Goodpasture syndrome is an autoimmune vascular disease associated with kidney and lung failure, with pathogenic circulating autoantibodies targeted to a set of discontinuous epitope sequences within the noncollagenous domain-1 (NC1) of the α3 chain of type IV collagen (α3(IV)NC1), the Goodpasture autoantigen. We demonstrate that basement membrane extracted NC1 domain preparations from Caenorhabditis elegans, Drosophila melanogaster, and Danio rerio do not bind Goodpasture autoantibodies, while Xenopus laevis, chicken, mouse and human α3(IV)NC1 domains bind autoantibodies. The α3(IV) chain is not present in C elegans and Drosophila melanogaster, but is first detected in the Danio rerio. Interestingly, native Danio rerio α3(IV)NC1 does not bind Goodpasture autoantibodies. Next, we cloned, sequenced, and generated recombinant Danio rerio α3(IV)NC1 domain. In contrast to recombinant human α3(IV)NC1 domain, there was complete absence of autoantibody binding to recombinant Danio rerio α3(IV)NC1. Three-dimensional molecular modeling from existing x-ray coordinates of human NC1 domain suggest that evolutionary alteration of electrostatic charge and polarity due to the emergence of critical serine, aspartic acid, and lysine residues, accompanied by the loss of asparagine and glutamine, contributes to the emergence of the 2 major Goodpasture epitopes on the human α3(IV)NC1 domain, as it evolved from the Danio rerio over 450 million years.
Introduction
Type IV collagen is the most abundant protein within all basement membranes (BMs).1 This collagen is present in most multicellular organisms, and thus is an evolutionary constant.2,3 In the BM, type IV collagen forms a network, which other basement membrane molecules attach to or become sequestered within.1,4 Type IV collagen in humans and mice is composed of 6 distinct α-chain gene products, α1(IV)-α6(IV). The α1(IV) and α2(IV) chains are ubiquitously present in most organs, while the α3(IV)-α6(IV) chains exhibit more restricted distributions and are present mostly in BMs with special functional roles, such as the kidney glomerular basement membrane (GBM).1,5-7
The α3-chain of type IV collagen is found in the kidney GBM, the alveolar BM of the lung, the BMs of the inner ear and the testis,1,8 and is present in most mammals.9 Mice deficient in the α3(IV) chain die due to renal failure, as this protein is required for the proper function of the GBM.10,11 This phenotype resembles human Alport syndrome, a disease affecting kidneys and the inner ear, which results from mutations in the α3 chain, the α4 chain, or the α5 chain of type IV collagen.8,10 Pathogenic autoantibodies directed against the NC1 domain of the α3 chain of type IV collagen (Goodpasture autoantigen) are associated with human Goodpasture syndrome (GP), a disease characterized by rapidly progressive glomerulonephritis and lung hemorrhage.8,12-13 These autoantibodies are targeted to specific amino acid sequences within the NC1 domain of the α3(IV) chain.8,14 By using mutagenesis analysis, it is known that a majority of the autoantibodies are directed against 2 distinct epitopes on the α3(IV)NC1 domain, namely epitopes EA and EB.15-18 Epitope EA appears to be the immunodominant epitope, as the majority of patients' antibodies are directed against this site.18 While the production of immunoglobulin G1 (IgG1) and IgG3 subclasses of α3(IV) autoantibodies is considered to be the pathogenic feature of Goodpasture syndrome, there is also strong evidence now for the role of T cells in the initiation of the disease.19-26 Therefore, in the past few years, Goodpasture syndrome has emerged as a classic autoimmune vascular disease mediated by B cells27-29 and T cells,19-26 and immunosuppression in conjunction with plasmapheresis remains the most effective therapy.30
Since the vast majority of autoantibodies bind to the immunodominant epitope EA and epitope EB, several studies have sought to elucidate the key residues in these sites. Such studies have been primarily focused on epitope EA because this site is capable of inducing disease in rodents.14 The method most commonly used has been to recombinantly produce chimeric proteins.15-18,31-32 Many of these chimeric proteins are comprised of amino acids of the epitope EA from the α3(IV)NC1 domain with site-specific substitutions of analogous residues from the α1(IV)NC1 domain.17,31-32 Such proteins were subsequently tested for their binding to autoantibodies from Goodpasture patients, thus identifying putative critical amino acids that comprise the Goodpasture epitope.
While others have employed site-directed mutagenesis and chimeric protein analysis as a way to identify the critical amino acids within the Goodpasture epitopes, we hypothesized that evolutionary changes over 450 million years33 in the amino acids within the α3(IV)NC1 domains might offer insight into the unique amino acid residues that comprise the B-cell epitope for Goodpasture autoantibodies. Comparative type IV collagen NC1 sequence analysis between 9 species from Caenorhabditis elegans to humans, BM extraction, NC1 domain analysis from different species, cloning, and recombinant protein production were used to identify the critical amino acids within the B-cell epitopes of the Goodpasture autoantigen. This study provides a strategy to identify critical amino acids responsible for Goodpasture autoantibody binding and offers new insights into potential therapeutic alternatives. A precise identification of amino acids responsible for autoimmunity will help design cell-mediated therapies to potentially impart systemic tolerance to pathogenic epitopes associated with this devastating disease.
Materials and methods
Animals and tissues
Wild-type C57BL/6 mouse kidney, wild-type Danio rerio strain AB* kidney, and wild-type domestic chicken kidney (Pel-Freez Clinical Systems, Rogers, AR) were used for all procedures. Wild type Xenopus laevis kidneys were the kind gift of Dr Sergei Sokol (Beth Israel Deaconess Medical Center/Harvard Medical School). Wild-type strains CS and W1118Drosophila melanogaster were the kind gifts of Dr Mel Feany (Brigham and Women's Hospital/Harvard Medical School). Wild-type C elegans, strain NL, were the kind gift of Dr Monica Colaiacovo (Department of Genetics, Harvard Medical School).
Cloning, primers, and sequencing
Total RNA was isolated from Danio rerio embryos 24 hours after fertilization (hpf) using TRIzol reagent (Invitrogen, Carlsbad, CA). A collection of cDNAs was created using Superscript II reverse transcriptase (Invitrogen) and oligo dT primers (Invitrogen). Polymerase chain reaction (PCR) amplification of Danio rerio α3(IV)NC1 domain was performed using AccuPrime Pfx DNA polymerase (Invitrogen) and the specific primers 5′ CACCAGGTGCAAAAGGTCCACAAC 3′ and 5′ ACGATTTTGTGTCAGGGTCAGAA 3′, designed from sequence data obtained from the Project Ensembl database and software system (http://www.ensembl.org), maintained by the European Molecular Biology Laboratory-European Bioinformatics Institute and the Sanger Institute. The resulting cDNA clones were subcloned into the pCRII 4.0-TOPO vector (Invitrogen), and sequenced by the Beth Israel Deaconess Medical Center sequencing facility using the M13 (-20) forward and M13 reverse primers. Sequences were analyzed using the Mac Vector 6.0 (Oxford Molecular Group, Cambridge, United Kingdom) and BLAST programs (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/). The cloned sequences were deposited in GenBank under the accession number AY954909.1.
Recombinant protein production
The following PCR primers were designed to amplify a cDNA encoding the full-length Danio rerio α3(IV)NC1, which we have termed zα3(IV)NC1; 5′ TCA CCA GGT GCA AAGCTT CCA CAA 3′ and 5′ TCT GTT CTCGAG TTT GGC GGT TTG 3′. The underlined sequence indicates the HindIII and XhoI restriction sites that were used for subsequent cloning of the generated cDNA fragment. The HindIII/XhoI fragment of the amplified product was cloned into the pcBFT expression vector containing the cytomegalovirus (CMV) promoter, the BM-40 signal peptide, and a FLAG (octapeptide DYKDDDDK) tag.34 293 human embryonic kidney cells were transfected with the plasmid using the 293 Fectin-system (Invitrogen) according to manufacturer's protocol. Recombinant zα3(IV)NC1 was secreted into the culture supernatant as a fusion protein with the FLAG epitope. Supernatant containing protein was collected 24 hours after transfection. Recombinant human α3(IV)NC1 production was described earlier.34
Histology and immunofluorescence
Adult Danio rerio were embedded in Tissue-Tek optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA) and snap-frozen on liquid nitrogen, as well as fixed in formalin and embedded in paraffin for further histologic analysis. Paraffin sections (5 μm) were stained with hematoxylin and eosin (H&E) according to routine histologic practice. Frozen sections (5 μm) were fixed with acetone, blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), incubated with the anti-T7 antibody (raised in rabbit against the T7 peptide of human α3(IV)NC1 domain)35 at 1:75 dilution, and the anti-human α3(IV)-36mer antibody at 1:200 dilution,32 followed by an antirabbit FITC-conjugated secondary antibody at 1:100 dilution (Jackson Immunoresearch, Bar Harbor, ME). The sections were mounted using the Vectashield mounting media containing DAPI for nuclear staining (Vector Laboratories, Burlingame, CA). Histologic images were captured using a Zeiss brightfield and fluorescence microscope, a 40×/1.3 numeric aperture Plan-Neofluar objective with Immersol oil, an AxioCam HRc digital camera, and Axiovision software version 4.3 (all from Zeiss, Oberkochen, Germany).
Isolation of native type IV collagen NC1 domains
Kidneys dissected from mouse, chicken, Xenopus laevis, and Danio rerio were snap-frozen in liquid nitrogen followed by homogenization by mortar and pestle. In the case of Drosophila and C elegans, the whole organisms were subjected to homogenization. Tissue homogenates were centrifuged and resuspended in PBS containing Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN), thoroughly homogenized again, centrifuged, and resuspended in 1 M NaCl supplemented with DNase I (80 μg/mL). Following a third centrifugation, samples were resuspended in 2% deoxycholate/protease inhibitor solution and centrifuged a final time. Samples were resuspended in bacterial collagenase solution (1 U/μL; Worthington Biochemical, Lakewood, NJ) to digest proteins with collagenous regions, in order to liberate soluble NC1 domains of type IV collagen.
Western blotting and antibodies
All Western blots were performed on either 10% or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad, Hercules, CA) under reducing or nonreducing conditions. Gels were transferred to nitrocellulose and incubated with sera and primary and secondary antibodies at the appropriate dilutions. The following primary antibodies were used: anti-α3(IV)-36mer antibody at 1:7500 dilution;32 2 separate α3(IV)NC1 antibodies;27 3 confirmed Goodpasture patient sera at 1:20 dilution;36 and anti-FLAG M2 antibody at 1:1000 dilution (Sigma, St Louis, MO). In addition, 2 normal human sera were used as negative controls at the same dilutions as the Goodpasture sera (data not shown). Anti-human IgG and anti-rabbit IgG horseradish peroxidase (HRP; Sigma)-conjugated secondary antibodies were used at 1:1000 dilutions. Enhanced chemiluminescence (ECL) reagent for peroxidase substrate (Amersham Biosciences, Freiburg, Germany) was used to visualize antibody binding by exposure to x-ray film (Denville, Metuchen, NJ).
Molecular graphics
The structures of the [(α1)2α2]2 NC1 hexamers from human placenta basement membranes (Protein Data Bank ID 1LI1)37 were rendered using the molecular graphics visualization program YASARA (YASARA Biosciences, Graz, Austria) and analyzed to generate modeled structures using the molecular graphics software InsightII (Accelrys, San Diego, CA).
Results
The emergence of the Goodpasture epitope
To study the binding of Goodpasture autoantibodies, we extracted NC1 domains from BM extracts using bacterial collagenase from the following species: mouse, chicken, Xenopus laevis, Danio rerio, Drosophila, and C elegans. For the higher vertebrate organisms, kidney tissue was used as a source of BM, whereas for Drosophila and C elegans whole-organism BM extracts were used. Type IV collagen NC1 domains were successfully extracted from all organisms as shown by SDS-PAGE analysis and Western blot evaluation under reducing conditions (Figure 1A-B). When analyzing Goodpasture autoantibody reactivity to the isolated BM extracts under nonreducing conditions, positive binding of sera to α3(IV) chain dimers and monomers could be observed in mouse, chicken, and Xenopus (Figure 1C). Interestingly, binding to dimers was stronger than to monomers, as others have described9 (Figure 1C). The Goodpasture epitope is considered to be sensitive to conformational changes and thus complete reduction of disulfide bonds with 10% β-mercaptoethanol leads to a total loss of antibody binding (Figure 1E), while incomplete reduction with 1.5% β-mercaptoethanol leads to stronger binding to α3(IV)NC1 monomers in all species (Figure 1D). Reactivity with Goodpasture sera could not be observed with NC1 domains extracted from Danio rerio, C elegans, and Drosophila (Figure 1C). In Drosophila and C elegans both α1(IV)-like and α2(IV)-like genes have been described,38-40 but our genomic database analysis and cloning attempts suggest that an α3(IV) gene does not appear to be present. Interestingly, our database search indicates that Danio rerio has 6 type IV collagen α chains similar to higher organisms (B.A.M., M.S., R.K., unpublished data). However, while the Danio rerio α3(IV) gene was identified in our database search, the native NC1 domain extracts do not bind to Goodpasture autoantibodies (Figure 1C).
Cloning, sequencing, and recombinant expression of the Danio rerio α3(IV) chain NC1 domain
Our results suggest that while the α3(IV) chain is present in the Danio rerio (by inference from genomic database sequences), the NC1 domain extract from BM preparations of Danio rerio renal tissue does not bind to Goodpasture autoantibodies. To further evaluate the capacity of the Danio rerio α3(IV)NC1 domain to bind these antibodies, we attempted to clone and generate recombinant protein.
In order to clone the Danio rerio α3(IV)NC1 domain (zα3(IV)NC1), we isolated total RNA from Danio rerio embryos and generated a collection of cDNAs by reverse transcription (RT), using oligo dTs as primers for the reaction. These cDNAs were subsequently used as a template to obtain the zα3(IV)NC1 domain by PCR. Gene-specific PCR primers were designed using sequence data information obtained from the project Ensembl database. We successfully generated an 821-bp cDNA fragment, and sequencing verified that the cloned cDNA fragment contained 23 bp of the collagenous region, 702 bp containing the entire NC1 domain, and 96 bp of noncoding sequence. The nucleotide and amino acid sequence of zα3(IV)NC1 is shown in Figure 2A. The 5 exons encoding the overall nucleotide and amino acid sequence of the zα3(IV)NC1, when compared with human α3(IV)NC1, have 60% and 74% homology, respectively. The 12 cysteines known to be essential for the appropriate conformational folding of the type IV collagen NC1 structure41 are all conserved in zα3(IV)NC1 (Figure 2A). Interestingly, a methionine and a lysine residue, potentially involved in a novel nondisulfide covalent crosslink37 are also conserved in the zα3(IV)NC1 (Figure 2A).
Extraction of basement membrane, bacterial collagenase solubilization and the analysis of native type IV collagen NC1 domains from different species. (A) Native protein (60 μg per lane) from the 6 different species and 250 ng of recombinant human α3(IV)NC1, (rh-α3(IV)NC1)35 was run on a 15% SDS-PAGE under extreme reducing (10% β-mercaptoethanol) conditions, and incubated with the anti-36mer antibody.23 Monomer forms (M) of NC1s are detected in all lanes. Although subjected to extreme reduction, significant amounts of the isolated NC1 remain as dimers (D). (B) C elegans NC1 in dimeric form (D) was visualized after increased film exposure. (C) The same samples were run on a 15% SDS-PAGE under nonreducing conditions and incubated with 3 different Goodpasture sera (1:20 dilution). A representative blot is shown. As expected, sera from Goodpasture patients bound recombinant human α3(IV)NC1 (lane 1). Mouse, chicken, and frog kidney BM extracts show strong reactivity with Goodpasture sera (lanes 2-4). NC1 in monomer form (M) could only be detected for chicken without the use of a reducing agent, whereas for the other species the Goodpasture sera detected dimers (D) under nonreducing conditions. Interestingly, no reactivity to Danio rerio, Drosophila, and C elegans NC1 was found with any of the analyzed Goodpasture sera (lanes 5-7). (D) Reduction of the samples eliminates the conformational structure of the Goodpasture epitope. With slight reduction (1.5% β-mercaptoethanol) some monomer (M) from each of the 3 positive species (Mus, Gallus, Xenopus) native NC1s was recognized by the Goodpasture sera. (E) Under extreme reducing conditions (10% β-mercaptoethanol), binding to all monomer and dimer forms of NC1 by Goodpasture sera was eliminated, as expected and as previously described.17
Extraction of basement membrane, bacterial collagenase solubilization and the analysis of native type IV collagen NC1 domains from different species. (A) Native protein (60 μg per lane) from the 6 different species and 250 ng of recombinant human α3(IV)NC1, (rh-α3(IV)NC1)35 was run on a 15% SDS-PAGE under extreme reducing (10% β-mercaptoethanol) conditions, and incubated with the anti-36mer antibody.23 Monomer forms (M) of NC1s are detected in all lanes. Although subjected to extreme reduction, significant amounts of the isolated NC1 remain as dimers (D). (B) C elegans NC1 in dimeric form (D) was visualized after increased film exposure. (C) The same samples were run on a 15% SDS-PAGE under nonreducing conditions and incubated with 3 different Goodpasture sera (1:20 dilution). A representative blot is shown. As expected, sera from Goodpasture patients bound recombinant human α3(IV)NC1 (lane 1). Mouse, chicken, and frog kidney BM extracts show strong reactivity with Goodpasture sera (lanes 2-4). NC1 in monomer form (M) could only be detected for chicken without the use of a reducing agent, whereas for the other species the Goodpasture sera detected dimers (D) under nonreducing conditions. Interestingly, no reactivity to Danio rerio, Drosophila, and C elegans NC1 was found with any of the analyzed Goodpasture sera (lanes 5-7). (D) Reduction of the samples eliminates the conformational structure of the Goodpasture epitope. With slight reduction (1.5% β-mercaptoethanol) some monomer (M) from each of the 3 positive species (Mus, Gallus, Xenopus) native NC1s was recognized by the Goodpasture sera. (E) Under extreme reducing conditions (10% β-mercaptoethanol), binding to all monomer and dimer forms of NC1 by Goodpasture sera was eliminated, as expected and as previously described.17
Cloning and sequencing of the Danio rerio α3(IV)NC1 and characterization of recombinant Danio rerio α3(IV)NC1 domain. (A) Nucleotide and peptide sequence of the NC1 domain of Danio rerio type IV collagen α3-chain, (zα3(IV)NC1). The 2 regions homologous to Goodpasture epitope, EA and EB, are boxed. The conserved methionine and lysine (diamonds) residues, potentially involved in a conserved novel nondisulfide covalent crosslink,27 are shown. The circles indicate the 12 conserved cysteines. (B) FLAG-tagged Danio rerio α3(IV)NC1 was recombinantly produced and an expected band at 29.4 kDa is recognized by the anti-FLAG antibody (lane 1), as well as with the anti-human α3(IV) NC1 antibody (lane 2). However, no binding with Goodpasture sera is observed (lane 3), despite prolonged exposure of film. (C) Native Danio rerio kidney extract (30 μg per lane) digested with bacterial collagenase was run on 15% SDS-PAGE under both reducing and nonreducing (NR) conditions. Under nonreducing conditions, only dimers (D) and trimers (T) can be observed with the anti-human α3(IV) NC1 antibody antibody (lane 1). Under reducing conditions, native α3(IV)NC1 monomers (M) are observed at the expected molecular weight (lane 3). FLAG-tagged recombinant human α3(IV)NC1 (rh-α3(IV)NC1; 250 ng) was used as positive control (lane 2).35 (D) No reactivity with Goodpasture sera with native Danio rerio kidney extract can be seen (lane 1). FLAG-tagged rh-α3(IV)NC1 was used as positive control (lane 2).
Cloning and sequencing of the Danio rerio α3(IV)NC1 and characterization of recombinant Danio rerio α3(IV)NC1 domain. (A) Nucleotide and peptide sequence of the NC1 domain of Danio rerio type IV collagen α3-chain, (zα3(IV)NC1). The 2 regions homologous to Goodpasture epitope, EA and EB, are boxed. The conserved methionine and lysine (diamonds) residues, potentially involved in a conserved novel nondisulfide covalent crosslink,27 are shown. The circles indicate the 12 conserved cysteines. (B) FLAG-tagged Danio rerio α3(IV)NC1 was recombinantly produced and an expected band at 29.4 kDa is recognized by the anti-FLAG antibody (lane 1), as well as with the anti-human α3(IV) NC1 antibody (lane 2). However, no binding with Goodpasture sera is observed (lane 3), despite prolonged exposure of film. (C) Native Danio rerio kidney extract (30 μg per lane) digested with bacterial collagenase was run on 15% SDS-PAGE under both reducing and nonreducing (NR) conditions. Under nonreducing conditions, only dimers (D) and trimers (T) can be observed with the anti-human α3(IV) NC1 antibody antibody (lane 1). Under reducing conditions, native α3(IV)NC1 monomers (M) are observed at the expected molecular weight (lane 3). FLAG-tagged recombinant human α3(IV)NC1 (rh-α3(IV)NC1; 250 ng) was used as positive control (lane 2).35 (D) No reactivity with Goodpasture sera with native Danio rerio kidney extract can be seen (lane 1). FLAG-tagged rh-α3(IV)NC1 was used as positive control (lane 2).
The cloned cDNA fragment was subsequently transferred to the pcBFT expression vector and transfected into 293 human embryonic kidney cells to produce recombinant zα3(IV)NC1 using a BM-40 signal peptide to enable extracellular secretion. Recombinant FLAG-tagged zα3(IV)NC1 fusion protein in the culture supernatants was analyzed by SDS-PAGE. Western blot analysis using antibodies directed against the FLAG tag and the human α3(IV)NC1 verified the recombinant expression of zα3(IV)NC1 monomers at the predicted molecular weight of approximately 29.4 kDa (zα3(IV)NC1+FLAG tag; Figure 2B). However, zα3(IV)NC1 did not bind to Goodpasture autoantibodies from 3 different patients with clinical disease (Figure 2B), despite prolonged incubation with high concentrations of the autoantibodies to detect even weak binding. However, as shown earlier, strong binding of anti-human α3(IV)NC1 polyclonal antibodies to isolated native zNC1 domain monomers, dimers, and trimers from BM extracts were observed (Figure 2C). Under reducing conditions the higher-molecular-weight trimers were lost and monomers of Danio rerio α3(IV)NC1 revealed stronger binding (Figure 2C). However, binding with the Goodpasture sera to the isolated Danio rerio NC1 domains could be not observed, as shown earlier (Figure 2D). Together, these results demonstrate that although Danio rerio contains the encoding gene and the α3 chain of type IV collagen protein, the protein does not possess the capacity to bind Goodpasture autoantibodies.
Expression of the α3(IV) chain in Danio rerio tissues
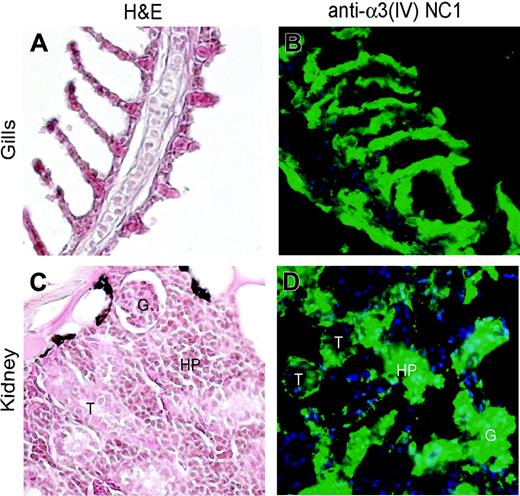
Since this report is the first description of the α3 chain of type IV collagen in the Danio rerio, and human α3(IV) chain is localized to the GBM of the kidney and the alveolar BMs of the lung, we performed immunohistochemistry to evaluate the expression of the zα3(IV)NC1 in gills and kidney tissue of Danio rerio. Immunohistochemical localization of the zα3(IV) chain in tissues was performed using 2 separate antibodies directed against the human α3(IV)NC1 domain. Identical results were obtained with both antibodies (Figure 3 and data not shown). The α3(IV) chain in Danio rerio is strongly expressed in the gills and the kidney (Figure 3B-D). Gills are the fish equivalent of lungs, and the lungs and kidney are the sites of high expression of the α3(IV) chain in mouse and humans.1,5-7 On the other hand, we could not observe any expression of the α3(IV) chain in Danio rerio testis (data not shown), although in mammals it is a site with high α3(IV) expression.1,5-7,42 In the gills the α3(IV) chain appeared to be expressed in the BM underlying the epithelial cell layer (Figure 3A-B). In the Danio rerio, the kidney morphology is distinct from that of higher organisms as it is also the site of hematopoiesis, in addition to its function in ultrafiltration (Figure 3C). In Danio rerio kidney the α3(IV) chain is expressed in both the BM of the glomeruli as well as some tubules (Figure 3D). Interestingly, we could also observe strong staining of the α3(IV) chain in the hematopoietic tissue (Figure 3D). In higher organisms, the α3(IV) chain is an important component of the GBM in the kidney and is also expressed in some tubular BMs.6 However, the potential role of this molecule in hematopoiesis is unknown. As observed in humans and other mammals, we were unable to detect zα3(IV)NC1 in liver and stomach basement membranes.
Expression of type IV collagen α3 chain in the Danio rerio. Adult Danio rerio tissues were stained with anti-human α3(IV) antibodies and H&E. (A-B) In Danio rerio the gills are the equivalent of the lung. Strong expression of α3(IV) can be seen in the Danio rerio gills (green). DAPI staining is used to stain the nuclei of cells (blue). (C-D) In Danio rerio the kidney is the main hematopoietic organ and thus morphologically distinct from that of higher organisms (C). Strong expression of α3(IV) can be seen in both the GBM (G) and the tubular basement membranes (T) as well as in the hematopoietic tissue (HP) (D).
Expression of type IV collagen α3 chain in the Danio rerio. Adult Danio rerio tissues were stained with anti-human α3(IV) antibodies and H&E. (A-B) In Danio rerio the gills are the equivalent of the lung. Strong expression of α3(IV) can be seen in the Danio rerio gills (green). DAPI staining is used to stain the nuclei of cells (blue). (C-D) In Danio rerio the kidney is the main hematopoietic organ and thus morphologically distinct from that of higher organisms (C). Strong expression of α3(IV) can be seen in both the GBM (G) and the tubular basement membranes (T) as well as in the hematopoietic tissue (HP) (D).
Comparison of the Goodpasture epitope sequences within the α 3(IV)NC1 among different organisms
Goodpasture autoantibodies bind to human and chicken α3(IV)NC1 domains, but do not bind to the Danio rerio α3(IV)NC1 domain. In order to explain observed differences in Goodpasture autoantibody binding, we compared amino acid sequences at the 2 putative B-cell Goodpasture epitope regions, EA and EB, among different species (Tables 1, 2). The Danio rerio and chicken sequence analysis reveals that while most amino acids are conserved, some critical changes can be noticed, which are further preserved in the human sequence. Our results demonstrate that Goodpasture antibody binding at epitope EA (Table 1) on the human α3(IV)NC1 domain requires conservation of threonine-17 (T17) and the presence of serine-21 (S21). Binding at epitope EB (Table 2) requires threonine-127 (T127), loss of asparagine-128 (N128), substitution of proline or alanine for glutamine-131(Q131), and lysine-141 (K141). This analysis is the first to show that T127 and K141 are important residues for antibody binding at Goodpasture epitope EB.
Sequence comparison of the putative Goodpasture epitope EA
Species/chain . | GP+/– . | Epitope EA sequence . |
|---|---|---|
| Human α1 NC1 | – | I D D P Q C P S G T K I L Y H |
| Human α3 NC1 | + | T A I P S C P E G T V P L Y S |
| Bovine 3 NC1 | + | T A I - S - - E - - E P - - S |
| Rattus α3 NC1 | + | T A N - S - - E - - Q P - - S |
| Mus α3 NC1 | + | T A I - S - - E - - Q P - - S |
| Gallus α3 NC1 | + | T K I - S - - E - - S Q - - F |
| Xenopus α3 NC1 | + | Missing/unknown |
| Danio rerio α3 NC1 | – | T V I - E - - A - - K R - - T |
| Drosophila α1 NC1 | – | E T V - A - S A - H T E - W T |
| C elegans α1 NC1 | – | T A V - Q - - P - - S Q - W E |
| Residue number | 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
Species/chain . | GP+/– . | Epitope EA sequence . |
|---|---|---|
| Human α1 NC1 | – | I D D P Q C P S G T K I L Y H |
| Human α3 NC1 | + | T A I P S C P E G T V P L Y S |
| Bovine 3 NC1 | + | T A I - S - - E - - E P - - S |
| Rattus α3 NC1 | + | T A N - S - - E - - Q P - - S |
| Mus α3 NC1 | + | T A I - S - - E - - Q P - - S |
| Gallus α3 NC1 | + | T K I - S - - E - - S Q - - F |
| Xenopus α3 NC1 | + | Missing/unknown |
| Danio rerio α3 NC1 | – | T V I - E - - A - - K R - - T |
| Drosophila α1 NC1 | – | E T V - A - S A - H T E - W T |
| C elegans α1 NC1 | – | T A V - Q - - P - - S Q - W E |
| Residue number | 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
Dashes in sequences indicate conserved and therefore nonreactive residues identical to those found in both human α1 and human α3 NC1 domains. Residue numbers are based on human α3(IV)NC1 residue positions.
Sequence comparison of the putative Goodpasture epitope EB
Species/chain . | GP+/– . | Epitope EB sequence . |
|---|---|---|
| Human α1 NC1 | – | I Q I P P C P S G W S S L W I |
| Human α3 NC1 | + | T D I P P C P H G W I S W L K |
| Bovine α3 NC1 | + | T D - - - - - A - - I - - - K |
| Rattus α3 NC1 | + | T A - - - - - Q - - V - - - K |
| Mus α3 NC1 | + | T A - - - - - Q D - V - - - K |
| Gallus α3 NC1 | + | T A V - A - - G - - I - - - K |
| Xenopus α3 NC1 | + | Missing/unknown |
| Danio rerio α3 NC1 | – | I N - - Q - - V - - L - - - E |
| Drosophila α1 NC1 | – | I E V - D - - N - - E G - - I |
| C elegans α1 NC1 | – | T S V - Q - - Q - - S G M - T |
| Residue number | 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 |
Species/chain . | GP+/– . | Epitope EB sequence . |
|---|---|---|
| Human α1 NC1 | – | I Q I P P C P S G W S S L W I |
| Human α3 NC1 | + | T D I P P C P H G W I S W L K |
| Bovine α3 NC1 | + | T D - - - - - A - - I - - - K |
| Rattus α3 NC1 | + | T A - - - - - Q - - V - - - K |
| Mus α3 NC1 | + | T A - - - - - Q D - V - - - K |
| Gallus α3 NC1 | + | T A V - A - - G - - I - - - K |
| Xenopus α3 NC1 | + | Missing/unknown |
| Danio rerio α3 NC1 | – | I N - - Q - - V - - L - - - E |
| Drosophila α1 NC1 | – | I E V - D - - N - - E G - - I |
| C elegans α1 NC1 | – | T S V - Q - - Q - - S G M - T |
| Residue number | 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 |
Dashes in sequences indicate conserved and therefore nonreactive residues identical to those found in both human α1 and human α3 NC1 domains. Residue numbers are based on human α3(IV)NC1 residue positions.
Interestingly, while many species are positive for Goodpasture autoantibody binding, they do not contain all of the amino acids previously determined to be critical for such activity by various chimeric protein and site-directed mutagenesis studies.15-18,31-32 Amino acids thought to be crucial for binding at the epitope EA are T17, A18, I19, P20, V27, P28, and S31.31-32 Among all of the amino acids determined to be critical utilizing a chimeric protein approach, our results are in agreement with T17 of epitope EA.32 Previous work demonstrates that although necessary, this threonine is not sufficient for autoantibody recognition.17,32 Previous work also reinforces the importance of S21. By altering this residue in a chimeric construct, autoantibody recognition by 100% of patient serum samples decreases to 25%.17 At epitope EB, similar mutational analysis has not been described.
All of the α3(IV) sequences, whether positive or negative for Goodpasture sera binding, share the following sequence homology in epitope EA—TxxPxCPxGTxxLYx, (Table 1). Cross-species conservation indicates that this sequence motif is evolutionarily preserved for structure or function. A similar conserved sequence motif, TxxPxCPxGWxSLWx (Table 2), exists in epitope EB, regardless of Goodpasture antibody binding. The only exceptions to this sequence conservation are isoleucine (I) substituted for the N-terminal threonine (T) in Danio rerio, and aspartate (D) substituted for the middle glycine (G) in mouse. Consistent with epitope EA, conservation of the EB sequence motif indicates evolutionary preservation of the sequence for NC1 domain structure or function, and underscores the importance of the intervening nonconserved residues (x) within these conserved sequence motifs for antigenic recognition by Goodpasture autoantibodies directed against epitope EA and EB. The conserved EB sequence is distinguished from EA by tryptophan (W) residues (W136 and W140), rather than threonine (T) and tyrosine (Y), and one additionally conserved C-terminal serine residue (S138).
Molecular modeling for 3D analysis of the human, chicken, and Danio rerio α 3(IV)NC1 domains
In order to spatially evaluate charge, hydropathy and side-chain polarity of the epitope EA and EB sequence in the context of the entire NC1 domain, we used molecular modeling software and the existing human type IV collagen NC1 domain crystal structure to generate models (Figure 4A-G). From crystallographic data it has been shown that the α1(IV)NC1 and α2(IV)NC1 chains are folded virtually identically, and their topologies very closely resemble one another.37 Based on the high sequence identity between all type IV collagen NC1 domains, the α3(IV)NC1 domain is expected to fold with the same topology as the α1(IV)NC1.37 Therefore, by aligning the α3(IV)NC1 and α1(IV)NC1 sequences and using the known [(α1)2α2]2 NC1 hexamer structure (Protein Data Base ID 1LI1), we have modeled amino acids that are divergent in the Danio rerio, chicken, and human EA and EB epitope regions on a backbone homology α3(IV)NC1 structure (Figure 4B-D). Amino acid differences in epitope EA (blue) and EB (green) regions in chicken compared to Danio rerio are indicated by side chain rendering in yellow, while further amino acid differences in human compared to chicken are shown by side chain rendering in red (Figure 4E-G).
Species-specific sequence changes in the emergent EA and EB epitope regions of the α3(IV)NC1 domain. (A) Alignment of epitopes and EA and EB of Danio rerio, chicken, and human. The 6 variable (nonscaffold) residues of epitope EA and the 5 variable (nonscaffold) residues of EB are numbered. The color coding of residues (which is identical to that in panels B-D) denotes the chemical properties of residues, and is explained in the colored key at right. The numbered keys delineate the emergence of these epitopes as they arose from immunonegative Danio rerio to immunopositive chicken and human by defining the evolutionarily induced changes in chemical properties and their effects on Goodpasture autoantibody binding. (B-G) Using the known [(α1)2α2]2 NC1 hexamer crystal structure (PDB ID 1LI1), we have modeled the species-specific amino acids in the EA and EB regions for Danio rerio (B,E), chicken (C, F) and human (D,G) onto a α3(IV)NC1 backbone-homology structure. (B-D) The molecular surface and amino acid character of nonconserved residues in both the EA and EB epitope regions for Danio rerio (B), chicken (C), and human (D) are rendered in a color-coded, semitransparent surface representation: neutral (gray), strongly hydrophilic (green), moderately hydrophilic (cyan), basic (blue), and acidic (red) amino acid character. The remainder of the α3(IV)NC1 domain backbone is shown in ribbon representation (gray). (E-G) The peptide backbone from residues 17-31 (using human α3(IV) numbering) in the α3(IV) chain corresponding to the EA epitope region is colored blue, and from residues 127-141 corresponding to the EB epitope region is colored green. Nonconserved residues in EA and EB are shown with rendered side chains, and identified by single-letter residue code. Sequence differences between the Danio rerio and chicken EA and EB epitopes are highlighted in yellow on the chicken homology α3(IV)NC1 domain structure. Additionally, sequence differences between the chicken and human EA and EB epitope regions are highlighted further in red on the human homology α3(IV)NC1 domain structure. The Q57 critical in forming the conformational, discontinuous epitope EA is colored magenta.
Species-specific sequence changes in the emergent EA and EB epitope regions of the α3(IV)NC1 domain. (A) Alignment of epitopes and EA and EB of Danio rerio, chicken, and human. The 6 variable (nonscaffold) residues of epitope EA and the 5 variable (nonscaffold) residues of EB are numbered. The color coding of residues (which is identical to that in panels B-D) denotes the chemical properties of residues, and is explained in the colored key at right. The numbered keys delineate the emergence of these epitopes as they arose from immunonegative Danio rerio to immunopositive chicken and human by defining the evolutionarily induced changes in chemical properties and their effects on Goodpasture autoantibody binding. (B-G) Using the known [(α1)2α2]2 NC1 hexamer crystal structure (PDB ID 1LI1), we have modeled the species-specific amino acids in the EA and EB regions for Danio rerio (B,E), chicken (C, F) and human (D,G) onto a α3(IV)NC1 backbone-homology structure. (B-D) The molecular surface and amino acid character of nonconserved residues in both the EA and EB epitope regions for Danio rerio (B), chicken (C), and human (D) are rendered in a color-coded, semitransparent surface representation: neutral (gray), strongly hydrophilic (green), moderately hydrophilic (cyan), basic (blue), and acidic (red) amino acid character. The remainder of the α3(IV)NC1 domain backbone is shown in ribbon representation (gray). (E-G) The peptide backbone from residues 17-31 (using human α3(IV) numbering) in the α3(IV) chain corresponding to the EA epitope region is colored blue, and from residues 127-141 corresponding to the EB epitope region is colored green. Nonconserved residues in EA and EB are shown with rendered side chains, and identified by single-letter residue code. Sequence differences between the Danio rerio and chicken EA and EB epitopes are highlighted in yellow on the chicken homology α3(IV)NC1 domain structure. Additionally, sequence differences between the chicken and human EA and EB epitope regions are highlighted further in red on the human homology α3(IV)NC1 domain structure. The Q57 critical in forming the conformational, discontinuous epitope EA is colored magenta.
We found that in the context of the 3-dimensional (3D) α3(IV)NC1 domain homology structures, residues that we identified as important in Goodpasture autoantibody binding at epitopes EA and EB are localized to the outer loops of the epitopes (Figure 4E-G). In addition, many of these residue changes alter electrostatic potentials in these solvent exposed epitope regions. For example, S21 and K141, described as important for epitope EA and EB binding, respectively, are glutamate residues in the immunonegative Danio rerio.
The 3D homology structures enabled us to assess conserved α3(IV) NC1 residues immediately proximal to epitope EA. Glutamine 57 (Q57) is conserved in all immunopositive proteins, but absent from all immunonegative proteins, including Danio rerio α3(IV) NC1. Surprisingly, although 26 residues away from epitope EA, in the tertiary NC1 domain structure, Q57 is immediately adjacent to epitope EA (Figure 4F-G; rendered in magenta) and provides an additional strongly hydrophilic residue. Previous work demonstrates Q57 is as important as S21 for autoantibody binding to EA. Specifically, alteration of Q57 results in 80% loss of patient sera binding17 to this discontinuous and conformational immunodominant epitope.
Our combined findings prompted us to map the amino acid character and molecular surface of species-variable residues (nonconserved) onto the modeled Danio rerio, chicken, and human EA and EB epitope regions of our backbone homology α3(IV)NC1 structures (Figure 4B-D). Using color-coded amino acid character maps, it is readily apparent that in both EA and EB electrostatic charge and polarity changes are important in Goodpasture autoantibody binding. A significant loss of electrostatic potential (positive/basic residue character) accompanies autoantibody recognition in the EA epitope region. The adjacent K27, R28 residue pair in Danio rerio is either a hydrophilic/polar residue as in chicken or hydrophobic/nonpolar residue as in human. Overall, the immunopositive chicken and human EA domains have reduced electrostatic potential and greater hydrophilic character compared with the homologous region in Danio rerio (Figure 4B-D). In contrast, the gain of a positive/basic residue in the epitope EB C-terminus is important for autoantibody binding (Figure 4A-G). K141 is 1 of 2 residue changes in epitope EB in chicken compared with Danio rerio that is present in humans; the other is T127. The substitution of threonine (T127) decreases the nonpolar character of the epitope EB N-terminus. In summary, these comparisons demonstrate that evolutionary alterations of electrostatic charge and polarity contribute to Goodpasture autoantibody binding in the α3(IV)NC1 EA and EB epitopes.
Interestingly, we found that the high hydrophobic character of both the EA and EB domains is largely conserved from Danio rerio to chicken to human. In previous studies, both V27 and P28 were shown to be important for antibody binding and critical residues of the EA autoepitope,17,31,43 through mutagenic and chimeric analysis between α1(IV)NC1 and α3(IV)NC1 sequences (in human α1(IV) these residues are lysine and isoleucine). Examination of the corresponding residues in chicken, serine and glutamine, lessens the idea that V27 and P28 are important in epitope EA for hydrophobic contributions, and suggests that the reduced antibody binding observed in mutagenic/chimeric analyses possibly reflects the effects of basic charge insertion and side-chain steric clash (Figure 4A-D). Of note, 1 variable (nonconserved) hydrophobic residue at position 131 appears important for the EB antibody recognition site (Figure 4A-D). In Danio rerio a glutamine occurs at this position, while there is a transition to alanine in chicken and proline in human. Interestingly, a nonpolar/hydrophobic residue type was conserved at position 137 (isoleucine/leucine/valine) in epitope EB between Danio rerio, chicken, and human. The immunonegative sequences of bovine α1(IV)NC1 and Drosophila and C elegans α1(IV)NC1 and α2(IV)NC1 contain glutamate or serine at position 137. This finding suggests that a nonpolar/hydrophobic residue at position 137 may be involved in autoantibody binding, but is not sufficient in the context of the Danio rerio EB region to impart antibody binding.
Discussion
Our results demonstrate that there has been an evolutionary pressure to obtain the α3 chain of type IV collagen in the Danio rerio. This could be linked to the required homeostasis and physiology of vertebrates to exercise greater regulation of fluid and gas exchanges. Interestingly, the Goodpasture epitope is not present within the α3(IV)NC1 domain of Danio rerio. The emergence of this epitope (in frogs) coincides with the transfer from aquatic to terrestrial living. Last, our study demonstrates the importance of Danio rerio in elucidating the evolutionary changes within the α3(IV)NC1 domain, which is the only species to possess an α3 chain of type IV collagen not recognized by Goodpasture autoantibodies. This insight into the critical amino acids required for binding autoantibodies will potentially lead to cell-mediated therapies to impart systemic tolerance to the pathogenic epitopes that cause this devastating disease. Future studies will hopefully shed more light on the physiologic need for such modification in the α3 chain of type IV collagen.
Prepublished online as Blood First Edition Paper, October 27, 2005; DOI 10.1182/blood-2005-05-1814.
Supported by National Institutes of Health (NIH) grants DK55001 and DK62987, and funds from the Center for Matrix Biology at the Beth Israel Deaconess Medical Center. M.S. is funded by the Sigrid Juselius Foundation, the Maud Kuistila Foundation, the Finnish Medical Society Duodecim, and the Emil Aaltonen Foundation. M.A.G. is funded by American Heart Association grant 05030348N. L.I.Z. is supported by the NIH and the Howard Hughes Medical Institute (HHMI). K.L.P. is supported by the Albert J. Ryan Fellowship.
R.K., B.A.M., M.S., and M.A.G. designed research; B.A.M., M.S., M.A.G., and K.H. performed research; K.L.P. and L.I.Z. contributed vital new reagents; R.K., B.A.M., M.S., and M.A.G. analyzed data; and R.K., B.A.M., and M.S. wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Ramani Ramchandran for his initial help in searching the Danio rerio genomic databases for type IV collagen genes.



![Figure 4. Species-specific sequence changes in the emergent EA and EB epitope regions of the α3(IV)NC1 domain. (A) Alignment of epitopes and EA and EB of Danio rerio, chicken, and human. The 6 variable (nonscaffold) residues of epitope EA and the 5 variable (nonscaffold) residues of EB are numbered. The color coding of residues (which is identical to that in panels B-D) denotes the chemical properties of residues, and is explained in the colored key at right. The numbered keys delineate the emergence of these epitopes as they arose from immunonegative Danio rerio to immunopositive chicken and human by defining the evolutionarily induced changes in chemical properties and their effects on Goodpasture autoantibody binding. (B-G) Using the known [(α1)2α2]2 NC1 hexamer crystal structure (PDB ID 1LI1), we have modeled the species-specific amino acids in the EA and EB regions for Danio rerio (B,E), chicken (C, F) and human (D,G) onto a α3(IV)NC1 backbone-homology structure. (B-D) The molecular surface and amino acid character of nonconserved residues in both the EA and EB epitope regions for Danio rerio (B), chicken (C), and human (D) are rendered in a color-coded, semitransparent surface representation: neutral (gray), strongly hydrophilic (green), moderately hydrophilic (cyan), basic (blue), and acidic (red) amino acid character. The remainder of the α3(IV)NC1 domain backbone is shown in ribbon representation (gray). (E-G) The peptide backbone from residues 17-31 (using human α3(IV) numbering) in the α3(IV) chain corresponding to the EA epitope region is colored blue, and from residues 127-141 corresponding to the EB epitope region is colored green. Nonconserved residues in EA and EB are shown with rendered side chains, and identified by single-letter residue code. Sequence differences between the Danio rerio and chicken EA and EB epitopes are highlighted in yellow on the chicken homology α3(IV)NC1 domain structure. Additionally, sequence differences between the chicken and human EA and EB epitope regions are highlighted further in red on the human homology α3(IV)NC1 domain structure. The Q57 critical in forming the conformational, discontinuous epitope EA is colored magenta.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/5/10.1182_blood-2005-05-1814/2/m_zh80050691760004.jpeg?Expires=1769305964&Signature=Na7wp7rMxW4lM1G3Yx86ROr~TxaELqe0kMtB9hrqVIAQmJUXgku9Z14b1yi1b78pA-bfuJtyvQjQunlcxwLuU~iFXxacwpsPBrL~~hWC7P0WB55XzMoBZffZE1OfRZ7sCd9pCaQEna8Idc52dMIQEHVgK2WHM~JgrJqaXpqQ5StMRvK4Ls5Z1oadb9~UokUosYUkUoFxRNNxmYAo4~upeLfk3jMj9DCEygH~~PCYC9iAp6PuVz6CnjJN9v7NmwyKMgXVl90dJh-7oKlYzYZg9uy~Xacqt0EP6QTcBSiZ9P6Ga6ctUL2pXthcQh9B17N2NnoZzklX4ZNfJea99UhJug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal