Abstract
Hodgkin/Reed-Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL) display unique characteristics that discriminate cHL from other B-cell lymphomas and normal B cells. Therefore, comparative gene expression profiling of Hodgkin and non-Hodgkin B cells could lead to the identification of candidate genes that are critical for the pathogenesis of cHL. We performed microarray analysis of Hodgkin and non-Hodgkin cell lines and identified activating transcription factor 3 (ATF3), a member of the cyclic AMP response element binding protein (CREB)/ATF family, as a differentially expressed candidate gene. Extensive analysis of a large panel of cell lines, primary tumor samples, and normal tissues revealed that high expression of ATF3 is found in nearly all cases of cHL and is almost exclusively restricted to it. Selective knock-down of ATF3 by RNA interference suppressed proliferation and strongly reduced viability of Hodgkin cells. Thus, overexpression of ATF3 is a molecular hallmark of cHL that contributes to the malignant growth of HRS cells. (Blood. 2006;107:2536-2539)
Introduction
Classical Hodgkin lymphoma (cHL) is a common malignant lymphoma characterized by the presence of typical malignant cells, the mononuclear Hodgkin and multinuclear Reed-Sternberg (HRS) cells.1 HRS cells, which represent only a minor cellular fraction of the affected lymphoid tissue, are thought to be derived from germinal or postgerminal center B cells.2,3 Although the precise molecular mechanisms underlying the malignant transformation of HRS cells are only partially understood, several observations indicate that defects in transcriptional regulation play a pivotal role in the pathogenesis of cHL. In line with this idea, we have previously identified high constitutive activity of the transcription factor nuclear factor (NF)-κB as a common characteristic of HRS cells that contributes to their survival and proliferation.4,5 In nonmalignant cells, NF-κB is only transiently activated, particularly by stress signals and in immune and inflammatory signaling events.6 Another hallmark of cHL is the aberrant expression of the AP-1 transcription factor family members c-Jun and JunB,7 which are usually activated in response to mitogenic stimuli and cellular stress signals.
HRS cells display unique morphologic and molecular characteristics that clearly discriminate cHL from B-cell non-Hodgkin lymphomas and normal B cells.8 Therefore, gene expression profiling is a promising approach to identify genes that contribute to malignant growth and survival in cHL. We performed oligonucleotide microarray analysis of Hodgkin and non-Hodgkin cell lines and identified activating transcription factor 3 (ATF3) as a differentially expressed candidate gene with high expression in HRS cells. ATF3 is a member of the ATF/CREB family of basic region leucine zipper transcription factors, which is involved in the cellular stress response.9 ATF3 expression is strongly induced by cellular damage in a variety of organ systems, including liver, heart, and neurons as well as in cultured cells following exposure to UV light, ionizing radiation, proteasome inhibitors, or interference with protein synthesis. Furthermore, ATF3 has been reported to be associated with cellular proliferation and survival.10-12 We therefore decided to analyze the role of ATF3 in cHL in more detail.
Study design
Cell lines, electroporation, and purification of transfected cells
Cell lines, culture conditions, and electroporation have been described previously.7 Where indicated, an additional purification step following transfection was included, employing either fluorescence-activated cell sorting (FACS) or magnetic cell sorting (MACS) as detailed in Mathas et al7 and Chatterjee et al.13
Oligonucleotide microarray analysis
RNA processing and hybridization to U95A GeneChip microarrays was performed according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Primary data files were analyzed with the Microarray Suite software (Affymetrix). Hierarchical clustering analysis was carried out with the Cluster and TreeView software (M. Eisen, Berkeley, CA; http://rana.lbl.gov/EisenSoftware.htm).
Northern and Western blot analysis
Northern and Western blotting were performed according to standard protocols as described previously.7 The following primary antibodies were used: anti-ATF3 (sc-188; Santa Cruz Biotechnology, Santa Cruz, CA), anti-E2F-4 (clone 4E2F04; Neomarkers, Fremont, CA), and anti-α-tubulin (MCA78A; Serotec, München, Germany).
Immunohistochemistry
All cases were retrieved from the files of the Institute of Pathology, Charité, Campus Benjamin Franklin, Berlin, Germany. Diagnosis was established after histologic and immunohistologic analyses according to the criteria of the World Health Organization classification. ATF3 immunohistochemistry was performed with a polyclonal rabbit anti-ATF3 antibody (sc-188; Santa Cruz Biotechnology) after heat pretreatment of the deparaffinized tissue sections in a pressure cooker for 2 minutes in citrate buffer (10 mM). Subsequently, a mouse anti-rabbit antibody (DakoCytomation, Glostrup, Denmark) was applied followed by development employing alkaline phosphatase and fast red as substrate. Approximately two-thirds of the tissue sections were arranged in tissue microarrays, whereas the remaining cases have been stained as single tissue sections.
Construction of siRNA expression plasmids
Vectors for transient expression of small interfering (si) RNAs were derived from the pSUPER plasmid according to the guidelines described in Brummelkamp et al.14 The episomal plasmid vector pRep-H1 (M.T., Uschi Luz, Reinhold Schäfer, and C.H., manuscript in preparation) was constructed by insertion of an H1-promoter-based PolIII transcription unit into the vector pTIP.HA1 × 3.EGFP that contains a puromycin selectable marker, a truncated EBNA gene, and an Epstein-Barr virus (EBV) oriP sequence. The following target sequences were chosen: EGFP 5′-GCAGCACGACTTCTTCAAG-3′, Luc (directed against firefly luciferase; pGL2 vectors; Promega, Madison, WI) 5′-CGTACGCGGAATACTTCGA-3′, E2F-4 5′-GCGGCGGATTTACGACATT-3′, ATF3_1 5′-GAGCTGAGGTTTGCCATCC-3′, ATF3_2 5′-GAGGCGACGAGAAAGAAAT-3′, and ATF3_3 5′-GAAGAAGGAGAAGACGGAG-3′.
Proliferation assay and analysis of apoptosis
DNA synthesis was determined by [3H]-thymidine incorporation assays as described in Lentzsch et al.15 To assess the percentage of viable and apoptotic cells, annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double-staining was performed using a human annexin V-FITC kit (Bender MedSystems, Eching, Germany). Cells were processed as described in the manufacturer's protocol and analyzed by flow cytometry.
Image acquisition
Images of the immunohistological stainings for ATF3 were captured with an Olympus AX70 microscope with plan-apochromatic objective lenses (4 ×/0.13 numeric aperture [NA]; 10 ×/0.40 NA; 20 ×/0.70 NA; 40 ×/0.95 NA) coupled to a digital camera (KY-F40; JVC, Friedberg, Germany). The images were stored and processed with the Diskus Program (version 4.20; Hilgers Technisches Buero, Koenigswinter, Germany). Original magnifications for Figure 1B: top row, from left to right: × 160, × 400, × 160; bottom row, from left to right: × 250, × 250, × 25.
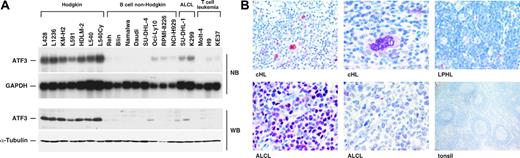
Hodgkin cell lines and primary HRS tumor cells are characterized by high constitutive ATF3 expression. (A) ATF3 mRNA and protein expression in Hodgkin and non-Hodgkin cell lines was determined by Northern (NB) analysis and Western (WB) blot analysis, respectively. As loading controls, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA and α-tubulin protein expression were included. (B) Immunostaining for ATF3 in primary tissue samples (cHL, classical Hodgkin lymphoma; LPHL, lymphocyte-predominant Hodgkin lymphoma; ALCL, anaplastic large cell lymphoma).
Hodgkin cell lines and primary HRS tumor cells are characterized by high constitutive ATF3 expression. (A) ATF3 mRNA and protein expression in Hodgkin and non-Hodgkin cell lines was determined by Northern (NB) analysis and Western (WB) blot analysis, respectively. As loading controls, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA and α-tubulin protein expression were included. (B) Immunostaining for ATF3 in primary tissue samples (cHL, classical Hodgkin lymphoma; LPHL, lymphocyte-predominant Hodgkin lymphoma; ALCL, anaplastic large cell lymphoma).
Results and discussion
To identify candidate genes that are involved in the pathogenesis of cHL, we performed oligonucleotide microarray analysis of Hodgkin and non-Hodgkin B-cell lines. Hierarchical clustering separated Hodgkin from non-Hodgkin cell lines of distinct B-cell differentiation stages. One of the most pronounced up-regulated genes in Hodgkin cell lines was the transcription factorATF3 (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). To validate the microarray data on ATF3 expression, Northern and Western blot analyses were carried out on a broad panel of cell lines. All Hodgkin cell lines demonstrated strong overexpression of ATF3 mRNA and protein compared with non-Hodgkin cell lines (Figure 1A). ATF3 expression in primary malignant and nonmalignant lymphoid tissue was determined by immunohistochemistry. A total of 415 lymphoma samples were immunostained for ATF3, comprising 71 cases of cHL and 26 cases of lymphocyte-predominant HL (LPHL) as well as 239 B-cell non-Hodgkin lymphomas, 46 T-cell non-Hodgkin lymphomas, and 33 anaplastic large cell lymphomas (ALCL). In addition, 20 specimens of reactive lymphoid tissue were included (Figure 1B and Table 1). Strong nuclear expression of ATF3 was detected in nearly all cases of cHL analyzed (69/71). NuclearATF3 staining was restricted to HRS cells and could not be detected in surrounding bystander cells (Figure 1B). In contrast, nuclear ATF3 immunoreactivity was observed in only a few samples of B- or T-cell non-Hodgkin lymphomas (6/239 and 2/46, respectively), including diffuse large B-cell lymphoma (2/80), plasmacytoma (4/20), and angioimmunoblastic T-cell lymphoma (2/10) (Table 1). In these cases, positive staining was found in only a varying, usually small proportion of the tumor cell population with generally weaker signal intensity compared with cHL and ALCL. In addition, no nuclear staining was observed in reactive lymphoid tissue (Figure 1B). Interestingly,ATF3 expression could not be demonstrated in the nuclei of LPHL tumor cells (Figure 1B). This is in agreement with the notion that cHL and LPHL represent independent entities that differ in biology and clinical course. On the other hand, besides cHL a proportion of ALCL cases demonstrated ATF3 staining (24/33; Figure 1B), which did not correlate with the expression of ALK fusion proteins (data not shown). ATF3 expression in cHL and ALCL is in line with the observation that these lymphomas share several features, including CD30 expression as well as high Notch-1 and AP-1 activity.7,16,17 It remains to be determined whether this pattern reflects common pathogenetic pathways in both lymphoma types. Taken together, our immunohistochemical analysis demonstrated that nuclear expression of ATF3 is almost exclusively restricted to HRS cells of cHL and to a fraction of ALCL.
ATF immunostaining in primary malignant and nonmalignant lymphoid tissue
. | ATF3 nuclear staining, no. cases . | . | |
|---|---|---|---|
. | Positive . | Negative . | |
| Classical Hodgkin lymphoma | 69 | 2 | |
| Lymphocyte-predominant Hodgkin lymphoma | 0 | 26 | |
| B-NHL | 6* | 233 | |
| T-NHL | 2† | 44 | |
| Anaplastic large cell lymphoma (ALCL) | 24 | 9 | |
| Reactive lymphatic tissue | 0 | 20 | |
. | ATF3 nuclear staining, no. cases . | . | |
|---|---|---|---|
. | Positive . | Negative . | |
| Classical Hodgkin lymphoma | 69 | 2 | |
| Lymphocyte-predominant Hodgkin lymphoma | 0 | 26 | |
| B-NHL | 6* | 233 | |
| T-NHL | 2† | 44 | |
| Anaplastic large cell lymphoma (ALCL) | 24 | 9 | |
| Reactive lymphatic tissue | 0 | 20 | |
Summary of ATF3 immunohistochemistry results. The group of B-cell non-Hodgkin lymphomas (B-NHLs) includes 21 mantle cell lymphomas (MCLs), 27 B-cell chronic lymphocytic leukemias (B-CLLs), 72 follicular lymphomas (FLs), 80 diffuse large B-cell lymphomas (DLBCLs, including 6 primary mediastinal B-cell lymphomas), 19 Burkitt lymphomas (BLs), and 20 plasmacytomas. T-cell non-Hodgkin lymphomas (T-NHLs) comprise 20 peripheral T-NHLs (PTCLs), 10 angioimmunoblastic T-NHLs (AILTs), 5 enteropathy-associated T-NHLs, 4 NK/T-cell lymphomas, and 7 cases of mycosis fungoides.
Two cases of DLBCL (one centroblastic variant and one primary mediastinal B cell lymphoma), 4 cases of plasmacytoma.
Two cases of angioimmunoblastic T-cell lymphoma (AILT), nuclear ATF3 staining in malignant T cells.
To assess the functional significance of ATF3 overexpression in HRS cells, we selectively blocked ATF3 expression by RNA interference. Three independent ATF3 siRNA sequences were chosen that resulted in a significant down-regulation of ATF3 protein level after transfection into L428 and L540Cy Hodgkin cells (Figure 2A). Several control siRNA sequences for irrelevant targets had no effect on ATF3 expression (Figure 2A). Next, we asked whether siRNA-mediated knock-down of ATF3 in Hodgkin cells would affect proliferation and tumor cell viability. Therefore, we transiently transfected Hodgkin cells with ATF3-directed siRNA expression plasmids and performed [3H]-thymidine incorporation assays. Down-regulation of ATF3 resulted in a marked reduction of DNA synthesis compared with control-transfected cells (Figure 2B). To investigate the effects of ATF3 down-regulation over an extended period of time, we used vector-based siRNA constructs that are propagated by extrachromosomal replication and carry a puromycin resistance gene permitting the selection of siRNAexpressing cells. Transfection of L428 and L540Cy Hodgkin cells with ATF3-directed siRNA plasmids and subsequent antibiotic selection resulted in a significant and sustained loss of viable cells, as determined by annexin V-FITC/PI double-staining and flow cytometry (Figure 2C). This effect could be induced with different ATF3-targeting constructs, but not with control siRNA plasmids directed against irrelevant targets. Our observations on the role of ATF3 in HRS cells are in good accordance with previous reports that describe a function of ATF3 in the control of cell cycle progression and apoptosis resistance in other cellular systems. Ectopic expression of ATF3 in hepatocytes stimulates cyclin D1 expression and proliferation.10 In addition, ATF3 has been shown to have antiapoptotic functions in cardiomyocytes and neuronal cells.11,12,18 In line with its cell cycle-promoting and antiapoptotic properties, ATF3 has been shown to partially transform chicken embryo fibroblasts, indicating an intrinsic oncogenic potential.19 Thus, in conjunction with previously published observations on its role in proliferation and survival, our data suggest an important oncogenic role of constitutive ATF3 expression in cHL.
Down-regulation of ATF3 by RNA interference impairs proliferation and compromises viability of Hodgkin cells. (A) L428 and L540Cy Hodgkin cells were transfected with control and ATF3-directed pSUPER siRNA expression plasmids, purified, and harvested 48 hours after electroporation. Whole-cell lysates were subjected to Western blot analysis using the indicated antibodies. (B) L540Cy HRS cells were transfected with the indicated pSUPER siRNA constructs, purified, and pulsed with [3H]-thymidine for 24 hours before harvesting and assessing incorporated radioactivity. Data are presented as cpm of [3H]-thymidine incorporation (mean ± SD of 5 measurements). (C) L428 and L540Cy HRS cells were transfected with control and ATF3-directed pRepH1 siRNA expression constructs and selected for siRNAexpressing cells by addition of puromycin 24 hours after electroporation. The percentage of viable and apoptotic cells was determined by annexin V-FITC/propidium iodide (PI) staining and flow cytometry. Viable L428 (left panel) and L540Cy (right panel) cells after 14 days in culture. The fraction of viable cells, negative for both annexin V-FITC and PI, is shown as percentage of total cells. Measurements were performed in triplicate. Error bars indicate SD.
Down-regulation of ATF3 by RNA interference impairs proliferation and compromises viability of Hodgkin cells. (A) L428 and L540Cy Hodgkin cells were transfected with control and ATF3-directed pSUPER siRNA expression plasmids, purified, and harvested 48 hours after electroporation. Whole-cell lysates were subjected to Western blot analysis using the indicated antibodies. (B) L540Cy HRS cells were transfected with the indicated pSUPER siRNA constructs, purified, and pulsed with [3H]-thymidine for 24 hours before harvesting and assessing incorporated radioactivity. Data are presented as cpm of [3H]-thymidine incorporation (mean ± SD of 5 measurements). (C) L428 and L540Cy HRS cells were transfected with control and ATF3-directed pRepH1 siRNA expression constructs and selected for siRNAexpressing cells by addition of puromycin 24 hours after electroporation. The percentage of viable and apoptotic cells was determined by annexin V-FITC/propidium iodide (PI) staining and flow cytometry. Viable L428 (left panel) and L540Cy (right panel) cells after 14 days in culture. The fraction of viable cells, negative for both annexin V-FITC and PI, is shown as percentage of total cells. Measurements were performed in triplicate. Error bars indicate SD.
Prepublished online as Blood First Edition Paper, November 1, 2005; DOI 10.1182/blood-2005-07-2694.
Supported by grants from the Deutsche Forschungsgemeinschaft (Klinische Forschergruppe KFO 105, Sonderforschungsbereich 366), the National Genome Research Network (NGFN), and the Deutsche Krebshilfe (10-2225-Ja 1).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Reuven Agami (Amsterdam, The Netherlands) for the gift of the pSUPER siRNA expression plasmid.

![Figure 2. Down-regulation of ATF3 by RNA interference impairs proliferation and compromises viability of Hodgkin cells. (A) L428 and L540Cy Hodgkin cells were transfected with control and ATF3-directed pSUPER siRNA expression plasmids, purified, and harvested 48 hours after electroporation. Whole-cell lysates were subjected to Western blot analysis using the indicated antibodies. (B) L540Cy HRS cells were transfected with the indicated pSUPER siRNA constructs, purified, and pulsed with [3H]-thymidine for 24 hours before harvesting and assessing incorporated radioactivity. Data are presented as cpm of [3H]-thymidine incorporation (mean ± SD of 5 measurements). (C) L428 and L540Cy HRS cells were transfected with control and ATF3-directed pRepH1 siRNA expression constructs and selected for siRNAexpressing cells by addition of puromycin 24 hours after electroporation. The percentage of viable and apoptotic cells was determined by annexin V-FITC/propidium iodide (PI) staining and flow cytometry. Viable L428 (left panel) and L540Cy (right panel) cells after 14 days in culture. The fraction of viable cells, negative for both annexin V-FITC and PI, is shown as percentage of total cells. Measurements were performed in triplicate. Error bars indicate SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/6/10.1182_blood-2005-07-2694/4/m_zh80060692450002.jpeg?Expires=1769096526&Signature=FBU-SpKCYCrdwJVdm6Um1p3jFimZtwvK6sRjuBMHRMRKhnk3Hp0jn6cF0VY9n9VL1PrD2TZGg1898e30pahGBVNG0aFTWTx9pcvUarO8I~pWBoxOkfDlpEebd-etmW12ayIotRB~iX9Pn1XHQnihuMXriGZwCeE0G3acZh~xaMlF78WPU8zaSDscTbaUgG271rk2tHXvWupdFH8yvP9PVCHdmuxYkOXvIwBMQB5lQKLar5h0OenffhCuYw-AV1-3XyaX1Whkah7fRP2e-zl9c-JvaDGr60xYhKX6-AB8nPma6Q7gTHR-L5fj8aMNVwpJ84L8r9ktI1c1q23HisX1ZA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal