Krüppel-like factor 6 (KLF6) is a member of a growing family of transcription factors that share a common 3 C2H2 zinc finger DNA binding domain and have broad activity in regulating proliferation and development. We have previously established that Klf6 is expressed in neuronal tissue, hindgut, heart, lung, kidney, and limb buds during midgestation. To explore the potential role of Klf6 in mouse development, we analyzed Klf6-/- mice and found that the homozygous mutation is embryonic lethal by embryonic day (E) 12.5 and associated with markedly reduced hematopoiesis and poorly organized yolk sac vascularization. Additionally, mRNA levels of Scl and Gata1 were reduced by approximately 80% in Klf6-/- yolk sacs. To further analyze this phenotype, we generated Klf6-/- embryonic stem (ES) cells by homologous recombination, and compared their capacity to differentiate into the hematopoietic lineage with that of either Klf6+/- or Klf6+/+ ES cells. Consistent with the phenotype in the early embryo, Klf6-/- ES cells displayed significant hematopoietic defects following differentiation into EBs. Prolongation of epiblast-like cells and delays in mesoderm induction were also observed in the Klf6-/- EBs, associated with delayed expression of Brachyury, Klf1, and Gata1. Forced expression of KLF6 using a tet-inducible system enhanced the hematopoietic potential of wild-type EBs. Collectively, these findings implicate Klf6 in ES-cell differentiation and hematopoiesis.
Introduction
KLF6 is a C2H2 transcription factor belonging to the family of Krüppel-like zinc finger transcription factors, which currently includes at least 15 members that regulate remarkably diverse processes including cell proliferation, signal transduction, and differentiation.1-5 Klf genes are highly conserved evolutionarily, with homologs expressed in zebrafish,6 Drosophila,7 and Xenopus.8 All KLF members possess a tightly conserved C-terminal, 81-amino acid zinc finger DNA-binding domain that can interact with “GC-box” or “CACCC-box” DNA motifs in responsive promoters, with each KLF having a distinct N-terminal activation domain.1 KLFs are phylogenetically organized into 4 subgroups and are now designated by number (eg, KLF1-15).3
While the DNA binding domains of KLFs are highly homologous, their divergent transcriptional interaction domains account for a remarkably broad range of biologic activities. Indeed, few generalizations can be made about the family apart from the structural similarity of its members, while the breadth of the members' known biologic features continues to expand. Some are transcriptional activators, others are repressors, while still others, for example KLF6, are bifunctional in transrepressing some genes while transactivating others.3 KLFs can homodimerize or heterodimerize with any of the KLFs9 or with heterologous transcription factors.10,11 KLFs may be tissue specific as in the case of KLF1 (formerly EKLF)12 or, more commonly, are ubiquitous, for example KLF6 and KLF7.13,14 Inducers and repressors of KLF activity are less well characterized than their transcriptional targets, but these stimuli include injury, cellular stress, and cytokines3 ; autocrine transcriptional regulation of KLFs may also be important.15
KLFs play key roles in several developmental pathways based on studies in both lower species6 and mammals. All Klf knock-out mice generated to date have a lethal phenotype. These models have revealed roles of KLFs in blood vessel stability (KLF2),16 beta globin synthesis during erythropoiesis (KLF1),17,18 and epithelial barrier integrity (KLF4).19 Studies using chimeric mice derived from Rag2-/- and Klf2-/- animals have also indicated a role of Klf2 in T-cell activation.20 These developmental activities are ascribed to both those KLFs that are tissue restricted, as well as those that are ubiquitously expressed. For example, Klf4 and Klf6 are both widely expressed, yet their developmental expression is restricted.7,21-23 Of importance, even heterozygous Klf gene-deleted mice may reveal an abnormal phenotype, as in the case of Klf5+/- animals, which have a defect in arterial wall remodeling when stressed.24
Among the subfamilies of Krüppel-like factors, the Klf6 gene is most closely related to Klf7, and they share a common progenitor in Drosophila, the Luna gene, whose inactivation using siRNA leads to defects in organogenesis and terminal differentiation.25 Despite its ubiquitous expression in adult tissues, developmental expression of Klf6 is somewhat restricted and distinct from Klf7. Whereas Klf7 mRNA is primarily expressed in neuronal tissue, Klf6 transcripts are also found in several nonneural sites including hindgut, heart, lung, kidney, and limb buds.7
Based on these findings, we analyzed the role of Klf6 in mouse development. Klf6-/- mice die by embryonic day (E) 12.5 and are characterized by markedly reduced hematopoietic differentiation in yolk sacs. Complementing this approach, we used a mouse embryonic stem (ES)-cell differentiation system to assess the capacity of murine Klf6-/- ES cells to differentiate in vitro.26,27
Materials and methods
Klf6 targeting vector construction and generation of Klf6+/- and Klf6-/- ES cells
Construction of the targeting vectors and homologous recombination were performed using methods previously described.28 In brief, 2 targeting constructs were generated in which the second exons of the Klf6 gene were replaced by a neomycin/lac Z cassette or hygromycin cassette (Figure 1A). First, Klf6+/- ES cells were generated by homologous recombination with a Klf6 targeting vector containing the neomycin cassette. To obtain Klf6-/- ES cells, Klf6+/- clones were subjected to a second round of homologous recombination and selection with hygromycin. Klf6+/- ES cells were screened by Southern blotting using a 5′ outside probe (Probe 1, Figure 1A). Klf6-/- ES cells were screened by Southern blotting using a 3′ outside probe (Probe 2, Figure 1A). Genomic DNA digested with HindIII was hybridized with Probe 1 and genomic DNA digested with BamHI was hybridized with Probe 2. Genotypes of all ES-cell clones were confirmed by genomic polymerase chain reaction (PCR) amplifying exon 2 of Klf6, and the neomycin and hygromycin cassettes. Primers and conditions of the genomic PCR for exon 2 of Klf6 are the same as that of reverse-transcriptase (RT)-PCR (Table 1). Sequences of forward (F) and reverse (R) primers, annealing temperatures (Ta's), and product sizes of genomic PCRs for neomycin and hygromycin cassettes were as follows: neomycin (F) 5′-TGCTATTGGGCGAAGTGCCGGGGC-3′ and (R) 5′-AGCGTATGCAGCC-GCCGCATTGC-3′ (Ta, 62°C; product size, 100 bp); hygromycin (F) 5′-GAGCCTGACCTATT-GCATCT-3′ and (R) 5′-GTTTCCACTATCGGCGAGTA-3′ (Ta, 55°C; product size, 736 bp). Sequence confirmation of ES cells was followed by analysis of metaphase spreads to confirm a normal karyotype prior to further analysis. Two Klf6-/- clones with normal karyotype were termed “Clone NA” and “Clone NB” and analyzed independently in all ES-cell experiments.
Primers and conditions used for PCR analysis
Gene, PCR type . | Direction . | Primer . | Product size, bp . | Annealing temperature, °C . | Cycle no. . |
|---|---|---|---|---|---|
| β-actin | |||||
| RT-PCR | F | GAC ATG GAG AAG ATC TGG CA | 148 | 55 | 25 |
| R | GGG TGT TGA AGG TCT CAA AC | ||||
| Brachyury | |||||
| RT-PCR | F | CAT GTA CTC TTT CTT GCT GG | 313 | 60 | 35 |
| R | GGT CTC GGG AAA GCA GTG GC | ||||
| qPCR | F | CTC ACC AAC AAG CTC AAT GG | 129 | 57 | NA |
| R | GGT CTC GGG AAA GCA GTG GC | ||||
| Fgf5 | |||||
| RT-PCR | F | GTG TCT CAG GGG ATT GTA GGA ATA CG | 314 | 65 | 35 |
| R | GTG AAG GAA AGT TCC GGT TGC | ||||
| Flk1 | |||||
| RT-PCR + qPCR | F | CAC CTG GCA CTC TCC ACC TTC | 219 | 55 | 35 |
| R | GAT TTC ATC CCA CTA CCG AAA G | ||||
| Gata1 | |||||
| RT-PCR | F | CAT TGG CCC CTT GTG AGG CCA GAG A | 289 | 60 | 35 |
| R | ACC TGA TGG AGC TTG AAA TAG AGG C | ||||
| qPCR | F | CAA CAG TAT GGA GGG AAT TCC T | 161 | 57 | NA |
| R | GTG TCC AAG AAC GTG TTG TTG C | ||||
| Gata2 | |||||
| RT-PCR + qPCR | F | GCA CCT GTT GTG CAA ATT GT | 195 | 57 | 35 |
| R | GCC CCT TTC TTG CTC TTC TT | ||||
| Gapdh | |||||
| qPCR | F | CAA TGA CCC CTT CAT TGA CC | 145 | 55 | 35 |
| R | GAT CTC GCT CCT GGA AGA TG | ||||
| Klf1 | |||||
| RT-PCR + qPCR | F | TCC ATC AGT ACA CTC ACC AC | 92 | 57 | 35 |
| R | AAA TCC TGC GTC TCC TCA GA | ||||
| Klf6 | |||||
| RT-PCR | F | TAT CTT CAG GAT GAG CCC TGC TAC | 276 | 60 | 30 |
| R | AGA CTT CAC CAA TGG GAT CAG AGG | ||||
| qPCR | F | ACC CGA CAT GGA TGT GCT CCC AAT | 150 | 57 | NA |
| R | GCA GGG CTC ACT CTG AAG ATA | ||||
| Klf7 | |||||
| RT-PCR | F | CAG CAT GGA CGT GTT GGC TAG TTA | 276 | 60 | 33 |
| R | TCT TCT CGC AGA TGG TGG CTT CCA | ||||
| p21 | |||||
| RT-PCR | F | GCA GAT CCA CAG CGA TAT CCA G | 332 | 60 | 33 |
| R | GAG GAA GTA CTG GGC CTC TTG T | ||||
| Rex1 | |||||
| RT-PCR + qPCR | F | CGT GTA ACA TAC ACC ATC CG | 129 | 60 | 35 |
| R | GAA ATC CTC TTC CAG AAT GG | ||||
| Scl | |||||
| RT-PCR + qPCR | F | ATA GCC TTA GCC AGC CGC TC | 153 | 55 | 33 |
| R | CGC CGC ACT ACT TTG GTG TG |
Gene, PCR type . | Direction . | Primer . | Product size, bp . | Annealing temperature, °C . | Cycle no. . |
|---|---|---|---|---|---|
| β-actin | |||||
| RT-PCR | F | GAC ATG GAG AAG ATC TGG CA | 148 | 55 | 25 |
| R | GGG TGT TGA AGG TCT CAA AC | ||||
| Brachyury | |||||
| RT-PCR | F | CAT GTA CTC TTT CTT GCT GG | 313 | 60 | 35 |
| R | GGT CTC GGG AAA GCA GTG GC | ||||
| qPCR | F | CTC ACC AAC AAG CTC AAT GG | 129 | 57 | NA |
| R | GGT CTC GGG AAA GCA GTG GC | ||||
| Fgf5 | |||||
| RT-PCR | F | GTG TCT CAG GGG ATT GTA GGA ATA CG | 314 | 65 | 35 |
| R | GTG AAG GAA AGT TCC GGT TGC | ||||
| Flk1 | |||||
| RT-PCR + qPCR | F | CAC CTG GCA CTC TCC ACC TTC | 219 | 55 | 35 |
| R | GAT TTC ATC CCA CTA CCG AAA G | ||||
| Gata1 | |||||
| RT-PCR | F | CAT TGG CCC CTT GTG AGG CCA GAG A | 289 | 60 | 35 |
| R | ACC TGA TGG AGC TTG AAA TAG AGG C | ||||
| qPCR | F | CAA CAG TAT GGA GGG AAT TCC T | 161 | 57 | NA |
| R | GTG TCC AAG AAC GTG TTG TTG C | ||||
| Gata2 | |||||
| RT-PCR + qPCR | F | GCA CCT GTT GTG CAA ATT GT | 195 | 57 | 35 |
| R | GCC CCT TTC TTG CTC TTC TT | ||||
| Gapdh | |||||
| qPCR | F | CAA TGA CCC CTT CAT TGA CC | 145 | 55 | 35 |
| R | GAT CTC GCT CCT GGA AGA TG | ||||
| Klf1 | |||||
| RT-PCR + qPCR | F | TCC ATC AGT ACA CTC ACC AC | 92 | 57 | 35 |
| R | AAA TCC TGC GTC TCC TCA GA | ||||
| Klf6 | |||||
| RT-PCR | F | TAT CTT CAG GAT GAG CCC TGC TAC | 276 | 60 | 30 |
| R | AGA CTT CAC CAA TGG GAT CAG AGG | ||||
| qPCR | F | ACC CGA CAT GGA TGT GCT CCC AAT | 150 | 57 | NA |
| R | GCA GGG CTC ACT CTG AAG ATA | ||||
| Klf7 | |||||
| RT-PCR | F | CAG CAT GGA CGT GTT GGC TAG TTA | 276 | 60 | 33 |
| R | TCT TCT CGC AGA TGG TGG CTT CCA | ||||
| p21 | |||||
| RT-PCR | F | GCA GAT CCA CAG CGA TAT CCA G | 332 | 60 | 33 |
| R | GAG GAA GTA CTG GGC CTC TTG T | ||||
| Rex1 | |||||
| RT-PCR + qPCR | F | CGT GTA ACA TAC ACC ATC CG | 129 | 60 | 35 |
| R | GAA ATC CTC TTC CAG AAT GG | ||||
| Scl | |||||
| RT-PCR + qPCR | F | ATA GCC TTA GCC AGC CGC TC | 153 | 55 | 33 |
| R | CGC CGC ACT ACT TTG GTG TG |
qPCR indicates quantitative real-time PCR; F, forward; R, reverse; and NA, not applicable.
Targeted disruption of the mouse Klf6 gene. (A) Genomic structure of the mouse Klf6 gene and targeting vectors. (B) Genotyping of Klf6+/+ and Klf6+/- ES cells by Southern blot analysis using Probe 1 following HindIII digestion of genomic DNA. (C) Genotyping of Klf6+/- and Klf6-/- ES cells by Southern blot analysis using Probe 2 following BamHI digestion. (D) Genotyping of ES cells by PCR. Exon 2 of the Klf6 gene and the neomycin and hygromycin genes were amplified. Klf6-/- ES cells express only the neomycin and hygromycin sequences, but not Klf6 exon 2. In these analyses, genomic DNAs were extracted after depletion of mouse embryonic feeder cells. (E) Expression of the Klf6 gene. After depletion of mouse embryonic feeder cells, total RNAs were extracted and subjected to RT-PCR.
Targeted disruption of the mouse Klf6 gene. (A) Genomic structure of the mouse Klf6 gene and targeting vectors. (B) Genotyping of Klf6+/+ and Klf6+/- ES cells by Southern blot analysis using Probe 1 following HindIII digestion of genomic DNA. (C) Genotyping of Klf6+/- and Klf6-/- ES cells by Southern blot analysis using Probe 2 following BamHI digestion. (D) Genotyping of ES cells by PCR. Exon 2 of the Klf6 gene and the neomycin and hygromycin genes were amplified. Klf6-/- ES cells express only the neomycin and hygromycin sequences, but not Klf6 exon 2. In these analyses, genomic DNAs were extracted after depletion of mouse embryonic feeder cells. (E) Expression of the Klf6 gene. After depletion of mouse embryonic feeder cells, total RNAs were extracted and subjected to RT-PCR.
Generation of Klf6-/- mice
Two independently targeted Klf6+/- clones were used to bring the null mutation into the germ line. Chimeric mice were produced by microinjection of targeted ES-cell clones into C57BL6 blastocysts, which were then transferred to pseudopregnant mothers. Highly chimeric males were mated with C57BL6 females, and germ-line transmission of the mutated allele was verified by Southern blotting and PCR analysis of tail DNA from F1 offspring. PCR genotyping was performed using 2 primer pairs amplifying the Klf6 exon 2 and the neomycin coding sequences. Two lines of Klf6+/- mice were obtained, analyzed, and found to display an indistinguishable phenotype from each other. All animal studies were performed with Institutional Animal Care and Use Committee (IACUC) approval and conformed to National Institutes of Health (NIH) guidelines.
Whole-mount PECAM1 staining
Yolk sacs with embryos were fixed in 4% paraformaldehyde/PBS overnight. After washing with PBS with 0.2% Triton X-100, samples were blocked by 3% instant skim milk, 0.1% Triton X-100 in PBS, and incubated with 1:100 dilution of anti-PECAM1 antibody (Pharmingen, San Diego, CA) overnight at 4°C. After washing with blocking solution, samples were incubated with 1:1000 dilution of antirabbit fluoresceinated antibody for 30 minutes and washed with PBS with 0.1% Triton X-100. Yolk sacs were spread on glass slides and mounted before observation.
Culture and differentiation of ES cells
ES cells were maintained on irradiated mouse embryonic fibroblast (MEF) feeder cells in DMEM supplemented with 15% FCS, penicillin, streptomycin, and 2% leukemia inhibitory factor (LIF) and 1.5 × 10-4 M mono-thioglycerol (MTG; Sigma-Aldrich, St Louis, MO). Prior to the initiation of differentiation, ES cells were passaged twice on gelatin-coated dishes in the same media to deplete the population of MEF feeder cells.
For the generation of embryoid bodies (EBs), ES cells were trypsinized into single-cell suspensions and plated at variable densities (300-4500 cells/mL) into differentiation media containing Iscoves modified Dulbecco medium (IMDM) supplemented with 15% FCS, 2 mM l-glutamine (Gibco/BRL, Grand Island, NY), 0.5 mM ascorbic acid (Sigma-Aldrich), and 4.5 × 10-4 M MTG in 60-mm Petri grade dishes. Cultures were maintained in a humidified chamber in a 5% CO2/air mixture at 37°C. EBs grown in differentiation medium were harvested and trypsinized for approximately 3 minutes. After trypsinization, EBs were gently passed through a 20-gauge needle several times to generate a suspension of single cells that was recounted prior to plating. To compare the proliferation of ES cells, 105 cells per well were seeded in a 6-well plate, and cell numbers were counted approximately 72 hours later.
RT-PCR and real-time quantitative PCR
Total RNA was extracted from undifferentiated or differentiated ES cells using RNeasy kit (Qiagen, Valencia, CA), and cDNAs were synthesized from 1 μg total RNA using an RT kit (Promega, Madison, WI). Sequences of PCR primers, annealing temperatures, and number of cycles for each target are indicated in Table 1.
Real-time quantitative PCR was performed using the PCR primers indicated in Table 1 on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). All experiments were done in triplicate and normalized to GAPDH mRNA expression. Fluorescent signals were analyzed during each of up to 40 cycles consisting of denaturation (95°C, 15 seconds), annealing (56°C, 15 seconds), and extension (72°C, 40 seconds). Relative quantitation was calculated using the comparative threshold cycle method (CT; as described in the User Bulletin no. 2,29 ABI PRISM 7900HT Sequence Detection System). CT indicates the fractional cycle number at which the amplified gene amounts to a fixed threshold within the linear phase of amplification. Median CT of triplicate measurements was used to calculate ΔCT as the CT of each gene. ΔCT of each gene for each sample was compared with ΔCT of GAPDH for the same sample expressed as ΔΔCT. Relative quantification was depicted as fold-change expression for each gene, compared with GAPDH mRNA, with the formula 2-ΔΔCT. Denaturing curves and agarose gel electrophoresis of PCR product for each gene were used to confirm homogeneity of the DNA products.
Fluorescence activated cell sorting (FACS) analysis
FACS analysis of Flk1-positive cells was performed as previously described.30 Briefly, cells were counted and incubated with a biotinylated antibody to Flk1 (Pharmingen) for 20 minutes and then stained with a secondary reagent (streptavidin R-phycoerythrin conjugate; BioSource International, Camarillo, CA). Cells were then washed again and analyzed on a FACScan (Becton Dickinson, San Jose, CA).
Methylcellulose colony assay
For the generation of blast-cell colonies, EB cells were counted and plated in IMDM containing 1.0% methylcellulose, 10% FCS, 5 ng/mL VEGF, 5 ng/mL IL-6, and 25% D4T endothelial-cell conditioned medium.31 Colonies were scored following 4 days of culture. For the growth of hematopoietic precursors, cells of EBs or yolk sacs of E 9.5 embryos were counted and plated in IMDM containing 1.0% methylcellulose, 10% plasma-derived serum (PDS; Antec, Tyler, TX), 2 mM glutamine, 1000 U/mL IL-1, 1% supernatant containing IL-3, 5 ng/mL IL-6, 25 ng/mL IL-11, 30 ng/mL G-CSF, 5 ng/mL M-CSF, 3 ng/mL GM-CSF, 1% supernatant containing c-Kit ligand, 5 ng/mL VEGF, 10 ng/mL LIF, 2 U/mL erythropoietin, and 5% protein-free hybridoma medium (PFHM-II; Gibco/BRL). Cells were cultured at 37°C in 5% CO2/air in 35-mm dishes, for approximately 3 to 4 days for EryP from EBs and EryD from yolk sacs, or 6 to 8 days for Mac and Mix colonies. Colony types were scored on the basis of morphologic criteria.32
Klf6 retrovirus construction
Full-length mouse Klf6 was obtained by RT-PCR, sequenced, and subcloned into the pCR2.1-TOPO plasmid (Invitrogen, Carlsbad, CA). Following sequence confirmation, the insert was excised with EcoRI and subcloned into the MSCVpac plasmid. The insert orientation was confirmed by digestion with PstI. Using lipofectamine 2000 (Invitrogen), the plasmids with or without Klf6 were transfected into Phoenix cells for the production of retroviruses. Up-regulation of p21 in Klf6 retrovirus-infected NIH3T3 cells was observed,33 which confirmed that KLF6 produced by this retrovirus transactivates a known KLF6 target gene. The viruses were harvested and 6 mL undiluted virus plus polybrene were used to infect 60% confluent ES cells cultured on gelatinized 10-cm dishes. After infection, ES cells were subjected to generation of EBs without selection.
Generation of ES cells with doxycycline-inducible KLF6 expression
A KpnI-XbaI fragment of full-length mouse Klf6 in pCR2.1-TOPO was ligated into pLox (a gift from Dr Michael Kyba, Whitehead Institute for Biomedical Research, Cambridge, MA), which was then coelectroporated along with pSalk-CRE into the targeting cell line Ainv16 (both also gifts from Dr Michael Kyba). The targeting ES cell line and targeting plasmid, pLox, have been described previously.34 The resulting cell line was selected and expanded in 400 μg/mL G418 (Sigma). To induce KLF6 expression in ES cells, 2 μg/mL doxycycline (Sigma) was added to the culture medium.
Results
Generation of Klf6+/- and Klf6-/- ES cells
Two targeting constructs were generated in which the second exon of Klf6 was replaced by a neomycin or hygromycin cassette to generate Klf6+/- and Klf6-/- ES cells, respectively (Figure 1A). Screening by Southern blot analysis confirmed both the first (Figure 1B) and second (Figure 1C) homologous recombination events. Genomic PCR analysis confirmed that Klf6+/- cells harbored only the neomycin cassette, whereas Klf6-/- cells contained both the neomycin and hygromycin cassettes, and Klf6+/+ cells did not harbor either resistance gene (Figure 1D). RT-PCR analysis revealed expression of Klf6 in Klf6+/+ and Klf6+/- ES cells but not in 2 clones of Klf6-/- ES-cell clones (Clone NA and Clone NB) (Figure 1E).
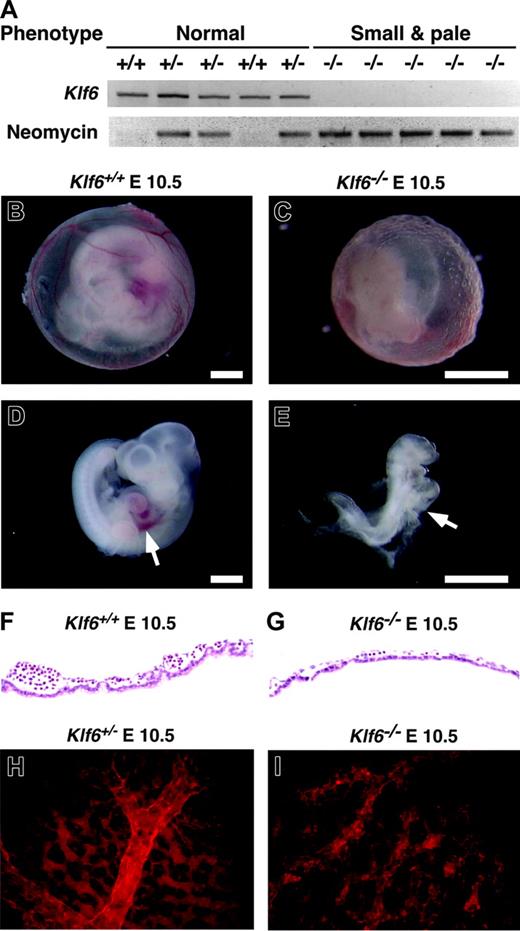
Comparison of Klf6-/-, Klf6+/-, and Klf6+/+ embryos. (A) Ten random E 10.5 embryos derived from Klf6+/- × Klf6+/- crosses were classified according to size and color and genotypes by PCR for Klf6 and neomycin. Klf6-/- embryos are small and pale (see also E-F), whereas no significant differences were detected between Klf6+/- and Klf6+/+ embryos. Compared with the appearance at E 10.5 of the Klf6+/+ yolk sac (B) and embryo (D), Klf6-/- yolk sac (C) and embryos (E) are smaller and paler, and vascular structures in the Klf6-/- yolk sac are poorly developed. The Klf6-/- embryo lacks a definable liver (arrow), although a beating heart was observed at the time of death (not shown). (Scale: bar = 1 mm.) Sections of yolk sacs reveal that Klf6-/- yolk sacs (G) have fewer hematopoietic cells than Klf6+/+ yolk sacs (F). Whole-mount immunohistochemistry of yolk sacs using anti-PECAM1 demonstrates that vascular formation in Klf6-/- yolk sacs (I) is much less organized than KLlf6+/- yolk sacs (H), while expression of PECAM1 is apparent. Images in Figure 2B-E were obtained using a Leica MZ APO stereomicroscope (Leica Microsystems, Bannockburn, IL) equipped with a 1 ×/0.09 objective lens at 12 × (B,D) or 25 × (C,E) total original mangification. A Sony 3CCD DXC-970MD camera (Sony, New York, NY) and NIH Scion public domain software (National Institutes of Health, Bethesda, MD) were used to photograph and capture images. Images in Figure 2F-I were obtained using a Nikon Eclipse E300 microscope equipped with a 40 ×/0.75 objective lens (Nikon Instruments, Melville, NY); they were photographed with a Spot RT Slider camera and then processed with Spot Image Capture for Mac OS 3.5.4 software (Diagnostic Instruments, Sterling Heights, MI).
Comparison of Klf6-/-, Klf6+/-, and Klf6+/+ embryos. (A) Ten random E 10.5 embryos derived from Klf6+/- × Klf6+/- crosses were classified according to size and color and genotypes by PCR for Klf6 and neomycin. Klf6-/- embryos are small and pale (see also E-F), whereas no significant differences were detected between Klf6+/- and Klf6+/+ embryos. Compared with the appearance at E 10.5 of the Klf6+/+ yolk sac (B) and embryo (D), Klf6-/- yolk sac (C) and embryos (E) are smaller and paler, and vascular structures in the Klf6-/- yolk sac are poorly developed. The Klf6-/- embryo lacks a definable liver (arrow), although a beating heart was observed at the time of death (not shown). (Scale: bar = 1 mm.) Sections of yolk sacs reveal that Klf6-/- yolk sacs (G) have fewer hematopoietic cells than Klf6+/+ yolk sacs (F). Whole-mount immunohistochemistry of yolk sacs using anti-PECAM1 demonstrates that vascular formation in Klf6-/- yolk sacs (I) is much less organized than KLlf6+/- yolk sacs (H), while expression of PECAM1 is apparent. Images in Figure 2B-E were obtained using a Leica MZ APO stereomicroscope (Leica Microsystems, Bannockburn, IL) equipped with a 1 ×/0.09 objective lens at 12 × (B,D) or 25 × (C,E) total original mangification. A Sony 3CCD DXC-970MD camera (Sony, New York, NY) and NIH Scion public domain software (National Institutes of Health, Bethesda, MD) were used to photograph and capture images. Images in Figure 2F-I were obtained using a Nikon Eclipse E300 microscope equipped with a 40 ×/0.75 objective lens (Nikon Instruments, Melville, NY); they were photographed with a Spot RT Slider camera and then processed with Spot Image Capture for Mac OS 3.5.4 software (Diagnostic Instruments, Sterling Heights, MI).
Lethal hematopoietic defects in Klf6-/- mice
Lethality by E 12.5 was apparent in Klf6-/- embryos, which were readily distinguished from Klf6+/+ and Klf6+/- embryos by their small and pale appearance (Figure 2A-F). No obvious differences were observed between Klf6+/+ and Klf6+/- embryos, and similarly, there was no difference in body weight or hematologic parameters between Klf6+/+ and Klf6+/- mice (data not shown). Klf6-/- embryos at E 10.5 were small and pale with no definable liver (Figure 2C,E), precluding analysis of hematopoiesis in this site. Klf6-/- yolk sacs were much thinner and disorganized compared with Klf6+/+ and Klf6+/- yolk sacs, and they contained very few blood cells (compare Figure 2F with 2G). Whole-mount immunohistochemistry using an antibody to PECAM1 displayed a poorly defined vasculature in Klf6-/- yolk sacs (compare Figure 2H with 2I). Collectively, these findings indicated that hematopoietic differentiation and vasculogenesis might be impaired in Klf6-/- embryos.
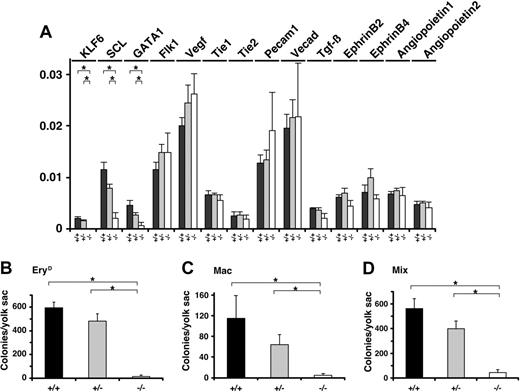
Hematopoietic differentiation in Klf6-/- yolk sacs is impaired. (A) Total RNAs were extracted from E 9.5 yolk sacs and corresponding cDNAs were analyzed by real-time PCR. The expression of indicated genes is shown. Each data point represents the average of 3 independent embryos. Error bars represent SE. (B-D) The cells from E 9.5 yolk sacs were subjected to a hematopoietic colony formation assay. Numbers of (B) EryD, (C) Mac, and (D) Mix colonies were counted, and each data point represents the average number of colonies per yolk sac. The hemoglobinized colonies were confirmed to be those of definitive erythrocytes but not primitive erythrocytes, based on lack of expression of the BH1 gene (not shown). Each data point represents the average colony number derived from at least 3 experiments (Klf6+/+, n = 6; Klf6+/-, n = 9; Klf6-/-, n = 3). Error bars represent SE. *P < .05 (Student t test).
Hematopoietic differentiation in Klf6-/- yolk sacs is impaired. (A) Total RNAs were extracted from E 9.5 yolk sacs and corresponding cDNAs were analyzed by real-time PCR. The expression of indicated genes is shown. Each data point represents the average of 3 independent embryos. Error bars represent SE. (B-D) The cells from E 9.5 yolk sacs were subjected to a hematopoietic colony formation assay. Numbers of (B) EryD, (C) Mac, and (D) Mix colonies were counted, and each data point represents the average number of colonies per yolk sac. The hemoglobinized colonies were confirmed to be those of definitive erythrocytes but not primitive erythrocytes, based on lack of expression of the BH1 gene (not shown). Each data point represents the average colony number derived from at least 3 experiments (Klf6+/+, n = 6; Klf6+/-, n = 9; Klf6-/-, n = 3). Error bars represent SE. *P < .05 (Student t test).
To further characterize these morphologic defects, we analyzed expression of hematopoietic and vascular marker genes (Scl, Gata1, Flk1, VEGF, Tie1, Tie2, Pecam1, VE cadherin, Tgfb, Ephrin B2, Ephrin B4, Angiopoietin 1, Angiopoietin 2) in yolk sacs by real-time quantitative PCR. Among the genes we examined, mRNA levels of Scl and Gata1 were reduced by approximately 80% in Klf6-/- yolk sacs (Figure 3A). Consistent with these observations, yolk sacs from the mutant embryos contained dramatically reduced numbers of hematopoietic colony-forming cells (progenitors) when compared with yolk sacs from Klf6+/+ or Klf6+/- embryos. The numbers of all progenitors including definitive erythroid (EryD), macrophage (Mac), and multipotential (Mix) were reduced (∼ 10% that of Klf+/+ colonies) (Figure 3B-D).
Hematopoietic and vasculogenic defects in Klf6-/- ES cells
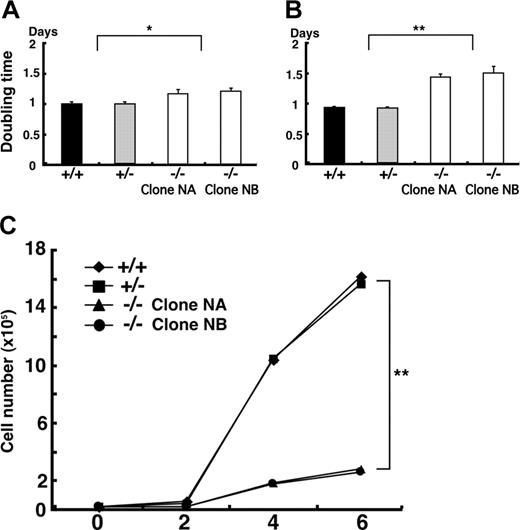
In order to complement these in vivo data, which implicated Klf6 in hematopoiesis and vasculogenesis, we compared the capacity of Klf6+/- and Klf6-/- ES cells to proliferate, generate EBs, and undergo hematopoietic differentiation in vitro. First, we cultured undifferentiated ES cells on mouse embryonic fibroblast (MEF) feeder cells and assayed their doubling times. The doubling times of Klf6-/- ES cells were about 20% greater than those of Klf6+/+ and Klf6+/- ES cells (Figure 4A). Second, we compared the proliferation of ES cells on gelatinized dishes and found that the doubling time of Klf6-/- ES cells was approximately 50% longer than that of the Klf6+/+ and Klf6+/- ES cells (Figure 4B). Finally, we compared the growth of the developing EBs. On day 6, cell numbers of Klf6-/- EBs were reduced by approximately 80% when compared with Klf6+/+ and Klf6+/- EBs (Figure 4C); the differences in cell numbers became obvious after 2 days.
Klf6-/- ES cells proliferate more slowly than Klf6+/+ ES cells. ES cells were plated on either MEF feeder cells (A) or gelatinized dishes (B) at the concentration of 1 × 105 cells/well of 6-well plates. Undifferentiated ES cells were counted after 72 hours of culture, and doubling times were calculated. Averages of 9 wells from 3 independent experiments are indicated. Error bars represent SE. *P < .05; **P < .01 (Student t test). (C) To calculate the growth curve of EBs, 2 × 104 cells were cultured in differentiation medium. EBs were harvested and cell numbers were counted at each time point. Each symbol represents the average of 3 plates.
Klf6-/- ES cells proliferate more slowly than Klf6+/+ ES cells. ES cells were plated on either MEF feeder cells (A) or gelatinized dishes (B) at the concentration of 1 × 105 cells/well of 6-well plates. Undifferentiated ES cells were counted after 72 hours of culture, and doubling times were calculated. Averages of 9 wells from 3 independent experiments are indicated. Error bars represent SE. *P < .05; **P < .01 (Student t test). (C) To calculate the growth curve of EBs, 2 × 104 cells were cultured in differentiation medium. EBs were harvested and cell numbers were counted at each time point. Each symbol represents the average of 3 plates.
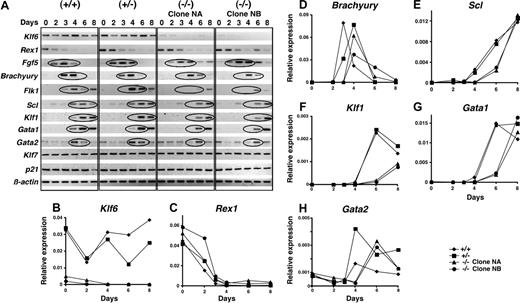
Next, patterns of gene expression were compared in differentiating EBs by semiquantitative RT-PCR. Throughout differentiation, Klf6 was detectable in Klf6+/+ EBs (Figure 5A) (the Klf6 mRNAs detected in Klf6-/- derive from mRNA from MEF feeder cells). The expression of Rex1, encoding a zinc-finger transcription factor expressed in totipotent ES cells and down-regulated upon their differentiation,35 and FGF5, a marker of the epiblast stage,36 was prolonged in Klf6-/- ES cells. The onset of expression of Brachyury, a marker of mesoderm,37,38 was delayed by approximately 24 hours in Klf6-/- EBs compared with the wild-type EBs. Sequential expression of Flk1,30,39,40 Scl,41-43 Klf1,9,17,18 Gata1,44-47 and Gata2,47,48 all markers of hematopoiesis, was likewise delayed in Klf6-/- EBs. On the other hand, there were no differences in expression levels of Klf7, which is the Klf that shares closest homology to Klf6,25 or of p21, which is a target gene of KLF6 in adult tissues, where KLF6 functions as a tumor suppressor gene.33
Expression of differentiation marker genes is delayed in Klf6-/- ES cells. (A) Differentiation into EBs of Klf6+/+ ES cells, Klf6+/- ES cells, and 2 clones of Klf6-/- ES cells was analyzed. EBs differentiated for 2, 3, 4, 6, or 8 days were harvested at each time point. Total RNAs were extracted and analyzed by RT-PCR. The expression of the indicated genes is shown. The cDNAs generated from the same mRNAs were also subjected to quantitative real-time PCR for (B) Klf6, (C) Rex1, (D) Brachyury, (E) Scl, (F) Klf1, (G) Gata1, and (H) Gata2.
Expression of differentiation marker genes is delayed in Klf6-/- ES cells. (A) Differentiation into EBs of Klf6+/+ ES cells, Klf6+/- ES cells, and 2 clones of Klf6-/- ES cells was analyzed. EBs differentiated for 2, 3, 4, 6, or 8 days were harvested at each time point. Total RNAs were extracted and analyzed by RT-PCR. The expression of the indicated genes is shown. The cDNAs generated from the same mRNAs were also subjected to quantitative real-time PCR for (B) Klf6, (C) Rex1, (D) Brachyury, (E) Scl, (F) Klf1, (G) Gata1, and (H) Gata2.
In addition to semiquantitative PCR, we quantified expression of mRNAs for Klf6, Rex1, Brachyury, Scl, Klf1, Gata1, and Gata2 by real-time quantitative PCR. The expression patterns of Klf6 in Klf6+/+ and Klf6+/- EBs during differentiation were initially similar. However, after more than 4 days of differentiation Klf6 expression in Klf6+/- EBs deviated from that of Klf6+/+ EBs (Figure 5B) (Klf6 signals detected in Klf6-/- ES clones were caused by contamination by mRNA from MEF feeder cells). Sustained expression of Rex1 was observed in Klf6-/- EBs (Figure 5C), whereas expression of Brachyury was delayed in Klf6+/- and Klf6-/- EBs although peak levels were similar (Figure 5D). While Scl expression was delayed by at least 2 days in Klf6-/- EBs compared with Klf6+/+ and Klf6+/- EBs, its peak expression levels were the same as in Klf6+/+ EBs at day 8 (Figure 5E). The expression of Klf1 and Gata1 in Klf6-/- EBs was also delayed by at least 2 days (Figure 5F-G). Of interest, the expression level of Gata2 mRNA in Klf6+/- and Klf6-/- EBs was higher than in Klf6+/+ EBs (Figure 5H), suggesting that Gata2 may compensate for reduced Klf6 in Klf6+/- and Klf6-/- EBs. Taken together, these findings indicate that loss of Klf6 function delays mesoderm induction and its specification to the hematopoietic lineages.
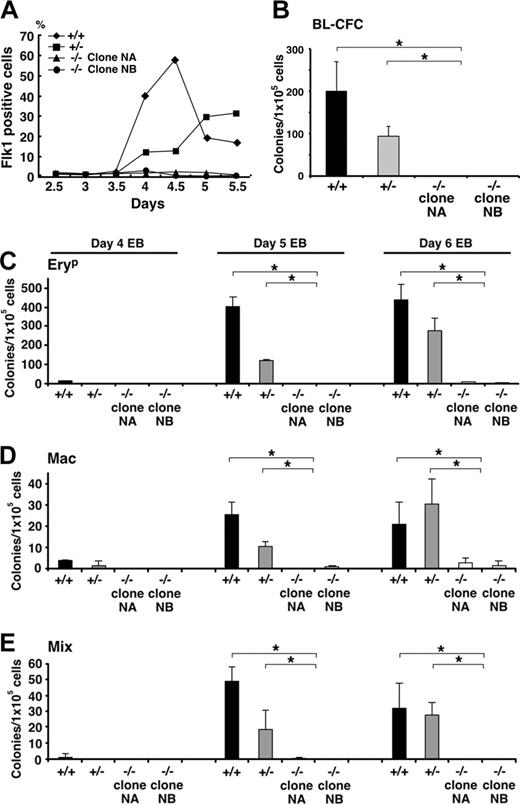
Evidence of a haploinsufficient phenotype in Klf6+/- EBs emerged from studies assessing levels of Flk1 cell-surface protein by antibody staining (Figure 6A). Whereas there was no difference in Flk1 mRNA in yolk sacs among the 3 genotypes, there was both delayed and reduced Flk1 expression by Klf6+/- EB cells, suggesting impaired differentiation of precursor cells of the hematopoietic and vascular lineages. This defect was even more apparent in Klf6-/- cells, where little Flk1 was detected throughout the 5-day differentiation period (Figure 6A). Consistent with this observation, Klf6-/- ES cells were significantly deficient in blast colony-forming cells (BL-CFCs), the in vitro equivalent of a hemangioblast (Figure 6B). Striking hematopoietic defects were also observed, since the Klf6-/- EBs contained only a fraction of the number of primitive erythrocyte (EryP), Mac, and Mix colonies compared with EBs generated from wild-type ES cells. Hematopoietic development in Klf6+/- EBs was either reduced or delayed, consistent with haploinsufficiency (Figure 6C-E).
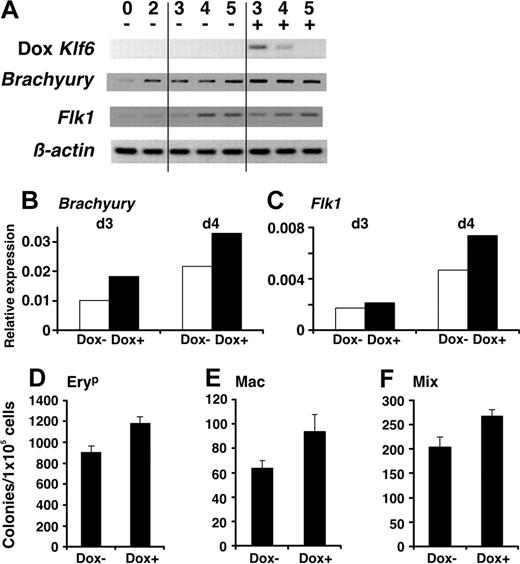
Finally, we examined the impact of reconstituting Klf6 expression in -/- ES cells. Wild-type Klf6 was introduced into Klf6-/- ES cells by retroviral infection, and its effects on proliferation and differentiation were assessed. Reconstitution of Klf6 decreased the doubling time of Klf6-/- ES cells by approximately 20%, and cell number at EB day 6 was approximately 40% greater (data not shown). As a final approach, we generated a cell line in which Klf6 expression could be induced in wild-type ES cells or EBs through the addition of doxycycline. When Klf6 was induced on day 2, Klf6 mRNA was about 40% higher than that of control cells. Moreover, this induction (Figure 7A) led to enhanced expression of Brachyury (Figure 7B) and Flk1 (Figure 7C) mRNAs, and enhanced the number of hematopoietic progenitors (Figure 7D-F). Of note, this effect was observed only when Klf6 was induced on day 2.
Discussion
In order to gain insight into the developmental function of Klf6, we used a combined in vivo and in vitro approach, using homozygous mutant Klf6 knock-out mice and ES cells, respectively. We found that midgestation Klf6-/- embryos are small and pale, and their yolk sacs display an impaired hematopoietic potential. These in vivo data are recapitulated by the in vitro experiments demonstrating that Klf6-/- ES cells proliferate more slowly than their wild-type counterparts and display a reduced ability to differentiate into hematopoietic and vascular cells. Our findings highlight the importance of KLF6 function in mesoderm induction and its specification into the hematopoietic lineage. This observed phenotype in Klf6-/- mice appears to result from multiple impairments in early hematopoietic development rather than from a maturational arrest of a precursor population.
Hematopoietic differentiation of Klf6-/- ES cells is impaired. (A) Cell-surface expression of Flk1 was analyzed by FACS. EBs differentiated for 2.5 to 5.5 days were stained with antibodies to Flk1, and the percentages of Flk1-positive cells are plotted. Reduced Flk1 expression is evident not only in Klf6-/- ES cells (for up to 5 days), but also in Klf6+/- cells. (B) Cells from day-4 EBs were subjected to blast colony-forming assay. EBs were trypsinized and 1 × 105 cells were plated into methylcellulose cultures containing VEGF. Numbers of blast cell colonies were counted 4 days later. (C-E) Hematopoietic colony-forming assays from days 4, 5, and 6 EBs were performed. Cells from EBs were trypsinized and 1 × 105 cells were plated into methylcellulose cultures containing a mix of cytokines (“Materials and methods”). Numbers of EryP (C), Mac (D), and Mix (E) colonies were counted. Bars represent SD of colony counts from 3 independent cultures. *P < .05 (Student t test).
Hematopoietic differentiation of Klf6-/- ES cells is impaired. (A) Cell-surface expression of Flk1 was analyzed by FACS. EBs differentiated for 2.5 to 5.5 days were stained with antibodies to Flk1, and the percentages of Flk1-positive cells are plotted. Reduced Flk1 expression is evident not only in Klf6-/- ES cells (for up to 5 days), but also in Klf6+/- cells. (B) Cells from day-4 EBs were subjected to blast colony-forming assay. EBs were trypsinized and 1 × 105 cells were plated into methylcellulose cultures containing VEGF. Numbers of blast cell colonies were counted 4 days later. (C-E) Hematopoietic colony-forming assays from days 4, 5, and 6 EBs were performed. Cells from EBs were trypsinized and 1 × 105 cells were plated into methylcellulose cultures containing a mix of cytokines (“Materials and methods”). Numbers of EryP (C), Mac (D), and Mix (E) colonies were counted. Bars represent SD of colony counts from 3 independent cultures. *P < .05 (Student t test).
The phenotype and embryonic lethality of Klf6-/- mice at E 12.5 are similar to those of Scl-/- mice.42,43 Of interest, Scl expression is reduced in Klf6-/- yolk sacs and also corresponds to down-regulation of Gata1, a possible target gene of SCL in Klf6-/- yolk sacs. Impaired hematopoietic development may therefore contribute to embryonic lethality of Klf6-/- mice since these animals, like Scl-/- and Gata1-/- mice, also die before E 12.5.42,43,45,46 However, down-regulation of Scl and Gata1 cannot solely explain the defects in Klf6-/- mice, since these animals also displayed other developmental defects, for example poorly defined liver (Figure 3E), incomplete neural tube closure (not shown), and impaired placental development (not shown). These multilineage abnormalities are consistent with the conclusion that defects in Klf6-/- ES cells are apparent before they differentiate into 3 germ layers.
These findings reinforce the conclusion that members of the Krüppel-like factor family broadly regulate developmental pathways. For example, KLF1 is critical for the development of erythrocytes,17,18 whereas KLF2 is essential for lung development and formation of tunica media of blood vessels,16 as well as for hematopoeisis.49 KLF4, on the other hand, is required for the differentiation of the skin barrier19 and goblet cells in colon.50 Most of these defects occur in late stages of development. In contrast, Klf5-/- mice die at an earlier stage, embryonic day 8.5,24 since this molecule may serve key roles in epithelial biology and cardiovascular regeneration.
Doxycycline-induced Klf6 restores hematopoietic differentiation. (A) Doxycycline was added to culture medium on day 2 of EB differentiation in cells expressing a doxycycline-regulable Klf6. Expression of Brachyury (B) and Flk1 (C) is increased in cells following induction of Klf6 and confirmed by real-time PCR. Cells from day-5 EBs were subjected to hematopoietic colony formation assay. Colony numbers of EryP (D), Mac (E), and Mix (F) were counted. Bars represent SE of colony counts from 3 independent cultures. *P < .05 (Student t test).
Doxycycline-induced Klf6 restores hematopoietic differentiation. (A) Doxycycline was added to culture medium on day 2 of EB differentiation in cells expressing a doxycycline-regulable Klf6. Expression of Brachyury (B) and Flk1 (C) is increased in cells following induction of Klf6 and confirmed by real-time PCR. Cells from day-5 EBs were subjected to hematopoietic colony formation assay. Colony numbers of EryP (D), Mac (E), and Mix (F) were counted. Bars represent SE of colony counts from 3 independent cultures. *P < .05 (Student t test).
The developmental importance of Klf6 has also been underscored by recent findings in Drosophila, where a common Klf6/Klf7 homolog, termed Luna, is involved in embryonic development and differentiation.25 Half of Luna-deficient organisms die prior to gastrulation, which is consistent with our in vitro analysis indicating that Klf6-/- EBs display a prolongation of the epiblast stage and a delay in mesoderm induction, indicating a potential role for KLF6 in gastrulation. Despite the partial sequence homology between Klf6 and Klf7 (identical 47 amino-terminal residues and DNA binding domains), KLF7 does not compensate for loss of KLF6 in Klf6 knock-out mice. Indeed, Klf7-/- mice suffer from a complex disruption of normal axon growth at multiple sites of the developing nervous system, a phenotype clearly distinct from Klf6 knock-out mice.51 Moreover, Klf7 mRNA was not overexpressed in Klf6-/- ES cells, suggesting that compensation of KLF6 function by KLF7 does not occur, or is very limited in Klf6-/- ES cells. On the other hand, Gata2 was up-regulated not only in Klf6-/- but also Klf6+/- EBs (Figure 5H). Although Gata2 is up-regulated in Gata1-/- EBs,52 this observation does not explain the induction of Gata2 in Klf6+/- EBs, since Gata1 expression in Klf6+/+ and Klf6+/- EBs is similar (Figure 5G). Our experiment with embryoid bodies suggests the possibility of compensation of KLF6 function by Gata1. Apart from regulating hematopoiesis, Klf6 contributes to vasculogenesis, as reflected in the dramatically reduced number of Flk1-positive cells and BL-CFCs in the Klf6-/- EBs. These findings further reinforce the emerging concept that the hemangioblast gives rise to both vascular and hematopoietic lineages.26,27,40,53,54 The deficiency in the development of the Flk1-positive population and BL-CFCs suggests that the one function of Klf6 may be to promote the specification of mesoderm to the hematopoietic and vascular lineages. Consistent with this interpretation is the demonstration that forced expression of KLF6 increases Flk1 expression and enhances hematopoietic potential in wild-type cells.
Klf6 has previously been identified as a tumor suppressor gene that is inactivated in a number of human cancers,33,55-58 in part through induction of the p21 gene as well as through regulation of cell cycle59 and apoptosis.60 Together with these published data, the findings described here indicate that KLF6 has critical, yet divergent, roles in regulating cellular proliferation and differentiation during embryonic development and adulthood. Of interest, although p21 is an established target gene of KLF6, it is not down-regulated in Klf6-/- embryos or differentiating ES cells. Thus, the activity and transcriptional targets of KLF6 are likely to differ depending on the development- and tissue-specific contexts. In support of this conclusion, whereas KLF6 inactivation promotes cell proliferation in adult tissues, Klf6-/- ES cells actually grew more slowly than Klf6+/+ cells. Other tumor suppressor genes display similarly divergent activities between developing and adult tissues. For example, Smad4/Dpc4-/- embryos are embryonic lethal by E 7.5 and show reduced proliferation in vivo and in vitro.61 Also, Pten-/- embryos die before E 7.5, Pten-/- EBs display impaired hemoglobinization,62 and Men1-/- ES cells exhibit impaired formation of EBs.63
In summary, our findings support a developmental role of Klf6 at several different stages in ES-cell differentiation in vitro and during murine development in vivo. In early stages of ES-cell differentiation, Klf6 promotes ES-cell proliferation and differentiation. Subsequently, Klf6 contributes to differentiation into mesoderm cells as defined by Brachyury expression. Finally, Klf6 plays a role in both hematopoiesis and vasculogenesis, reinforcing its potential contribution to hemangioblast development.
Prepublished online as Blood First Edition Paper, October 18, 2005; DOI 10.1182/blood-2005-05-1916.
Supported by National Institutes of Health grants DK37340 (S.L.F.), AR38648 (F.R.), and HL048834 (G.K.); the Bendheim Foundation Trust (S.L.F.); New York State Spinal Cord Injury Research (F.R.); and the St Giles Foundation (F.R.).
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Xiao Zhao and Sayaka Moriyama for technical contributions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal