiotargeting].”
A.L.H. and G.T. are senior coauthors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs R. Schwartz, M. Lenardo, M. Delong, and S. Kennedy for their support; Dr V. Kapoor and W. Telford for cell sorting and flow cytometry support; Dr G. P. Nolan for providing the retroviral vector; Ms L. Sierra, Dr L. Yao, and Dr M. Narazaki for technical support; and Drs N. Patel, S. Suchting, K. Tahtis, and R. Yarchoan for help on various aspects of this work.
C.K.W. is a National Institutes of Health-University of Oxford Health Science Scholar.

![Figure 2. Effects of Dll4 expression on endothelial cell proliferation. (A) HUVECs transduced 3 to 10 days earlier with either vector only or Dll4 (70%-90% of cells expressing EGFP) were cultured for 72 hours in medium alone or medium supplemented with VEGF-A (25 ng/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SEMs) from 5 separate experiments, each performed in triplicate cultures. *P < .001. (B) Dose dependency of VEGF-A (3-24 ng/mL) induced HUVEC proliferation 3 to 10 days after transduction with vector only or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate cultures (representative of 4 determinations). (C) Analysis of HUVEC proliferation in response to bFGF (3-28 ng/mL) 3 to 10 days after transduction with vector alone or Dll4 retrovirus. Cell proliferation was measured as described in “Materials and methods”; the results reflect the means (± SDs) of triplicate determinations (representative of 4 determinations).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/3/10.1182_blood-2005-03-1000/4/m_zh80030690190002.jpeg?Expires=1768394990&Signature=mpSKdvpdHl4hk1pxTpKsduhWLPkv3nObNlD7I7uvxFQ4SE7cmUCVRwu1t6uuT1ELsxrI5wngqLqI-AGeeaTEsflubPjXZGrYFP5jaHtzKMt8dDx2u6wuUtPTJU1OwH2D-jyl9u-fAbN9t5rgc6Lqk~JpgaiEQkh8roq0RudGe2aXAYQG~4emC7Q-enP6QvtV6nNQInDGditGABIdQecuXF70IjzaIanqGGUPVcwc1NyAS-APq6KH91wdePzl3mkbLOvp4sknmIelSA8PvHCNlfgHrZPJoHTEFS3rpY1ISvjfkHXYB2XBDk2lKeoN-642jUuocIdQBN43cbqPP9L83A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 6. Expression of Notch receptors in endothelial cells and Notch function in Dll4-transduced cells. (A) mRNAs for Notch receptors 1 to 4 detected by RT-PCR in HUVECs transduced with vector. (B) Quantitative RT-PCR analysis of HEY1, HEY2, ephrinB2, and EphB2 expression in control HUVECs transduced with vector only or Dll4-overexpressing HUVECs. The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4 versus control HUVECs from 2 to 11 separate determinations. *P < .05. (C) Effects of immobilized soluble recombinant human Dll4 (rhDLL4) on the proliferation of HUVECs. The endothelial cells were cultured for 72 hours in the presence of VEGF-A (25 ng/mL) on plates coated with rhDLL4 (1 μg/mL) or diluent (0.1% BSA); proliferation was measured by 3H thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .05. (D) Surface levels of VEGFR2 expression in HUVECs cultured for 48 hours onto rhDLL4-coated plates (1 μg/mL) (rhDLL4) or diluent-coated plates (gelatin) measured by flow cytometry. Control reflects background surface fluorescence staining with appropriate controls. Representative of 2 performed. (E) Effects of the γ-secretase inhibitor L-685458 (0.1-7.5 μM) on the proliferation of HUVECs cultured with rhDLL4-coated plates (1 μg/mL); proliferation was measured by [3H] thymidine deoxyribose uptake during the last 18 hours of culture. The results reflect the means (± SDs) from triplicate cultures. Representative experiment of 4 performed. *P < .01 (rhDLL4 with 4 μM L-685458 inhibitor versus VEGF alone). (F) Quantitative RT-PCR analysis of GAPDH and VEGFR2 expression in vector- and Dll4-transduced HUVECs after 72-hour culture with L-685458 (2 μM) or diluent only (0.02% DMSO). The results reflect relative mRNA levels (normalized to GAPDH) and are expressed as the mean (± SEM) fold mRNA change in Dll4-transduced versus control HUVECs from 4 separate experiments, in which levels of VEGFR2 expression in the absence of inhibitor were reduced by at least 5-fold in Dll4-transduced compared with control HUVECs (*P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/3/10.1182_blood-2005-03-1000/4/m_zh80030690190006.jpeg?Expires=1768394990&Signature=ai~3lskH0xRP273JuzN1QihDmB47Br3XvBqpcHoZYdhrUQKMJWZzgl-BZ35IveXUl3qVXBvoUdCuL18ASfNr7V3g5kMNRS5D8XyKdgdccRuHFXmIKXvquOXWadu3As5cawpnLLGgRY6CuKZdoevgNJnf4rHB0cMKaGhRi1PCCVE~1qMZg4aEr1MhgSVY6p8q~x9KlUqkM60pMmnb3v3k8N07LVBGDvy6ac3CVnF71aPgTUkFZ4tiTB9G4Q7KDNWHqj7KK0NlLJscWvF4jN8aVYV6FAEhmAXk~EhaRU~tNnAS56PcfDcx6J1u78OpH3ne2UndV-wA0~frRKzyGHwCTg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal