Abstract
Our considerable wealth of data concerning hematologic processes has come despite difficulties working with stem and progenitor cells in vitro and their propensity to differentiate. Key methodologies that have sought to overcome such limitations include transgenic/knock-out animals and in vitro studies using murine embryonic stem cells, because both permit investigation of the formation of hematopoietic tissue from nonhematopoietic precursors. Although there have been many successful studies in model animals for understanding hematopoietic-cell development, differences between lower vertebrates and humans have left gaps in our understanding. Clearly, human-specific strategies to study the onset of hematopoiesis, particularly the earliest events leading to the specification of both normal and abnormal hematopoietic tissue, could bring an investigational renaissance. The recent availability of human embryonic stem (hES) cells suggests that such a system is now at hand. This review highlights the potential of hES cells to model human hematologic processes in vitro with an emphasis on disease targets.
Fifty years of pluripotent stem-cell studies
Defined by their capacity to generate progeny of all 3 embryonic germ layers (ectoderm, endoderm, and mesoderm), pluripotent stem cells were first studied by Stevens and Little1 in experiments that involved isolation of the undifferentiated component of spontaneous testicular teratomas in the 129 inbred mouse strain. These embryonal carcinoma cells (ECCs) differentiated in vitro following aggregation into cystic “embryoid bodies” (EBs) where they again demonstrated the elaboration of all 3 germ layers.2 Although surprising considering their tumor origins, some lines of ECCs were further shown to contribute multiple tissues to chimeric mice following introduction into the murine blastocyst.3
An extension of these studies naturally sought to isolate the stem-cell component of “normal,” nontumor tissues including the early embryo. It would be another 27 years after Stevens and Little began the isolation of the ECCs before murine embryonic stem (mES) cells were first derived from the day 2.5 postfertilization murine blastocyst.4,5 This was followed by the description of yolk sac–like blood islands containing embryonic globin-expressing, nucleated megaloblasts in murine ES cell–derived EBs,6 thereby setting the stage for an entirely new area of investigation, namely, the in vitro formation of hematopoietic tissue from nonhematopoietic precursors, tissue that could be further studied in vivo following transplantation.
mES cells possess a robust capacity for hematopoietic specification in vitro and further lend themselves to the study of specific genetic lesions in vivo.7-9 Modest, short-term hematopoietic reconstitution of lethally irradiated murine hosts was first shown possible with mES cells differentiated in vitro for 4 days,10 suggesting that true hematopoietic stem cells (HSCs) might be capable of being derived from ES cells. These studies were followed by focused attempts to augment the inherent hematopoietic capacity of mES cells. Early experiments made use of the BCR/ABL fusion gene to promote hematopoietic proliferation in mES cells,11-13 although experimental animals succumbed to leukemia. Hypothesizing that downstream effectors of BCR/ABL might stimulate the formation of hematopoietic elements from mES cells yet spare the animals from overt leukemia, other transgenes, including STAT5,14 have been shown capable of increasing colony-forming capacity in vitro, as well as supporting limited engraftment in vivo. Among these, HoxB415 has demonstrated perhaps the greatest capacity to promote hematopoiesis from an mES-cell starting point.16
mES cells stimulated with HoxB4 following coculture on the macrophage colony-stimulating factor (M-CSF)–deficient OP9 stromal cell line17 are capable of long-term engraftment in irradiated hosts.18 This system could also rescue a murine model of immunodeficiency when combined with gene replacement using recipient-specific mES cells created via nuclear transfer (NT)19 (see “Source and utility of disease-specific hES cell lines”). Finally, the HoxB4/OP9 system has recently been used in combination with expression of the caudal type homeobox transcription factor 4 (Cdx4)20 to specify the functional equivalent of the HSC, namely, the capacity to clonally reconstitute primary and secondary transplant recipients in both myeloid and lymphoid lineages.21
These studies represent the fruition of many years of work aimed at creating a rigorous system to investigate hematopoietic lineage specification. Strains of mice (and other animals) bearing spontaneous hematopoietic anomalies (eg, the W mouse22 ) have previously enabled the study of mammalian hematopoietic ontogeny on a fine scale, under the microscope, and in a manner permitting biochemical analysis. The derivation of laboratory animals carrying defined alterations of genes with hematopoietic activities has broadened the field of hematology research even further.
Murine models
Murine ES cells and genetically modified mice have taught us volumes about blood. Powerful methodologies such as analysis of mid-gestation embryos, flow cytometry, and methylcellulose colony-forming assays have combined to reveal intricate relationships between genes and their capacity to direct blood-cell fates.
Among the earliest knock-out mouse models with clear hematopoietic defects were those investigating targeted disruptions of the β2-microglobulin (β2M)23-25 and erythroid transcription factor Gata-126 genes. Previous studies regarding the biology of major histocompatibility (MHC) class I positive cells suggested that at least some class I proteins had roles beyond T-cell maturation. However, while biallelic knock-outs for the β2M light-chain locus demonstrated clear deficiencies of CD4–8+ T-cells (and T-cell–mediated toxicity), the resulting animals were otherwise healthy and fertile.24,25 In a similar proof of the power of in vivo targeting studies, earlier expression analyses had suggested that loss of Gata-1 should produce a defective erythroid phenotype (eg, Tsai et al27 ). While Gata-1–/–cells were found in virtually all somatic lineages assayed including a minor contribution to leukocytes (as determined via glucose phosphate isomerase polymorphisms between the injected ES cells and the wild-type component of the host blastocyst), the resulting animals were clearly deficient in erythroid maturation.26 These examples serve to highlight the power of genetically engineered ES cells as experimental tools. The number of hematopoiesis-related genes that have been investigated using this methodology would prove far too numerous to list in a single review.
A final example demonstrating the utility of ES-cell–based in vitro analysis to elucidate the details of genetic anomalies was recently reported in von Hippel-Lindau (VHL) disease.28 Although cloned over a decade ago29 and previously implicated in a cohort of diseases including Chuvash polycythemia,30,31 the VHL tumor suppressor gene function remains somewhat ambiguous despite a great deal of biochemical information regarding its role in oxygen-dependent transcriptional regulation.32-34 This is due in part to the fact that studies in primary patient tumor material are obscured by additional genetic lesions and that the knock-out mouse model is embryonic lethal.35 It had been shown that loss of Vhl was sufficient to inhibit the down-regulation of oxygen-sensing transcriptional machinery, although insufficient to promote tumorigenesis as a single lesion.36 Building on these observations, the authors used a patient-specific VHL mutation knock-in strategy with Vhl–/– mES cells to interrogate the impact of disease-related base changes.28 A 3-pronged analysis including downstream gene expression, tumor (teratoma) formation, and tumor vascularity/fibronectin deposition demonstrated discrete relationships between pathologically significant mutations and their phenotypic impact, including the relative capacity to restore downstream transcription/protein production (VEGF) and promote tumor growth.28
One caveat of these studies is that the human VHL gene (and mutations) was imposed on mouse cells. Such a situation may not accurately predict what could be gleaned from a wholly human system. It has often been noted that the chain of events leading to mature blood cells is tightly conserved between mice and humans (for a review, see Lensch and Daley37 ). Although studies in the mouse have established the lion's share of our knowledge of mammalian blood development, gaps remain. Without a doubt, an ability to investigate the impact of specific mutations on the development of human blood diseases would be highly instructive, particularly for congenital conditions. In this manner, an in vitro human system comparable to murine embryonic stem cells would be a powerful resource to study hematopoiesis (and the formation of other tissue). What prevents such human-specific studies?
Turning the microscope toward ourselves
Immortalized cell lines represent a reliable and abundant resource for human-specific in vitro studies. However, they are a better substrate for analysis of cellular biology as opposed to developmental biology for at least 2 reasons. First, the process of transformation, whether achieved in vivo as part of malignancy prior to isolation or afterward using effectors such as Epstein-Barr virus,38 telomerase,39 SV40,40 or other means, imposes its own unique biology on the cell and represents a constant source of uncertainty when attempting to model a cellular process as it occurs in normal tissue. Secondly, a research study using differentiated, even fetal, tissue can at most speak to biologic events distal to the specification of the cell in use. An in vitro experimental system that could answer questions relating to tissue genesis would require a primitive, unspecialized cell line that could be coaxed to differentiate into the tissue under study. In 1998, with the first description of hES cells,41 such studies into human lineage-specification became possible.
Human embryonic stem cells in the study of hematopoiesis
Isolated from the preimplantation blastocyst approximately 5 to 7 days following in vitro fertilization (IVF), hES cells can differentiate into representatives of all 3 embryonic germ layers (endoderm, mesoderm, and ectoderm)41 as well as cells with properties of extraembryonic trophectoderm.42-44 Methodologies allowing for directed differentiation of hES cells have been reported for a variety of key cellular types, including cardiac myocytes,45 neural progenitors,46-48 and hematopoietic tissue.49 Early experiments with hES cells have lacked the experimental wizardry of contemporary murine studies due to the dearth of key platform technologies. This is changing rapidly given reports demonstrating genetic modification of hES cells using retro/lentiviral transgenesis,44,50,51 homologous recombination,52 and RNA interference (RNAi).44,53-55 An experimental target gene has not yet been reported that is comparable to the ROSA-26 locus56 in mice, a region of chromatin permissive for the constitutive expression of transgenes and useful for expressing factors such as transactivators in conditional gene expression systems.57 However, gene-trapping studies are under way in hES cells.58
Despite the need to develop basic technologies that enable their manipulation, hES cells have already proven quite useful in the study of hematopoiesis. In a cohort of manuscripts, Bhatia's group has reported serum-based cultivation conditions with mesoderm/hematopoietic cell-inducing cytokines (BMP-4, VEGF165, IL-3, IL-6, SCF, FLT-3L, and G-CSF) that lead to a high percentage of blood cells in culture.59-65 However, this hematopoietic tissue is limited in its capacity to engraft in animal transplantation models due in part to an abnormal expression profile for Hox A- and B-cluster genes.63 Other groups have also studied the elaboration of lympho-myeloid hematopoietic tissue from hES cells via different culture methodologies,66-71 although again, no hematopoietic engraftment has been demonstrated. What is clear, however, is that a trajectory of discovery similar to that seen in comparable murine studies is present in the hES cell research, with an arc that points to engraftable hematopoietic cells likely being generated in the next few years. The present inability to demonstrate engraftment aside, hES cell-based studies are likely to prove particularly valuable for research aimed at understanding genetic diseases of the blood.
Filling in the gaps
A host of model systems in the mouse have fallen short in achieving the goal of illuminating human disease. An unfortunate and not uncommon aspect of mouse engineering is that successful gene targeting often leads to a limited or indiscernible phenotype in the resulting animals. A good example of this is the murine model of the DNA repair disease Fanconi anemia (FA), a recessive condition that manifests as congenital anomalies, aplastic anemia/pancytopenia progressing to bone marrow failure, and predisposition to solid tumors and acute myelogenous leukemia (AML).72 Acquired losses of FA proteins may occur in adult AML.73 The condition demonstrates genetic heterogeneity with at least 11 complementation groups currently described.74 Mouse models for loss of the Fanconi complementation group C (FA-C) gene (Fancc)75 develop neither bone marrow failure nor leukemia, although they do manifest both a DNA repair defect as well as hypersensitivity to apoptosis-inducing, inhibitory cytokines in hematopoietic colony-forming assays.76 These sensitivities represent a considerable myelosuppressive stress and likely contribute to the development of progressive aplastic anemia, pancytopenia, and eventual bone marrow failure in patients with FA. Furthermore, such environmental pressures in the marrow would endow subsequent apoptosis-resistant, leukemic clones with a profound selective advantage,77 a hypothesis that has been met with some validation in animal studies.78 Some hematologic abnormalities including impaired competitive repopulating ability have been observed,79 but the fact that mice do not exhibit the bone marrow failure phenotype seen in human patients suggests that certain aspects of the phenotype remain poorly modeled in murine systems.
FA is a disease wherein clinically significant manifestations can arise in utero. Individuals with FA often bear congenital skeletal anomalies including agenesis of the radius and thumb, occasional aplastic anemia in the neonate, and in some patients, retention of embryonic/fetal globin expression. Hox genes are clearly important in the specification of both the appendicular skeleton (reviewed in Krumlauf80 ) as well as the hematopoietic system (reviewed in Lawrence et al81 ) indicating what may be related patterning defects. As shown in Table 1, FA is but one example of a group of genetic illnesses that may lend themselves to detailed study via disease-specific models in hES cells. Although this list is limited for the sake of space, it depicts conditions with defective maintenance of hematopoietic tissue and perhaps formation of blood as well.
Congenital hematologic diseases lending themselves to study using hES cells
Disease . | OMIM* . | Genes . | Blood phenotype . |
|---|---|---|---|
| Fanconi anemia | 227650 | Several including BRCA2 | Aplastic anemia, pancytopenia, MDS, AML |
| TAR syndrome | 274000 | Unknown | Pancytopenia, anemia, hypercellular marrow, granulocytosis |
| Severe congenital neutropenia | 202700 | Neutrophil elastase | Myeloid arrest, AML, agranulocytosis |
| Shwachman-Diamond syndrome | 260400 | SBDS | Anemia, thrombocytopenia, pancytopenia, MDS, AML |
| Diamond-Blackfan anemia | 205900 | Ribosomal protein S19, unknown | Macrocytic anemia, thrombocytopenia, fetal hemoglobin |
| Trisomy 21 | 190685 | N/A | Transient myeloproliferative disease, DS-AMKL |
| Chuvash polycythemia | 263400 | VHL | Erythrocytosis, increased erythropoietin |
| Neurofibromatosis type 1 | 162200 | Neurofibromin | JMML |
| Lesch-Nyhan syndrome | 300322 | HPRT1 | Megaloblastic anemia |
| SCID | 601457 | Several including ADA, RAG1/2 | Lymphopenia, lack of mature B and T cells |
| α-thalassemia | 141800 | α-globin | Methemoglobinemia, polycythemia, hemolysis |
| β-thalassemia | 141900 | β-globin | Anemia, hypochromic microcytes, splenomegaly |
| Sickle cell anemia | 603903 | β-globin | Anemia, septicemia, microvascular occlusion |
Disease . | OMIM* . | Genes . | Blood phenotype . |
|---|---|---|---|
| Fanconi anemia | 227650 | Several including BRCA2 | Aplastic anemia, pancytopenia, MDS, AML |
| TAR syndrome | 274000 | Unknown | Pancytopenia, anemia, hypercellular marrow, granulocytosis |
| Severe congenital neutropenia | 202700 | Neutrophil elastase | Myeloid arrest, AML, agranulocytosis |
| Shwachman-Diamond syndrome | 260400 | SBDS | Anemia, thrombocytopenia, pancytopenia, MDS, AML |
| Diamond-Blackfan anemia | 205900 | Ribosomal protein S19, unknown | Macrocytic anemia, thrombocytopenia, fetal hemoglobin |
| Trisomy 21 | 190685 | N/A | Transient myeloproliferative disease, DS-AMKL |
| Chuvash polycythemia | 263400 | VHL | Erythrocytosis, increased erythropoietin |
| Neurofibromatosis type 1 | 162200 | Neurofibromin | JMML |
| Lesch-Nyhan syndrome | 300322 | HPRT1 | Megaloblastic anemia |
| SCID | 601457 | Several including ADA, RAG1/2 | Lymphopenia, lack of mature B and T cells |
| α-thalassemia | 141800 | α-globin | Methemoglobinemia, polycythemia, hemolysis |
| β-thalassemia | 141900 | β-globin | Anemia, hypochromic microcytes, splenomegaly |
| Sickle cell anemia | 603903 | β-globin | Anemia, septicemia, microvascular occlusion |
TAR indicates thrombocytopenia-absent radius; SCID, severe combined immunodeficiency; BRCA2, breast cancer type 2 gene; MDS, myelodysplastic syndrome; AML, acute myelogenous leukemia; SBDS, Shwachman-Bodie-Diamond syndrome gene; DS-AMKL, Down syndrome-associated acute megakaryoblastic leukemia; VHL, von Hippel-Lindau gene; JMML, juvenile myelomonocytic leukemia; HPRT, hypoxanthine guanine phosphoribosyltransferase 1 gene; ADA, adenosine deaminase; RAG, recombination activating gene.
Online Mendelian Inheritance in Man, OMIM. McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), 2000. World Wide Web URL: http://www.ncbi.nlm.nih.gov/omim/.
Most entries in Table 1 are known single gene disorders but others, such as Down syndrome (DS) or trisomy 21, have a more complex genetic etiology. Chromosome 21 contains as many as 350 genes82 and although the exact role of each aberrantly expressed sequence is unknown in DS, the transient myeloproliferative disease (TMD) of the DS neonate as well as a DS-specific form of acute megakaryoblastic leukemia (DS-AMKL) are unambiguously associated with this additional chromosomal material. The most convincing data in support of this comes from the finding that although many DS patients are mosaic for trisomy 21, the proliferating blasts are those containing 3 copies of chromosome 21.83 Also, in DS-associated leukemia, leukemic evolution preferentially occurs in the trisomic cells rather than the euploid stem-cell population in the mosaic patient.84
As for FA, a murine model for DS is not completely informative. Human chromosome 21 is syntenic to regions of chromosomes 10, 16, and 17 in the mouse although the majority is represented on mouse 16. Trisomy 16 in the mouse is lethal85 and despite the creation of a minimal mouse model containing the DS-critical region, the system fails to recapitulate important aspects of the DS phenotype.86 Certain DS features have been more effectively modeled in another transgenic mouse system containing the majority of human chromosome 21.87
Sources and utility of disease-specific hES cell lines
How does one create ES cell lines to study hematopoietic diseases? Three potential routes are available: identification of embryos via preimplantation genetic diagnosis (PGD), modification from existing hES cell lines using genetic manipulation, or creation from patient tissues using nuclear transfer (NT).
PGD is routinely offered to families wherein a known, heritable illness segregates. The genotype of an embryo in question is determined via DNA analysis from the first or second polar bodies or by extracting a single blastomere following embryo cleavage.88 This method identifies a set of embryos that carries disease-associated genetic lesions, some of which have been donated to biomedical research and subsequently used for the creation of hES cell lines.89 Among these are 18 hES cell lines containing genetic anomalies known to cause a variety of conditions including adrenoleukodystrophy, Duchenne muscular dystrophy, FA, neurofibromatosis type 1 (NF1), β-thalassemia, and Huntington disease.89 The potential avenues of investigation are many for each of these lines in the study of normal and pathologic tissue formation.
Of particular hematologic interest in this set of hES cell lines is NF1, a single gene disorder wherein patients present with congenital anomalies including macrocephaly and benign neurofibromas. NF1 also predisposes patients to both solid tumors and hematologic disease, in this case juvenile myelomonocytic leukemia (JMML; for a review, see Emanuel90 ). Although stem and progenitor cells in JMML are known to be hypersensitive to granulocyte-macrophage colony-stimulating factor (GM-SCF) stimulation,91,92 this is insufficient to induce leukemia.93 NF1 and other reported hES cell lines derived following PGD will undoubtedly contribute to important basic research. However, limited donor participation, low frequencies for certain diseases presenting for PGD, and a strong white bias among individuals seeking assisted reproduction94 make it impractical to imagine PGD as a reliable means of modeling more than a small subset of human diseases.
Methods established for genetic modification of murine ES cells have been applied to hES cell lines. Homologous recombination has been demonstrated for the hypoxanthine phosphoribosyltransferase (HPRT) gene.52,95 Although HPRT defects are primarily associated with Lesch-Nyhan syndrome96 and gout, megaloblastic anemia has been noted in both patients with Lesch-Nyhan syndrome97 as well as Hprt–/– mice98 and likely results from the impaired purine metabolism inherent to this defect of nucleotide salvage. As previously noted, RNAi has been used in hES cells44,53-55 and is highly efficient, approximating a true gene knock-out effect.44 Cellular fusion has also been used to create ES cells with unique genetic properties.99,100 One caveat of such studies is that the resulting cells are, at present, unavoidably aneuploid. Although works using such cellular fusions are important advances toward a more detailed understanding of nuclear reprogramming, aneuploid products are of uncertain clinical utility. Many would be reluctant to administer anything other than euploid cells during cell-replacement therapies. Additionally, chromosomal aneuploidy is a routine finding in malignancy and has itself been considered an important component of multistep transformation.101 The engineering of existing cell lines using these and other technologies will likely become a routine aspect of hES-cell research.
The third method for obtaining disease-specific hES cell lines is NT, a methodology that has been used in vertebrate systems to study development for over 50 years. First used in the frog, Briggs and King observed that the transfer of a cellular nucleus isolated from a late stage blastocyst into an enucleated oocyte was capable of generating a normal tadpole,102 thereby indicating that “nuclear differentiation” was reversible and that the nuclei of somatic cells were equivalent. King and Briggs also noted that this potency declined as cells became more differentiated, observations that were confirmed by Gurdon and colleagues in work wherein fertile adult frogs were obtained using nuclei from the intestinal epithelium of feeding larvae-stage donors (though unsuccessful from adult nuclei).103,104 The most famous incarnation of such studies is undoubtedly “Dolly” the sheep, the first mammal cloned via NT from an adult somatic cell from the mammary epithelium.105 Although capturing the public imagination as a potential (though unlikely106 ) avenue leading to human reproductive cloning, NT has rather proven itself to be a valuable methodology to investigate the manner by which cellular specialization occurs, including mechanisms of epigenetic regulation (reviewed in Hochedlinger and Jaenisch107 ).
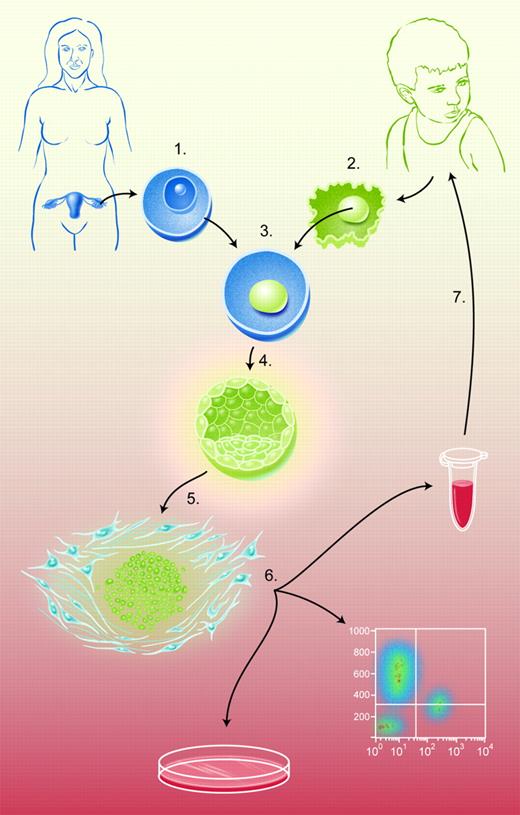
The figure summarizes the nuclear transfer (NT) process as used to create genetically defined hES cell lines. (1) Hyperovulation of female oocyte donors, surgical harvest of oocytes, and oocyte enucleation via micromanipulation to create an ooplast. (2) Research subject somatic-cell donation and enucleation via micromanipulation to isolate donor nucleus. (3) Microinjection of donor nucleus into ooplast to create a reconstituted oocyte. (4) Activation of reconstituted oocyte to induce mitosis. (5) Isolation of inner-cell mass from blastocysts via immunosurgery and establishment cultivation of hES cell line. (6) In vitro analysis/directed differentiation of hES cells. (7) Cellular therapy with in vitro–differentiated, genetically repaired hES-cell derivatives.
The figure summarizes the nuclear transfer (NT) process as used to create genetically defined hES cell lines. (1) Hyperovulation of female oocyte donors, surgical harvest of oocytes, and oocyte enucleation via micromanipulation to create an ooplast. (2) Research subject somatic-cell donation and enucleation via micromanipulation to isolate donor nucleus. (3) Microinjection of donor nucleus into ooplast to create a reconstituted oocyte. (4) Activation of reconstituted oocyte to induce mitosis. (5) Isolation of inner-cell mass from blastocysts via immunosurgery and establishment cultivation of hES cell line. (6) In vitro analysis/directed differentiation of hES cells. (7) Cellular therapy with in vitro–differentiated, genetically repaired hES-cell derivatives.
As shown in Figure 1, NT is a process wherein a hES cell line of known genetic makeup might be created from a specific patient and used to model disease, to develop histocompatible tissues for transplantation therapies, or both. NT allows scientists to model disease using cells taken from patients with a well-described medical history and, thus, the phenotypic consequences of specific genetic lesions would be unambiguous. A major stumbling block for creating models of human disease is the need for oocyte donation, itself a difficult process.
Of note, the veracity of certain reports describing the derivation of hES cell lines via NT has recently been called into question.108 Although the results of these individual studies (Hwang et al109 and Hwang et al110 ) have been retracted, the scientific goals of NT for the establishment of genetically defined hES cells remain unchanged. Indeed, work in other laboratories continues with these aims in mind.111
Although the potential utility of hES cell lines generated by any of the aforementioned techniques to study disease pathogenesis is profound, a major advantage of NT methodology is the creation of pluripotent cells that are immunologically compatible with patients. Such cells are uniquely valuable for deriving repopulating cells in vitro for transplantation therapy in vivo. In short, the laboratory could become the transplant donor and thereby work toward alleviating the vast shortfall that currently exists in transplantation registries worldwide. Although it is an understatement to say that a great deal of work remains between this notion and the availability of hES-cell–based therapy, proof-of-principle work in rodent models suggests it is, in fact, possible. What stands to be gained by working toward such a goal?
We have often heard the criticism that adult stem-cell–based therapies have saved many, whereas ES-cell–based therapies have saved none. Given that the field of hES-cell research is but 8 years old, whereas bone marrow transplantation has been practiced for decades, such comments represent an unfair critique. Furthermore, although it is indisputable that bone marrow, peripheral-blood stem cell, and cord-blood transplantation therapies have improved the lives of innumerable individuals, continued refinements of these methodologies are sorely needed to enhance their efficacy. Indeed, biomedical research is inspired in part by failures of clinical practice, by those therapies that remain disappointing despite their status as “standard of care.” To suggest that the current challenges of modern medicine would be best met with continued exploration of conventional therapies alone seems misguided. It is our hope that this review conveys the rationale for using hES cells to probe unknown areas of human development and disease, hopefully forging new therapies in the process. In this manner, we seek to complement the current armamentarium of effective modalities for treating hematologic disease.
Taking a closer look at our “successes” reveals that bone marrow transplantation therapies for malignancy allow some to enter into durable remission, whereas others are less fortunate and will experience disease relapse or graft failure. Many more still will perish while awaiting transplantation due to the unavailability of a suitably HLA-matched tissue donor. As many as two thirds of individuals who could potentially benefit from bone marrow, peripheral-blood stem cell, or cord-blood therapy will fail to locate a suitable donor, a figure that can be even worse for certain ethnic groups (for a review, see Grewal et al112 ). Furthermore, although allogeneic bone marrow transplantation has improved greatly since Thomas and colleagues reported their first attempt following myeloablative therapy in 1957,113 unrelated donor transplant survival rates remain low for most hematologic diseases with combined 3-year survivals (Kaplan-Meier) of approximately 33% ± 7% in one cohort of 211 patients.114 Nonmalignant morbidity including cataracts, thyroid dysfunction, and osteoporosis also continue to be significant, especially as long-term survival rates improve.115
Owing to the unavailability of matched siblings for many patients as well as a greater tolerance of HLA mismatches, partially HLA-matched cord blood is becoming an increasingly important transplantation resource116 not to mention an important platform for understanding basic disease mechanisms.117 In one multicenter study of childhood AML, the 2-year Kaplan-Meier estimate of overall leukemia-free survival following unrelated cord-blood transplantation was 42% ± 5% with infection being the major cause of nonleukemia-related mortality (18 of 95 patients).118 These are somewhat discouraging statistics although results in leukemia predisposition syndromes may fare even worse.
Conditioning is a delicate affair in patients with FA due to increased sensitivity to both ionizing radiation and cyclophosphamide (reviewed in Liu and Dokal119 ). Nevertheless, 17% to 30% of patients with FA will go on to receive hematopoietic stem-cell transplantation (reviewed in Alter et al120 ). FA patients are at increased risk for both acute graft-versus-host disease (aGvHD) and therapy-related malignancies, including primarily squamous cell carcinoma of the head and neck (53% 15-year incidence).121 One-year survival in a cohort of 29 FA patients after transplantation was 34% using unrelated or mismatched sibling donors.122 These numbers are more encouraging with HLA-matched sibling marrow and indicate greater than 70% overall survival (n = 50) at 5 years and just over 50% at 10 years (reviewed in Liu and Dokal119 ).
In contrast to these numbers, a recent cohort evaluating long-term survival in recipients of autologous transplants indicated an overall 10-year survival of 68.8% ± 1.8%.123 Although such numbers must be taken with a note of caution because they include data from multiple primary diagnoses, conditioning regimens, and stem-cell–harvest techniques, they nonetheless indicate the obvious superiority of a genetically identical transplant compared to allogeneic or even HLA-matched sibling alternatives. Reflecting on this observation alone, an ability to produce genetically identical hematopoietic tissue for transplantation therapy might reduce the morbidity of the transplant procedure, making it suitable for routine treatment of genetic diseases such as sickle cell anemia, diseases that are typically not considered for transplantation where complications such as aGvHD present an unacceptable risk. In this manner, diseases like sickle cell anemia and thalassemia would be among the first line of conditions poised to take advantage of improved transplantation modalities. For treatment of bone marrow diseases, malignant or otherwise, the use of NT to generate matched hES cells might alleviate the donor shortfall, enhance the quality of life in the patients given the transplant, and improve long-term survival rates. NT studies combined with in vitro protocols for the directed differentiation of the resulting hES cell lines into long-term repopulating HSCs aim squarely at this target. What's more, this suggestion does not even mention additional advances that might come from these technologies if used to produce more differentiated, infusible cell types in vitro including red-cell precursors,124 platelets,125 or defined components of the lymphoid lineage.
The recent availability of hES cells and the many research advances already made using such cells point toward a future wherein cells become increasingly important as medicine. Furthermore, new opportunities presented by hES cells to model normal and abnormal development offer great promise and excitement in the field of biomedical research. In recognition of this potential, the National Research Council has recently issued detailed guidelines for the conduct of hES-cell research.126 Certainly, there are hurdles to be overcome along the way, including the risk of hES-cell contamination by animal products used in tissue culture127 as well as a background propensity to develop chromosomal defects that promote adaptation to extended culture in vitro.128,129 We shall also undoubtedly see a refinement in the variety of hES cell lines used worldwide as it becomes obvious that certain lines hold greater affinity for study than others. The field would additionally welcome the emergence of more effective, animal product–free protocols for standardized hES-cell culture. These issues aside, the greatest scientific challenges remain in the area of defining pathways that endow the specification and isolation of long-term repopulating, multilineage hematopoietic tissue from an hES-cell starting point. Work in the mouse suggests that this goal is not unreasonable. Informed by the trajectory of previous studies and assuming adequate support to enable rigorous investigation, we expect equivalent goals to be met using hES cells within the next few years.
Prepublished online as Blood First Edition Paper, December 6, 2005; DOI 10.1182/blood-2005-07-2991.
Supported by grants from the National Institutes of Health (NIH) and the NIH Director's Pioneer Award of the NIH Roadmap for Medical Research. G.Q.D. is a recipient of the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research. M.W.L. is a Fellow of the Leukemia and Lymphoma Society and received financial support from the Eppley Foundation for Research and a Seed Grant from the Harvard Stem Cell Institute.
The authors wish to thank the members of the Daley laboratory for providing critical reviews of the manuscript, in particular Ms Kristy Rialon.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal