Genetic studies in mouse and zebrafish have established the importance of activin receptor-like kinase 1 (ALK1) in formation and remodeling of blood vessels. Single-allele mutations in the ALK1 gene have been linked to the human type 2 hereditary hemorrhagic telangiectasia (HHT2). However, how these ALK1 mutations contribute to this disorder remains unclear. To explore the mechanism underlying effect of the HHT-related ALK1 mutations on receptor activity, we generated 11 such mutants and investigated their signaling activities using reporter assay in mammalian cells and examined their effect on zebrafish embryogenesis. Here we show that some of the HHT2-related mutations generate a dominant-negative effect whereas the others give rise to a null phenotype via loss of protein expression or receptor activity. These data indicate that loss-of-function mutations in a single allele of the ALK1 locus are sufficient to contribute to defects in maintaining endothelial integrity.
Introduction
Hereditary hemorrhagic telangiectasia (HHT), or Osler-Rendu-Weber syndrome, is an autosomal dominant vascular dysplasia with characteristics of nasal and gastrointestinal hemorrhage. This vascular malformation is reminiscent of defects in vascular remodeling and maturation.1 Type 2 HHT (HHT2) is caused by mutations in the gene encoding ALK1 (activin receptor-like kinase 1),2 which is a type I receptor for transforming growth factor-β (TGF-β) and is predominantly expressed in endothelial cells.3 ALK1 is activated by the TGF-β type II receptor (TβRII) in the presence of TGF-β, and transduces its signal via Smad1/5.4-7 Targeted disruption of the Alk1 gene in mice results in embryonic lethality owing to abnormal vascular development, including excessive fusion of capillary plexes into cavernous vessels and hyperdilation of large vessels.6,8 Similarly, the homozygous mutation of zebrafish ALK1-violet beauregarde (vbg) leads to an abnormal circulation with dilated cranial vessels.9 These results strongly suggest an essential role for ALK1 in the establishment and maintenance of blood vessel integrity.
More than 100 different mutations, including frame-shift, nonsense, and missense mutations, have been identified in the ALK1 gene from patients with HHT2. These mutations are distributed throughout the protein, though most (about 70%) cluster in the intracellular kinase domain.10-14 How these mutations affect ALK1 function is poorly understood. To elucidate the mechanism underlying the contribution of ALK1 mutations to HHT, we constructed 11 ALK1 mutants and analyzed their activities in vitro and in vivo. Our results revealed that HHT mutations of ALK1 can cause either loss-of-function or dominant-negative effect, suggesting that the reduced level of functional ALK1 protein may contribute to the vascular malfunction related to HHT.
Study design
DNA constructs
ALK1 mutants were generated through site-directed mutagenesis by polymerase chain reaction (PCR) using appropriate primers with desired mutation and were subcloned into mammalian expression vector pCMV5. The validity of all mutations was confirmed by DNA sequencing. All the constructs were tagged at the C-terminus with the hemagglutinin (HA) epitope.
Cell lines and transfection
HEK293T and COS7 cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and Hep3B cells were maintained in MEM with 10% fetal bovine serum in 5% CO2 at 37°C. Transfection was performed with calcium phosphate for HEK293T cells or using LipofectAMINE (Invitrogen, Carlsbad, CA) for Hep3B and COS7 cells.
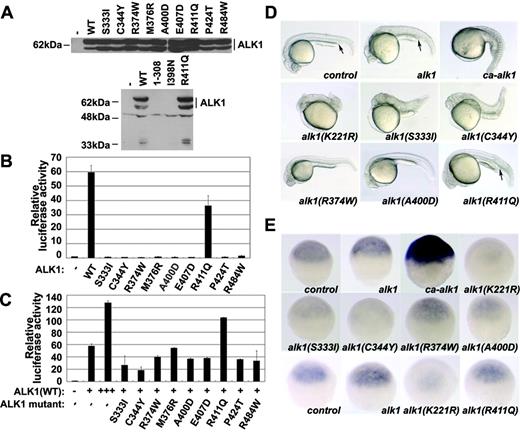
HHT2-related ALK1 mutants are defective in transcriptional activation and dorsoventral patterning of zebrafish embryos. (A) Plasmids encoding HA-tagged wild-type or mutant ALK1 were transfected into HEK293T cells (2 μg each plasmid per 60 mm dish of cells). Forty hours after transfection, the cells were harvested, and the protein expression levels of ALK1 were examined by immunoblotting with an anti-HA antibody. (B) Hep3B cells were transiently transfected with BRE4-luciferase (0.5 μg) and the ALK1 constructs (0.03 μg each) as indicated. Forty hours later, the cells were harvested and luciferase activity was measured. (C) Wild-type (+: 0.03 μg; +++: 0.09 μg) and individual mutant ALK1 (0.06 μg) were cotransfected into Hep3B cells together with the BRE4-luciferase reporter plasmid (0.5 μg) as indicated and the luciferase activity was measured 40 hours later. Reporter assays were performed in triplicate in the absence of TGFβ, and Renilla (20 ng) was cotransfected as an internal control. The data represent the mean plus or minus the standard deviation after normalization to Renilla activity. (D) mRNA encoding wild-type or various mutant ALK1 was injected into one-cell embryos with 1 ng per embryo. Because of severe embryo lethality, a lower amount of ca-ALK1 mRNA (150 pg) was used. The phenotype was observed at 24 hpf. The embryos shown are lateral views, with anterior to the left. The arrows indicate the intermediate cell mass. Control: no injection. (E) Expression of ventral ectoderm marker gata2 was examined by in situ hybridization at the shield stage. The embryos shown are ventral views with the animal pole at the top. Note that the ventralized embryos exhibit increased expression of gata2 (ca-alk1) whereas the dorsalized embryos show a reduction of gata2 expression (alk1(K221R)). Control: no injection.
HHT2-related ALK1 mutants are defective in transcriptional activation and dorsoventral patterning of zebrafish embryos. (A) Plasmids encoding HA-tagged wild-type or mutant ALK1 were transfected into HEK293T cells (2 μg each plasmid per 60 mm dish of cells). Forty hours after transfection, the cells were harvested, and the protein expression levels of ALK1 were examined by immunoblotting with an anti-HA antibody. (B) Hep3B cells were transiently transfected with BRE4-luciferase (0.5 μg) and the ALK1 constructs (0.03 μg each) as indicated. Forty hours later, the cells were harvested and luciferase activity was measured. (C) Wild-type (+: 0.03 μg; +++: 0.09 μg) and individual mutant ALK1 (0.06 μg) were cotransfected into Hep3B cells together with the BRE4-luciferase reporter plasmid (0.5 μg) as indicated and the luciferase activity was measured 40 hours later. Reporter assays were performed in triplicate in the absence of TGFβ, and Renilla (20 ng) was cotransfected as an internal control. The data represent the mean plus or minus the standard deviation after normalization to Renilla activity. (D) mRNA encoding wild-type or various mutant ALK1 was injected into one-cell embryos with 1 ng per embryo. Because of severe embryo lethality, a lower amount of ca-ALK1 mRNA (150 pg) was used. The phenotype was observed at 24 hpf. The embryos shown are lateral views, with anterior to the left. The arrows indicate the intermediate cell mass. Control: no injection. (E) Expression of ventral ectoderm marker gata2 was examined by in situ hybridization at the shield stage. The embryos shown are ventral views with the animal pole at the top. Note that the ventralized embryos exhibit increased expression of gata2 (ca-alk1) whereas the dorsalized embryos show a reduction of gata2 expression (alk1(K221R)). Control: no injection.
Luciferase assay
Forty hours after transfection, cells were harvested for determination of luciferase activity with a luminometer (Berthold Technologies, Baden-Wuerttenberg, Germany), as described previously.7 All assays were performed in triplicate.
Biotinylation of cell-surface receptors
Cells transfected with HA-tagged receptors were biotinylated with 0.5 mg/mL NHS-SS-biotin (Pierce, Rockford, IL) for 40 minutes at 4°C, as described previously.15 Biotinylated surface receptors were precipitated with ImmunoPure Streptavidin beads (Pierce) and analyzed by anti-HA immunoblotting.
Immunoprecipitation and immunoblotting
Forty-eight hours after transfection with the appropriate plasmids, HEK293T cells were washed once with DMEM containing 0.2% FBS and incubated in the presence of 200 pM TGFβ1 for 30 minutes. Protein complexes containing Flag-tagged Smad1(ΔCT) were immunoprecipitated with anti-Flag monoclonal antibody M2 (Sigma, St Louis, MO) and protein A-Sepharose beads (Zymed, San Francisco, CA). Protein complexes containing His-tagged TβRII were precipitated with Ni-NTA beads (Qiagen, Valencia, CA). The precipitated protein complexes were analyzed by immunoblotting with anti-HA, anti-Flag, or anti-His antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
For the Smad1 phosphorylation assay, transfected COS7 cells were harvested and phosphorylated Smad1 and total Smad1 proteins were detected by anti–phospho-Smad1 antibody (Chemicon, Temecula, CA) and anti-Smad1 antibody (Santa Cruz Biotechnology), respectively.
In vitro synthesis of mRNA, microinjection, and whole-mount in situ hybridization
Messenger RNA generated using the T7 Cap-Scribe Kit (Roche, Milan, Italy) was injected into one-cell stage zebrafish embryos (1 ng ALK1 mRNA in a volume of 0.003 μL per embryo, except for ca-ALK1, for which 150 pg mRNA was used due to severe embryo lethality). Digoxigenin–uridine 5′-triphosphate (UTP)–labeled antisense RNA probes were generated by in vitro transcription. The probes for eve1 and gata2 were described by Joly et al16 and Detrich et al,17 respectively. Whole-mount in situ hybridization followed the standard protocol with minor modifications.18 The live zebrafish were placed in 6% methylcellulose solution (Sigma, St Louis, MO); images were obtained at an original magnification of × 80 by using a 8 ×/0.14 numeric aperture (NA) Planapo objective lens. In situ–hybridized embryos were stained with purple alkaline phosphatase substrate (Roche, Milan, Italy) and placed in glycerol; images were obtained at an original magnification of × 110 by using a 11 ×/0.14 NA Planapo objective lens. All images were visualized using a Leica MZ16 microscope (Leica, Heidelberg, Germany) equipped with a Spot Insight 3.2.0 color camera (Diagnostic Instruments, Sterling Heights, MI). Images were processed with Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Effect of HHT2-related ALK1 mutations on the cell surface distribution of the receptor and its ability to activate Smad1. (A) HEK293T cells transfected with plasmids encoding HA-tagged wild-type or mutant ALK1 and His-tagged TβRII were treated with 200 pM TGFβ1 for 30 minutes. Protein complexes were isolated from the cell lysates by precipitation with Ni-NTA beads and detected by immunoblotting (IB) with anti-HA antibodies (top panel). Protein expression was confirmed by immunoblotting with anti-HA and anti-His antibodies (middle and bottom panels). (B) HEK293T cells transfected with plasmids encoding HA-tagged wild-type or mutant ALK1 and Flag-tagged Smad1 lacking the C-terminal 8 amino acids (shown to associate with active receptor7 ), were treated with 200 pM TGFβ1 for 30 minutes. Protein complexes were isolated from the cell lysates by immunoprecipitation (IP) with an anti-Flag antibody and detected by immunoblotting with an anti-HA antibody (top panel). Protein expression was confirmed by immunoblotting with anti-HA and anti-Flag antibodies (middle and bottom panels). (C) Cells transfected with HA-tagged wild-type or mutant ALK1 were biotinylated with 0.5 mg/mL NHS-SS-biotin for 40 minutes at 4°C. Biotinylated surface receptors were precipitated with ImmunoPure Streptavidin beads and analyzed by anti-HA immunoblotting. (D) COS7 cells transfected with the plasmids encoding HA-tagged ALK1 wild-type, ALK1 mutants, Flag-tagged Smad1, and His-tagged TβRII were treated with 200 pM TGFβ1 for 30 minutes. The cells were subsequently lysed and the phosphorylated Smad1 was detected by immunoblotting (IB) with anti–phospho-Smad1 antibodies (upper panel). Total protein expression was confirmed by immunoblotting with anti-Smad1 and anti-HA antibodies (middle and bottom panels). The active form of BMP type I receptor BMPRIB(QD) was used as a positive control for induction of Smad1 phosphorylation.
Effect of HHT2-related ALK1 mutations on the cell surface distribution of the receptor and its ability to activate Smad1. (A) HEK293T cells transfected with plasmids encoding HA-tagged wild-type or mutant ALK1 and His-tagged TβRII were treated with 200 pM TGFβ1 for 30 minutes. Protein complexes were isolated from the cell lysates by precipitation with Ni-NTA beads and detected by immunoblotting (IB) with anti-HA antibodies (top panel). Protein expression was confirmed by immunoblotting with anti-HA and anti-His antibodies (middle and bottom panels). (B) HEK293T cells transfected with plasmids encoding HA-tagged wild-type or mutant ALK1 and Flag-tagged Smad1 lacking the C-terminal 8 amino acids (shown to associate with active receptor7 ), were treated with 200 pM TGFβ1 for 30 minutes. Protein complexes were isolated from the cell lysates by immunoprecipitation (IP) with an anti-Flag antibody and detected by immunoblotting with an anti-HA antibody (top panel). Protein expression was confirmed by immunoblotting with anti-HA and anti-Flag antibodies (middle and bottom panels). (C) Cells transfected with HA-tagged wild-type or mutant ALK1 were biotinylated with 0.5 mg/mL NHS-SS-biotin for 40 minutes at 4°C. Biotinylated surface receptors were precipitated with ImmunoPure Streptavidin beads and analyzed by anti-HA immunoblotting. (D) COS7 cells transfected with the plasmids encoding HA-tagged ALK1 wild-type, ALK1 mutants, Flag-tagged Smad1, and His-tagged TβRII were treated with 200 pM TGFβ1 for 30 minutes. The cells were subsequently lysed and the phosphorylated Smad1 was detected by immunoblotting (IB) with anti–phospho-Smad1 antibodies (upper panel). Total protein expression was confirmed by immunoblotting with anti-Smad1 and anti-HA antibodies (middle and bottom panels). The active form of BMP type I receptor BMPRIB(QD) was used as a positive control for induction of Smad1 phosphorylation.
Results and discussion
To investigate the effect of HHT2-related ALK1 mutations on receptor activity, we generated 11 mutants, which represent the human mutations in the intracellular kinase domain.19-23 The residues S333, R374, M376, and P424 are conserved in most protein kinases. The residues C344, I398, A400, E407, R411, and R484 are conserved in the TGFβ type I receptor family. ALK1(1-308) is a truncated mutant that lacks most of the kinase domain.
To determine whether these mutations affect protein expression, we transfected these ALK1 mutant constructs into HEK293T cells and detected the mutant proteins by anti-HA immunoblotting. As shown in Figure 1A, expression of both ALK1(1-308) and ALK1(I398N) was not detected, indicating that these 2 mutations affect protein expression and/or destabilize the proteins or mRNA, whereas the other mutants were expressed at levels comparable to wild-type ALK1.
Next, we investigated the effect of these mutations on ALK1 activity, using a Smad1-responsive transcriptional reporter, BRE4-luciferase.24 The reporter assay was carried out in human hepatoma Hep3B cells, which do not express ALK1 (data not shown). In the absence of exogenously added TGF-β, overexpression of wild-type ALK1 was able to stimulate the expression of BRE4-luciferase, whereas most mutants were defective in activating the reporter gene, except for ALK1(R411Q), which exhibited a reduced activity (Figure 1B). Similar results were obtained in the presence of TGF-β.
As many HHT2-related ALK1 mutations occur in only one allele of the ALK1 locus, the mutations in the coding region of the ALK1 gene could lead to generation of alleles lacking receptor activity or alleles with dominant-negative functions. To test these possibilities, we cotransfected wild-type ALK1 and individual mutants into Hep3B cells and assayed the effect of the mutants on the ability of wild-type ALK1 to activate the transcriptional reporter. As shown in Figure 1C, the mutants ALK1(S333I) and ALK1(C344Y) apparently had dominant-negative effects by suppressing the activity of wild-type ALK1. ALK1(R411Q) was able to collaborate with wild-type ALK1 to activate the reporter, consistent with the result that this mutant retains a reduced receptor activity. However, the other mutants had little or no effect on wild-type ALK1.
ALK1 is structurally and functionally conserved between humans and zebrafish.9 To further analyze the functions of these human ALK1 mutants in vivo, the mRNAs encoding wild-type and mutant ALK1 were injected into single-cell zebrafish embryos. About 30% of embryos injected with 1 ng human wild-type ALK1 mRNA exhibited a weakly ventralized phenotype with expanded intermediate cell mass (equivalent to mammalian blood islands) at 24 hours after fertilization (hpf) (Figure 1D). When injected with the mRNA expressing the constitutively active form ca-ALK1, ALK1(Q201D),7 more than 90% of embryos died even at a dose of 150 pg mRNA, and all the surviving embryos were severely ventralized and characterized by an expanded intermediate cell mass and loss of both anterior structures and the notochord (Figure 1D). This result is consistent with the previous report that overexpression of zebrafish active ALK1 induces embryo ventralization.9 Conversely, the dorsalized phenotype was observed when a kinase-dead dominant-negative form ALK1(K221R)9 was overexpressed; about 80% of the surviving embryos exhibited an enlarged head and a short tail (Figure 1D). Overexpression of ALK1(S333I) and ALK1(C344Y) caused dorsalization of 47% and 29% of the surviving embryos, respectively (Figure 1D), whereas expression of ALK1(R374W) or ALK1(A400D) had no apparent effect.
The effect of these mutants on dorsoventral patterning of zebrafish embryos was supported by examination of the expression of the ventral ectoderm marker gata2 at the shield stage (∼6 hpf). Increased expression of gata2 indicates ventralization of the embryos.17 Both ALK1(S333I) and ALK1(C344Y), like the dominant-negative ALK1(K221R), reduced gata2 expression, whereas ALK1(R374W) and ALK1(A400D) had no obvious effect (Figure 1E, upper panel). Similarly, ALK1(S333I) and ALK1(C344Y) decreased the expression of eve1,16 a ventral mesoderm marker (data not shown). Taken together, these data suggest that both ALK1(S333I) and ALK1(C344Y) function in a dominant-negative manner to induce embryonic dorsalization. Consistent with our reporter assay data, overexpression of ALK1(R411Q) led to a ventralized phenotype in about 20% of embryos (Figure 1D) and slightly increased expression of gata2 (Figure 1E, lower panel). These results suggest that ALK1(R411Q) still retains some activity.
We next attempted to investigate the mechanism by which these mutations influence ALK1 activity. As the interaction of ALK1 with TβRII and Smad1/5 is essential for ALK1 signaling, we examined the ability of ALK1 mutants to associate with TβRII and Smad1. We found that all of the expressed mutants were able to interact with TβRII and Smad1 (Figure 2A-B). To investigate whether these mutations influence receptor trafficking to the cell surface, biotinylation analysis was performed to detect the surface expression of wild-type and mutant ALK1. We selected 5 mutants for further analysis that represented 3 types of function alterations: dominant-negative effect (S333I and C344Y), loss of function (R374W and A400D), and reduced activity (R411Q). Both wild-type and R411Q were highly expressed at the cell surface, whereas the other 4 mutants, S333I, C344Y, R374W, and A400D, were expressed at lower levels (Figure 2C), indicating that these mutations may influence receptor trafficking to the cell surface. Finally, we examined the effect of these mutants on Smad1 phosphorylation. Expression of wild-type ALK1 and R411Q enhanced Smad1 phosphorylation in the presence of TGF-β, whereas expression of other mutants did not alter levels of phosphorylated Smad1 (Figure 2D), consistent with the reporter assay results shown in Figure 1B.
Taken together, our functional analyses suggest that the HHT2-linked ALK1 mutations might contribute to the vascular disorder through different mechanisms of influencing ALK1 activity. Some mutations, such as S333I and C344Y, generate a dominant-negative effect, whereas the others give a null phenotype through loss of protein expression or function. However, it is unclear how R411Q contributes to HHT2 phenotypes, as ALK1(R411Q) still retains certain activity. It is possible that the reduced activity is sufficient to cause the disease phenotypes or that there exists another functional mutation in these patients. Because of limited information about the patients with HHT2 carrying these ALK1 mutations, it is difficult to correlate the types of alterations in ALK1 functions with the severity of HHT disease phenotypes. Nevertheless, these data strongly suggest that proper ALK1 signaling activity is required for its normal functions in endothelial cells, and its haploinsufficiency or dominant-negative effect resulting from the mutations of a single allele of the ALK1 locus may contribute to defects in vascular remodeling and integrity maintenance.
Prepublished online as Blood First Edition Paper, November 10, 2005; DOI 10.1182/blood-2005-05-1834.
Supported by grants from the Bugher Foundation (New York), the National Science Foundation of China (grant nos. 30 125 021, 30 270 681, 30 430 360), and the 973 Program (2004CB720 002).
Y.G. performed most of the experiments; P.J. performed the embryo assays with Y.G.; L.Z. performed the Smad1 phosphorylation experiments; X.Z. provided assistance with the reporter assays; X.G. assisted with the Smad1 phosphorylation experiments; Y.N. helped generate the mutant constructs; A.M. assisted with data analysis and discussion; and Y.-G.C. was involved in experimental design, data analysis, and manuscript writing.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Brant M. Weinstein for ALK1(K221R) plasmid, and Drs Aaron Gitler, Jing-Wei Xiong, and Jing-Dong Han for critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal