Molecular markers like IgVH mutational status, chromosomal abnormalities, and CD38 and ZAP-70 expression have prognostic value in B-cell chronic lymphocytic leukemia (B-CLL). These may be pathogenetic because of the coincidental expression of ZAP-70 and increased B-cell receptor (BCR) signaling and the signaling function of CD38 in CLL. This study shows that ZAP-70+ CLL B cells respond in vitro more readily than ZAP-70– CLL and normal B cells to chemokine migratory signals through enhanced surface CCR7 expression (P = .009; P < .001) and increased responsiveness to its ligands CCL19 and CCL21, demonstrated by F-actin polymerization (P < .05) and cellular migration (P < .01). In addition, ZAP-70+ CLL cells exhibit sustained ERK phosphorylation/activation following stimulation with CXCL12 (SDF1-α, a survival factor produced by stromal cells) compared with ZAP-70– cells (P = .004). Following coculture with nurse-like cells, the survival of ZAP-70+ but not ZAP-70– CLL cells is significantly enhanced by the addition of CXCL12 (P < .05), an effect that is partially blocked by the MEK inhibitor PD98059. These advantageous migratory and survival responses may promote easier access to and greater proliferation in pseudo-germinal centers and explain in part the more progressive nature of ZAP-70+ disease.
Introduction
Genomic screens examining the differences between expression profiles of mutated and unmutated IgVH genes, a good prognostic marker in chronic lymphocytic leukemia (CLL),1-3 have demonstrated that ZAP-70 expression in CLL B cells is strongly associated with mutational status of the IgVH gene.4,5 Several groups have since confirmed the observation that ZAP-70 positivity is strongly correlated to an unmutated IgVH gene status, whereas ZAP-70 negativity is associated with a mutated IgVH gene status.5,6 Other independent molecular markers in CLL include surface CD38 expression7,8 and the presence of specific chromosomal aberrations (trisomy 12, 11q23, 13q14, or 17p13 deletions).9,10 Recent evidence has suggested that ZAP-70 is a potent independent prognostic marker and potentially a more powerful predictor of the need for early treatment than mutational IgVH status.11
Progressive CLL disease is marked by an increasing accumulation of CD5+ lymphocytes within the blood, bone marrow, and secondary lymphoid organs, where the lymphocyte doubling time (LDT) is shorter than 1 year. It is clear, however, that CLL cells do not proliferate in the peripheral blood (PB). More than 99% of PB CLL B cells are arrested in the G0/G1 stage of the cell cycle.12 This has raised the question of where these cells proliferate. Histologic findings in lymph node (LN) and bone marrow (BM) have demonstrated the presence of “germinal-like” centers (pseudogerminal centers) or proliferation centers (PCs) in which CLL B cells are found associated with CD40L+/CD4+ T helper (Th) cells and follicular dendritic cells.13 It seems likely therefore that the CLL B cell must migrate from the PB to the LN or BM in order to proliferate and it is currently unclear how this happens.
CCR7 expression is known to regulate trafficking of normal B, T, and dendritic cells into the LN. The CCR7 ligands CCL19 (ELC/MIP3β) and CCL21 (SLC/6Ckine) are expressed on high endothelial venules, which mark the entry barrier into the LN.14-18 These chemokines regulate T-cell migration into and around LNs,19 are involved in B-cell LN entry following contact with antigen,20,21 and are crucial for dendritic cell migration to secondary lymphoid organs following uptake of antigen.16,19 CCR7 expression on PB CLL B cells is upregulated and the migratory responses of CLL B cells to CCL21 are increased in CLL patients with clinical lymphadenopathy.18,22 Increased expression of CCR7 may therefore be one mechanism through which the CLL B cells can move from PB to BM and LNs.
Within the LN and BM, CLL B cells appear not only to associate with a subset of CD4+ Th cells in PCs or pseudo-follicles but also to upregulate expression of mRNA for CCL17 and CCL22.23 The receptor for CCL17 and CCL22, CCR4, is expressed on CD4+/CD40L+ Th cells and these cells are found in involved tissues clustered around Ki67+-proliferating CLL B cells.23 Furthermore it appears that CD40 ligation of PB CLL B cells also induces CCL17 and CCL22 mRNA expression and CCL22 protein release into culture supernatant.23 These findings suggest that CLL B cells can attract CD4+ Th cells that in turn can induce further chemokine production by the CLL cell, promoting a cycle of Th-cell recruitment and CLL B-cell proliferation.
Entry to this favorable microenvironment can also lead to the exposure of the CLL B cell to additional survival signals. One such survival signal is mediated through contact of the CLL cell with the stromal-like cells found within the LN and BM. CLL B cells cultured in vitro rapidly undergo apoptosis24 but contact of CLL B cells with stromal cells inhibits this.25-27 Both stromal cells and nurse-like cells (NLCs), the stromal-like cells found in CLL PB, release high levels of the chemokine CXCL12/SDF-1α.28,29 The effects of CXCL12 are mediated through ligation of its receptor, CXCR4.30,31 Addition of exogenous CXCL12 to CLL B-cell cultures in the absence of NLCs partially inhibits apoptosis, and NLC-mediated protection against apoptosis can be blocked by an antibody to CXCL12.28 It is not yet clear how CXCL12-CXCR4 ligation induces this protective effect. However, rapid activation of p44/42 extracellular signal-regulated kinases (ERKs) is seen in normal T lymphocytes on exposure to CXCL12, and these kinases constitute a key pathway promoting cell survival through transcriptional-dependent and -independent mechanisms.28,32 In T lymphocytes, stimulation with CXCL12 results in prolonged activation of p44/42 ERK.33 This occurs following ligation of CXCR4 and requires both ZAP-70 and the Src homology 2 domain–containing protein of 76 kDa (SLP-76).33 SLP-76 is a protein normally expressed by T cells and myeloid cells but which has recently been identified as being expressed in CLL B cells with the 11q23 deletion, an adverse prognostic marker.34
As the chemokine receptors CCR7 and CXCR4 may have potential roles in regulating the movement, survival, and proliferation of CLL B cells, this investigation examined whether ZAP-70+ CLL cells have differing expression of CCR7 and CXCR4 compared with ZAP-70– CLL cells and whether ZAP-70 expression enables the cells to respond differently to their respective chemokine ligands, CCL19, CCL21, and CXCL12.
Patients, materials, and methods
Local research ethics committee approval was obtained for in vitro studies on blood samples from B-CLL patients and healthy volunteers who gave fully informed consent for blood samples to be taken. Approval was obtained from the South West Devon Research Ethics Committee for these studies. Table 1 shows the patients' stage of disease (Rai and Binet) and whether the patient has received treatment. Blood mononuclear cells were isolated by density-gradient centrifugation over Lymphoprep (Axis Shields, Oslo, Norway). Cells were either analyzed immediately or were resuspended in fetal bovine serum (FBS) containing 10% dimethyl sulfoxide (DMSO) for storage in liquid nitrogen. The viability of cells was assessed at the initiation of culture and was at least 90%, as determined by trypan blue exclusion. Cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin at 37°C/5% CO2.
Patient data, mutational status, VH gene usage, LDT, CD38, ZAP-70, and FISH results
Patient ID . | M/UM IgVH . | VH gene usage . | Clinical stage . | LDT . | CD38, % . | ZAP-70, % . | FISH . |
|---|---|---|---|---|---|---|---|
| 1* | UM | VH1-69, D3-22, JH6 | A/0 | <1 y | 40.7 | 56.0 | del(13)(q14) |
| 2 | M | VH1-8, D6-6 | A/0 | >1 y | 0.8 | 8.4 | del(13)(q14) |
| 3 | M | VH3-23 | A/0 | >1 y | 1.75 | 1.1 | del(13)(q14) |
| 4* | M | VH2-5, D4-17, JH6 | A/0 | >1 y | 0.1 | 12.2 | del(13)(q14)† |
| 5 | M | VH5-51, D7-27, JH5 | A/0 | >1 y | 0.1 | 14.5 | ND |
| 6* | UM | VH1-69, D3-10, JH6 | A/0 | <1 y | 33.7 | 53.6 | +12 |
| 7* | M | VH4-34, D4-17, JH4 | B/II | <1 y | 1.7 | 3.7 | del(13)(q14) |
| 8* | M | VH1-46, D3-22, JH6 | B/I | <1 y | 10.9; bi | 26.6 | Normal |
| 9 | M | VH4-39 | A/0 | >1 y | 19.55 | 12.9 | Normal |
| 10* | UM | VH3-53, D2-8 | A/0 | >1 y | 63.8 | 39.2 | Normal |
| 11 | M | VH3-35, D3-10, JH5 | A/0 | >1 y | 0.8 | 7.7 | Normal |
| 12* | M | VH1-2, D3-16, JH1 | A/1 | <1 y | 61.0 | 40.1 | del(13)(q14) |
| 15 | M | VH1-69, D5-12, JH5 | A/0 | >1 y | 15.3; bi | 25.1 | del(13)(q14) |
| 16* | M | VH2-5 | A/0 | >1 y | 2.6 | 3.7 | del(13)(q14)† |
| 17* | M | VH3-64, D3-3 | A/I | <1 y | 1.2 | 17.7 | del(17)(p13)‡ |
| 18* | M | VH3-21, JH6 | C/II | <1 y | 31.2; bi | 64.2 | Normal |
| 19* | UM | VH1-69 | A/I | <1 y | 58.0 | 24.5 | del(11)(q23)‡ |
| 20* | M | VH4-34, D3-22, JH4 | B/II | <1 y | 44.5 | 55.0 | del(17)(p13) |
| 21 | M | VH3-7, D2-2, JH6 | A/0 | <1 y | 3.6 | 31.8 | del(13)(q14) |
| 22* | M | VH3-33 | C/III | <1 y | 1.6 | 8.3 | del(13)(q14) |
| 23* | UM | VH1-69, D3-3, JH5 | A/I | <1 y | 14.9 | 46.4 | +12 |
| 25* | M | VH3-74, D5-12, JH5 | A/0 | >1 y | 95.3 | 8.5 | +12 |
| 27* | UM | VH1-69, D3-3 | A/I | <1 y | 3.55 | 43.1 | del(11)(q23)‡ |
| 28* | UM | VHI-69, D3-3 | A/0 | <1 y | 14.5; bi | 44.4 | del(13)(q14) |
| 29 | M | VH4-61, D3-10, JH6 | C/0 | >1 y | 5.1; bi | 42.3 | del(13)(q14) |
| 30 | UM | VH1-69, D3-22, JH2 | A/0 | <1 y | 8.5; bi | 38.1 | del(17)(p13)‡ |
| 31 | M | VH3-48 | A/0 | 1 y | 68 | 60.7 | del(17)(p13)‡ |
| 32 | M | VH1-2 | A/0 | >1 y | 1.1 | 4.7 | del(13)(q14)† |
| 33 | M | VH3-21 | A/0 | >1 y | 4.4 | 13.8 | ND |
| 34 | M | VH1-46 | A/0 | >1 y | 51.4; bi | 21.8 | +12 |
| 35* | M | VH2-5, D3-10, JH4 | A/II | >1 y | 24.4; bi | 12.7 | del(17)(p13)‡ |
| 36 | M | VH3-23, D1-1, JH5 | A/0 | >1 y | 4.9 | 7.7 | Normal |
| 37 | M | VH5-51 | A/0 | >1 y | 2.35 | 11.1 | del(13)(q14)† |
| 38 | M | VH3-33, D7-27, JH6 | A/0 | >1 y | 0.75 | 0.9 | del(13)(q14) |
Patient ID . | M/UM IgVH . | VH gene usage . | Clinical stage . | LDT . | CD38, % . | ZAP-70, % . | FISH . |
|---|---|---|---|---|---|---|---|
| 1* | UM | VH1-69, D3-22, JH6 | A/0 | <1 y | 40.7 | 56.0 | del(13)(q14) |
| 2 | M | VH1-8, D6-6 | A/0 | >1 y | 0.8 | 8.4 | del(13)(q14) |
| 3 | M | VH3-23 | A/0 | >1 y | 1.75 | 1.1 | del(13)(q14) |
| 4* | M | VH2-5, D4-17, JH6 | A/0 | >1 y | 0.1 | 12.2 | del(13)(q14)† |
| 5 | M | VH5-51, D7-27, JH5 | A/0 | >1 y | 0.1 | 14.5 | ND |
| 6* | UM | VH1-69, D3-10, JH6 | A/0 | <1 y | 33.7 | 53.6 | +12 |
| 7* | M | VH4-34, D4-17, JH4 | B/II | <1 y | 1.7 | 3.7 | del(13)(q14) |
| 8* | M | VH1-46, D3-22, JH6 | B/I | <1 y | 10.9; bi | 26.6 | Normal |
| 9 | M | VH4-39 | A/0 | >1 y | 19.55 | 12.9 | Normal |
| 10* | UM | VH3-53, D2-8 | A/0 | >1 y | 63.8 | 39.2 | Normal |
| 11 | M | VH3-35, D3-10, JH5 | A/0 | >1 y | 0.8 | 7.7 | Normal |
| 12* | M | VH1-2, D3-16, JH1 | A/1 | <1 y | 61.0 | 40.1 | del(13)(q14) |
| 15 | M | VH1-69, D5-12, JH5 | A/0 | >1 y | 15.3; bi | 25.1 | del(13)(q14) |
| 16* | M | VH2-5 | A/0 | >1 y | 2.6 | 3.7 | del(13)(q14)† |
| 17* | M | VH3-64, D3-3 | A/I | <1 y | 1.2 | 17.7 | del(17)(p13)‡ |
| 18* | M | VH3-21, JH6 | C/II | <1 y | 31.2; bi | 64.2 | Normal |
| 19* | UM | VH1-69 | A/I | <1 y | 58.0 | 24.5 | del(11)(q23)‡ |
| 20* | M | VH4-34, D3-22, JH4 | B/II | <1 y | 44.5 | 55.0 | del(17)(p13) |
| 21 | M | VH3-7, D2-2, JH6 | A/0 | <1 y | 3.6 | 31.8 | del(13)(q14) |
| 22* | M | VH3-33 | C/III | <1 y | 1.6 | 8.3 | del(13)(q14) |
| 23* | UM | VH1-69, D3-3, JH5 | A/I | <1 y | 14.9 | 46.4 | +12 |
| 25* | M | VH3-74, D5-12, JH5 | A/0 | >1 y | 95.3 | 8.5 | +12 |
| 27* | UM | VH1-69, D3-3 | A/I | <1 y | 3.55 | 43.1 | del(11)(q23)‡ |
| 28* | UM | VHI-69, D3-3 | A/0 | <1 y | 14.5; bi | 44.4 | del(13)(q14) |
| 29 | M | VH4-61, D3-10, JH6 | C/0 | >1 y | 5.1; bi | 42.3 | del(13)(q14) |
| 30 | UM | VH1-69, D3-22, JH2 | A/0 | <1 y | 8.5; bi | 38.1 | del(17)(p13)‡ |
| 31 | M | VH3-48 | A/0 | 1 y | 68 | 60.7 | del(17)(p13)‡ |
| 32 | M | VH1-2 | A/0 | >1 y | 1.1 | 4.7 | del(13)(q14)† |
| 33 | M | VH3-21 | A/0 | >1 y | 4.4 | 13.8 | ND |
| 34 | M | VH1-46 | A/0 | >1 y | 51.4; bi | 21.8 | +12 |
| 35* | M | VH2-5, D3-10, JH4 | A/II | >1 y | 24.4; bi | 12.7 | del(17)(p13)‡ |
| 36 | M | VH3-23, D1-1, JH5 | A/0 | >1 y | 4.9 | 7.7 | Normal |
| 37 | M | VH5-51 | A/0 | >1 y | 2.35 | 11.1 | del(13)(q14)† |
| 38 | M | VH3-33, D7-27, JH6 | A/0 | >1 y | 0.75 | 0.9 | del(13)(q14) |
Reagents and antibodies
CXCL12, CCL19, and CCL21 were obtained from Peprotech (London, United Kingdom). The following antibodies were used in this study for detection of human surface antigens: anti-CD5–FITC monoclonal antibody (mAb), anti-CD19–RPECy5 mAb (Dako, Glostrup, Denmark); anti-CD3–RPE, anti-CD56–RPE, anti-CD38–PE mAb, anti-CD5–PC5, and anti-CD19–ECD mAbs (Beckman Coulter, High Wycombe, United Kingdom); anti-CXCR4–PE mAb and anti-CCR7–PE mAb (R&D Systems, Abingdon, United Kingdom). The Intrastain cell permeabilization kit was purchased from Dako. The following antibodies were used in permeabilization studies: anti–human ZAP-70 mAb clone 2F3.2 (Upstate, Lake Placid, NY) or anti–human ZAP-70–FITC mAb clone 1E7.2 (Insight, Wembley, United Kingdom) and sheep anti–mouse immunoglobulin-FITC (NovoCastra Laboratories, Newcastle upon Tyne, United Kingdom). CD3 Dynabeads were used in cellular isolations (Dynal Biotech, Wirral, United Kingdom). For blocking studies and Western blotting the following antibodies were used: anti-CXCR4 (Fusin) mAb (R&D Systems); rabbit anti–phospho p44/42 MAPK (Thr202/Tyr204) and rabbit anti-p44/42 MAPK antibodies, goat anti–rabbit IgG-HRP secondary antibody (NEB, Hitchin, United Kingdom); rabbit anti–sheep IgG-HRP and goat anti–mouse IgG-HRP (Upstate). FITC-phalloidin, wortmannin, pertussis toxin (PTX), paraformaldehyde, 1-α-lysophosphatidylcholine (LPC), and HEPES were obtained from Sigma (Poole, United Kingdom).
Analysis of surface protein expression by flow cytometry
To determine the level of CXCR4 and CCR7 expression on CLL B cells or healthy donor B cells, 106 freshly isolated lymphocytes or cells from cryopreserved specimens were incubated with 5 μL of anti-CD5–FITC and anti-CD19–RPECy5 antibodies and with 10 μL anti-CXCR4–PE (clone 44717) or anti-CCR7 (clone 150503) or appropriate isotype control antibody for 40 minutes at 4°C. At least 5000 specifically labeled CD19+ healthy PB B cells and 10 000 PB CLL cells were analyzed.
Analysis of CD38 expression on B-CLL cells was carried out by incubating either whole blood or 106 cells from cryopreserved specimens with 5 μL of the antibodies anti-CD5–FITC, anti-CD38–PE, and anti-CD19–RPECy5 for 20 minutes and at least 10 000 cells were counted. Each sample was run with appropriate isotype control antibodies to define the negative-staining cells. The percentage of CD38+ cells was defined as the percentage of CD19+ CD5+ cells that were CD38+. The threshold for CD38 expression was set at 30%; less than 30% were defined as CD38– and greater than 30% as CD38+, as previously described.7
Analysis of ZAP-70 expression by flow cytometry
Isolated lymphocytes, as well as whole blood, were incubated with the following antibodies: anti-CD3–PE, anti-CD56–PE, anti-CD19–ECD, anti-CD5–PC5. Cells were then fixed using reagent A from the Dako Intrastain kit, per the manufacturer's instructions. Permeabilization with reagent B (Dako Intrastain) was carried out in the presence of either 1.5 μg anti–ZAP-70 antibody (Upstate) or an IgG2a isotype control antibody (Dako) or in later experiments with 2 μL anti–ZAP-70–FITC (Insight) or an IgG1 isotype control antibody for 40 minutes. Where appropriate, cells were washed and then incubated for 20 minutes with goat anti–mouse immunoglobulin–FITC. After a further wash step, cells were analyzed with a Beckman Coulter Epics MCL flow cytometer. Analysis of stained samples was carried out using the Expo 32 analysis software (Beckman Coulter). Lymphocytes were gated to avoid inclusion of doublets and debris. The CD3+ and CD56+ (T and NK) cells act as internal positive controls. The percentage of B cells positive for ZAP-70 was determined by gating the CD19+CD5+ population and examining on the FL-1 (ZAP-70–FITC) versus FL-2 (CD3/CD56-PE) plot, where the quadrants have been first set using the isotype control antibody (< 0.5% cells ZAP-70+). The threshold was set at 20%; less than 20% were defined as ZAP-70– and greater than 20% as ZAP-70+, as previously described.5,6
Immunoglobulin VH gene status
IgVH mutational status and gene usage investigations were performed using a multiplex polymerase chain reaction (PCR) method and BIOMED-2 standardized primers and protocol, as previously described.37 The sequences were analyzed using the National Institutes of Health (NIH) Ig blast database38 and a cutoff at 98% concordance with germ line sequences used to differentiate between mutated (< 98% homology) and unmutated (> 98% homology) IgVH status.37
Interphase FISH
Slides were prepared for interphase fluorescence in situ hybridization (FISH) from stored, DMSO preserved, frozen patient cells that were thawed, treated with hypotonic solution, and fixed with methanol/acetic acid according to standard cytogenetic procedures. Separate hybridizations were undertaken using single-locus probes for ATM (chromosome 11q23), p53 (chromosome 17p13), and D13S319 (chromosome 13q14) and an alpha satellite probe for chromosome 12 centromere. Hybridization was undertaken according to the manufacturer's protocol. Single-locus probes were cohybridized with a centromere probe from the same chromosome or, in the case of D13S319, with a further single-locus probe from 13q34. (All probes were manufactured by Vysis [Downers Grove, IL], supplied by Abbott Diagnostics [Maidenhead, United Kingdom].) A minimum of 200 interphase nuclei were assessed for each hybridization.
Cell isolations and treatments
For Western blotting analysis of ERK activation, patient samples with less than 95% CLL B cells were enriched by removing T cells, using CD3 Dynabeads, as per the manufacturer's instructions. The enriched population contained greater than 95% CLL B cells as assessed by staining with anti-CD5–FITC and anti-CD19–RPECy5 antibodies. For the blocking studies, cells were preincubated with 30 μg/mL anti-CXCR4 mAb (Fusin) or 500 nM wortmannin for 30 minutes at 37°C. For PTX pretreatment, cells were preincubated with 200 ng/mL PTX for 2 hours at 37°C. Following the incubation, cells were washed twice and incubated with CXCL12 (100 ng/mL) for varying times before protein was collected. Protein from untreated (t = 0) cells was collected for comparison.
Protein collection and Western blotting
Cells were lysed in a buffer (10 mM Tris-HCl, 67.5 mM NaCl, 0.75 mM MgCl2, 0.5 mM EGTA, 1% Triton X-100, 5% glycerol) containing 1:200 dilution of Calbiochem (Nottingham, United Kingdom) protease inhibitors 50 mM NaF and 1 mM Na3VO4. Protein concentrations were determined using the BioRad Dc Assay kit (BioRad, Hemel Hempstead, United Kingdom). For Western blotting, 20 μg of total protein was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a PVDF membrane. Membranes were incubated with the primary antibody overnight (rabbit anti-p42/44 ERK [T-ERK] or anti–phospho-p42/44 ERK [P-ERK; NEB]). After washing 3 times with TBS/Tween 20 (0.05%), membranes were incubated with appropriate secondary antibodies conjugated to HRP (NEB, Upstate). Following 3 further washes, the blots were developed using Enhanced Chemiluminescence Kit (Amersham Pharmacia, Buckingham, United Kingdom). Densitometric quantification was carried out using a BioRad Fluor S Multi-imager and readings were normalized by calculating the ratio of P-ERK to T-ERK.
Intracellular F-actin polymerization to determine responsiveness to chemotactic signals
Isolated lymphocytes were labeled with anti-CD5–PC5 and anti-CD19–ECD (Beckman Coulter) for 20 minutes at room temperature. Cells were washed and resuspended in RPMI containing 25 mM HEPES at 5 × 106 cells/mL and incubated at 37°C for 10 minutes. Actin polymerization was assessed in cells incubated with CXCL12/CCL19 or CCL21 (100 ng/mL, previously worked out to be an optimal concentration following a dose response; data not shown) for varying amounts of time (0, 15, 30, 45, and 60 seconds). At the indicated time points, 100 μL of the cell suspension was added to 400 μL F-actin buffer (PBS, 4% paraformaldehyde, 10–7 M FITC-labeled phalloidin, 125 μg/mL 1-α-lysophosphatidylcholine) and incubated at 37°C for 10 minutes. The fixed cells were then analyzed by flow cytometry on a Beckman Coulter EPICS-MCL and all time points were plotted relative to the mean relative fluorescence of the sample before addition of the chemokine.
Chemotaxis assay
The chemotaxis assay across bare polycarbonate was performed as described previously.39 Briefly, CLL cells were resuspended in RPMI 1640 with 0.5% BSA. A total of 100 μL, containing 5 × 105 cells, was added to the top chamber of a 6.5-mm diameter Transwell culture insert (Costar, VWR, Lutterworth, United Kingdom) with a pore size of 5 μm. Filters were then transferred into wells containing medium with the chemokines CCL19, CCL21 (1 μg/mL), or CXCL12 (100 ng/mL) or migration buffer alone. The concentrations of the chemokines were those that induced maximal migration as described previously.18 The chambers were incubated for 6 hours at 37°C in 5% CO2. After this incubation, the cells in the lower chamber were thoroughly resuspended and 100-μL aliquots were collected for counting with a Beckman Coulter Epics MCL flow cytometer. Transmigrated cells were counted by adding in a known number of Flow Check beads and counting 200 beads/sample. All the assays were performed in triplicate. To determine the CLL cell content of the transmigrated cells, an aliquot was stained for CD19 and CD5 and analyzed by flow cytometry. The migration index (MI; no. of CD19+CD5+ cells transmigrating with chemokine divided by the no. of cell transmigrating in the absence of chemokine) was then calculated. Student t test was used to determine the statistical significance of the results.
CLL survival studies
CLL cells were cultured for 14 days in complete media to allow the outgrowth of NLCs as previously described.28 After 14 days the CLL cells were removed from the NLCs and cultured in either the presence or absence of CXCL12 (500 ng/mL).28 To determine the involvement of ERK in CXCL12-mediated survival, cells were pretreated with the MEK inhibitor PD98059 (Calbiochem) at a concentration of 50 μM for 1 hour. Cells were washed once in complete media prior to the addition of CXCL12. Cellular viability was assessed at 24 hours using the annexin V–PE/PI kit (Becton Dickinson, Cowley, Oxford, United Kingdom) as per the manufacturer's instructions on duplicate samples from each treatment. CLL cell viability was presented as a percentage of the control value observed at t = 0 hours.
Statistics
Differences between chemokine receptor expression, responsiveness to chemokines, and levels of P-ERK activation were analyzed using a 2-sample, 2-tailed Student t test. Associations between ZAP-70 and LDT were determined using a chi-square test. Survival or viability differences following coculture experiments were examined using the mean plus or minus the standard error of the mean.
Results
ZAP-70 protein expression in CLL B cells and correlation with IgVH mutational status
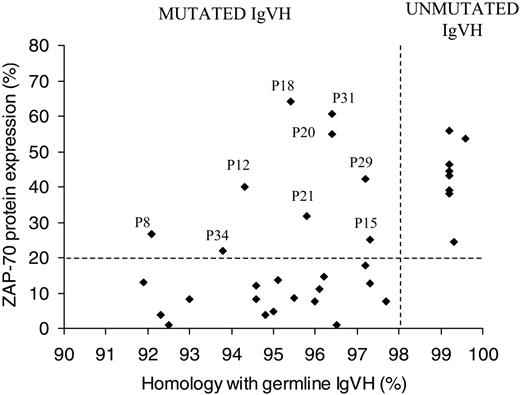
We examined the expression of ZAP-70 in 34 patients with B-CLL by flow cytometry and correlated this with IgVH mutational status. Patients were defined as being ZAP-70+ when expression was greater than 20%. Within our patients we found a clear association between ZAP-70 expression (> 20%) and unmutated IgVH genes (8/8; 100%; Figure 1). Seventeen of 26 (65%) patients with mutated IgVH genes had no/low ZAP-70 protein expression (< 20%). Nine patients (26%) were discordant for ZAP-70 expression and IgVH. Of these, all 9 were ZAP-70+ and had mutated IgVH. Closer analysis of these patients revealed that all had between 1 and 3 additional poor prognostic markers, for example, CD38 positivity/bimodal expression,8,35 unfavorable IgVH gene usage,40 unfavorable chromosomal aberrations (11q23 or 17p13 deletions, trisomy 12),9,10 and LDTs of less than a year (Table 1). In addition, 5 (55%) of 9 of these patients were diagnosed at stage B or stage I and above compared with 3 (17.6%) of 17 ZAP-70– mutated patients. ZAP-70 expression was strongly associated with LDT (χ2 = 9.3; degrees of freedom [df] = 1; P < .01; Table 2), in that 80% of ZAP-70+ patients have an LDT shorter than 1 year and 81% of ZAP-70– patients have an LDT longer than 1 year.
ZAP-70 expression strongly correlated with LDT
. | No. patients (%) . | . | |
|---|---|---|---|
| LDT . | ZAP-70+ . | ZAP-70- . | |
| Longer than 1 y | 3 (9.7) | 13 (41.9) | |
| 1 y or shorter | 12 (38.7) | 3 (9.7) | |
. | No. patients (%) . | . | |
|---|---|---|---|
| LDT . | ZAP-70+ . | ZAP-70- . | |
| Longer than 1 y | 3 (9.7) | 13 (41.9) | |
| 1 y or shorter | 12 (38.7) | 3 (9.7) | |
Correlation between ZAP-70 expression and LDT (chi-square = 9.3; df = 1; P < .01). Eighty percent of ZAP-70+ patients had an LDT shorter than 1 year and 81% of ZAP-70- patients had an LDT longer than 1 year.
ZAP-70+ CLL B cells express significantly higher levels of CCR7 than ZAP-70– CLL B cells but they express similar levels of CXCR4
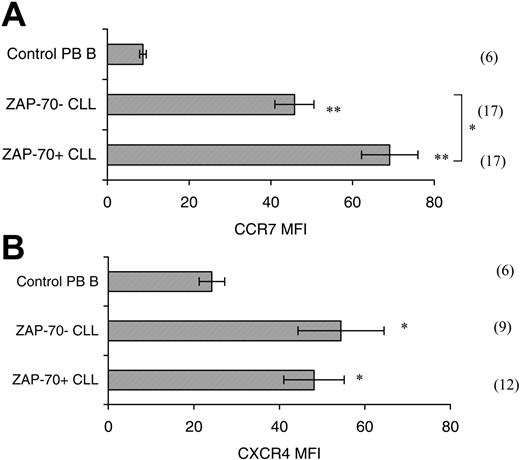
Examination of surface CCR7 levels revealed that CLL PB cells express significantly higher levels (5- to 7-fold increase in mean fluorescent intensity (MFI; P < .001) when compared with control PB B cells (Figure 2A). Comparison of CCR7 levels in ZAP-70+ (n = 17) and ZAP-70– (n = 17) CLL cells revealed that ZAP-70+ CLL B cells have significantly higher (MFI ± SEM; 69.1 ± 6.9) expression than ZAP-70– CLL cells (45.8 ± 4.8; Figure 2A; P = .009). The level of CXCR4 surface expression is significantly higher in CLL B cells when compared with B cells from healthy donors (P < .05; Figure 2B).41,42 Comparison of CXCR4 expression in ZAP-70+ (n = 12) and ZAP-70– patients (n = 9) revealed that expression levels did not differ significantly between the groups (P = .53). In both cases, however, the level of expression was significantly different from that seen in healthy donor B cells (ZAP-70+ vs healthy B, P = .016; ZAP-70– vs healthy B, P = .012).
Correlation of ZAP-70 expression with IgVH mutational status. Sixty-five percent of patients with mutated IgVH gene were ZAP-70– as assessed by flow cytometry and 100% of patients with unmutated IgVH genes were ZAP-70+. Twenty-six percent of patients were ZAP-70+ and mutated, of these patients the majority had additional bad prognostic markers (ie, CD38 positivity or unfavorable IgVH gene usage; Table 1).
Correlation of ZAP-70 expression with IgVH mutational status. Sixty-five percent of patients with mutated IgVH gene were ZAP-70– as assessed by flow cytometry and 100% of patients with unmutated IgVH genes were ZAP-70+. Twenty-six percent of patients were ZAP-70+ and mutated, of these patients the majority had additional bad prognostic markers (ie, CD38 positivity or unfavorable IgVH gene usage; Table 1).
Surface CCR7 and CXCR4 expression. (A) Surface CCR7 expression is increased on CLL B cells when compared with healthy PB B cells, and ZAP-70+ CLL B cells have a significantly higher surface expression than ZAP-70– CLL B cells. CLL PB B cells have a 5- to 7-fold higher level of surface CCR7 expression than control PB B cells (Student t test; **P < .001). ZAP-70+ CLL B cells have a significantly higher CCR7 expression than ZAP-70– cells (*P = .009). The n numbers are shown in parentheses. Isotype control antibodies staining MFI mean ± SEM; 0.3 ± 0.03. (B) Surface CXCR4 expression is significantly increased on CLL B cells compared with B cells from healthy controls. CXCR4 MFI for ZAP-70+ CLL B cells (n = 12), ZAP-70– CLL B cells (n = 9), and control PB B cells (n = 6). The MFI of both ZAP-70+ and ZAP-70– CLL samples differs significantly from the control PB B-cell MFI (Student t test; *P < .05). There was no significant difference between the CXCR4 MFI of ZAP-70+ and ZAP-70– CLL cells (P = .53).
Surface CCR7 and CXCR4 expression. (A) Surface CCR7 expression is increased on CLL B cells when compared with healthy PB B cells, and ZAP-70+ CLL B cells have a significantly higher surface expression than ZAP-70– CLL B cells. CLL PB B cells have a 5- to 7-fold higher level of surface CCR7 expression than control PB B cells (Student t test; **P < .001). ZAP-70+ CLL B cells have a significantly higher CCR7 expression than ZAP-70– cells (*P = .009). The n numbers are shown in parentheses. Isotype control antibodies staining MFI mean ± SEM; 0.3 ± 0.03. (B) Surface CXCR4 expression is significantly increased on CLL B cells compared with B cells from healthy controls. CXCR4 MFI for ZAP-70+ CLL B cells (n = 12), ZAP-70– CLL B cells (n = 9), and control PB B cells (n = 6). The MFI of both ZAP-70+ and ZAP-70– CLL samples differs significantly from the control PB B-cell MFI (Student t test; *P < .05). There was no significant difference between the CXCR4 MFI of ZAP-70+ and ZAP-70– CLL cells (P = .53).
ZAP-70+ CLL B cells are more responsive to CCL19 and CCL21 than ZAP-70– CLL cells but are equally responsive to CXCL12, as determined by changes in F-actin polymerization and cellular chemotaxis
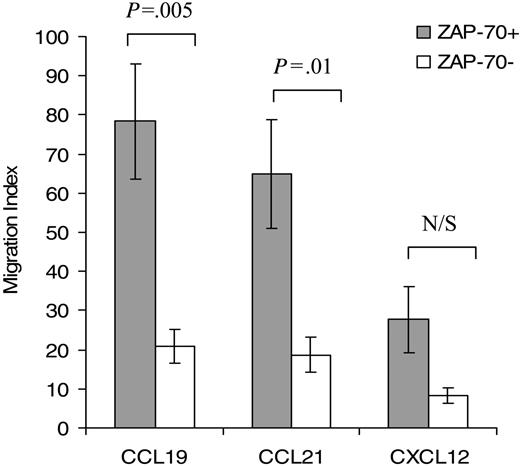
Both CCL19 and CCL21 treatment resulted in an increase in intracellular F-actin at 15 seconds, which declined thereafter toward pretreatment levels (Figure 3). In ZAP-70+ cells significantly greater responses to both CCL19 (P < .05) and CCL21 (P < .05) were observed when compared with ZAP-70– cells (Figure 3A and 3B, respectively). CXCL12 treatment resulted in increased intracellular levels of F-actin but there was no significant difference in response between ZAP-70+ and ZAP-70– patients (Figure 3C). CLL cell migration was also significantly greater in ZAP-70+ CLL cells toward CCL19 (P = .005) and CCL21 (P = .01) but not CXCL12 when compared with ZAP-70– CLL cells when assessed using a transwell chemotaxis assay (Figure 4).
ZAP-70+ CLL cells are more responsive to CCL19 and CCL21, but not CXCL12, when compared with ZAP-70– CLL. CLL B cells were treated for 0, 15, 30, 45, and 60 seconds with 100 ng/mL CCL19 (A), CCL21 (B), or CXCL12 (C). Changes in intracellular F-actin were measured using FITC-labeled phalloidin in CD19, CD5-prelabeled CLL cells. Results are shown as the percent change in intracellular F-actin relative to the level observed in untreated cells. Results for each chemokine are the mean ± SEM of at least 13 independent experiments. Student paired t test; *P < .05.
ZAP-70+ CLL cells are more responsive to CCL19 and CCL21, but not CXCL12, when compared with ZAP-70– CLL. CLL B cells were treated for 0, 15, 30, 45, and 60 seconds with 100 ng/mL CCL19 (A), CCL21 (B), or CXCL12 (C). Changes in intracellular F-actin were measured using FITC-labeled phalloidin in CD19, CD5-prelabeled CLL cells. Results are shown as the percent change in intracellular F-actin relative to the level observed in untreated cells. Results for each chemokine are the mean ± SEM of at least 13 independent experiments. Student paired t test; *P < .05.
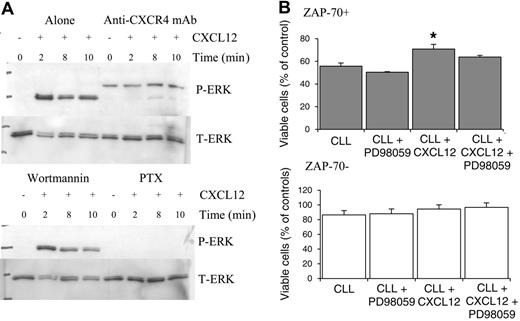
CXCL12 induces sustained activation of ERK in ZAP-70+ CLL cells
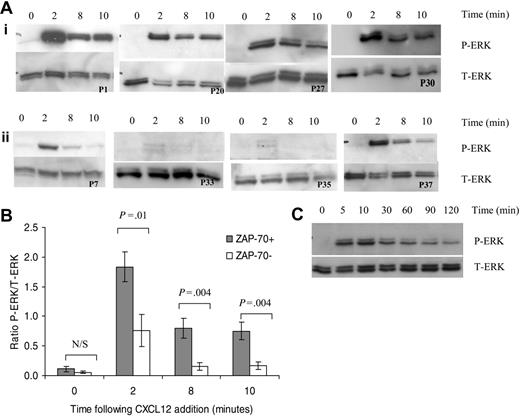
CXCL12 treatment induced rapid phosphorylation of ERK1 and ERK2 after 2 minutes in CLL B cells (Figure 4A). Prolonged activation of ERK was observed at 8 and 10 minutes following CXCL12 addition in 4 of 4 ZAP-70+ patients (Figure 4Ai) but not in those isolated from ZAP-70– patients (Figure 4Aii). Densitometric quantification of P-ERK (active form of ERK) and T-ERK Western blots was carried out for 7 ZAP-70+ and 7 ZAP-70– CLL patients. To compensate for differences in loading between lanes, the ratio of P-ERK readings to T-ERK readings was calculated. ERK activation was significantly increased in ZAP-70+ CLL patients when compared with ZAP-70– CLL patients at 2, 8, and 10 minutes (Figure 5B). Furthermore this ERK signaling was sustained for up to 2 hours in ZAP-70+ cells (Figure 5C).
CXCL12-mediated ERK activation is inhibited by the blocking CXCR4 monoclonal antibody and pertussis toxin but not wortmannin
To confirm that the changes in ERK activation in ZAP-70+ CLL cells were a CXCL12-specific response, anti-CXCR4 blocking antibody studies were carried out. Addition of the anti-CXCR4 mAb significantly inhibited the activation of ERK (Figure 6A). To further analyze the signaling pathways, the effect of the Gi-protein inhibitor pertussis toxin (PTX) and the broad-spectrum phosphatidylinositol-3-kinase (PI3K) inhibitor wortmannin were also assessed. Wortmannin had no effect on ERK activation. In contrast, PTX potently inhibited the changes in ERK activation following CXCL12 treatment.
CXCL12 promoted survival in ZAP-70+ CLL cells that was partially dependent on the presence of ERK
CLL cells when cultured in vitro undergo a process of spontaneous apoptosis. In ZAP-70+ CLL cells, CXCL12 treatment resulted in a significant increase in cell viability (n = 3; P < .05) and this effect could be partially inhibited by blocking ERK activation by pretreating cells with the MEK inhibitor PD98059. In ZAP-70– CLL cells, CXCL12 treatment did cause a slight increase in survival but this did not reach significance and inhibition of ERK was without effect.
Migration of ZAP-70+ CLL cells toward CCL19 and CCL21, but not CXCL12, is increased when compared with ZAP-70– CLL cells. PB CLL cells were assayed in the bare filter chemotaxis assay for migration in response to CCL19, CCL21 (1 μg/mL), or CXCL12 (100 ng/mL). CLL cell migration is expressed as a migration index as described in “Patients, materials, and methods” under “Chemotaxis assay.” The concentration of the chemokines were those that induced maximal migration.18 Input and transmigrated cells were stained with anti-CD19 and anti-CD5 antibodies to determine the percentages of input CLL B cells that migrated into the lower chambers. Results for each chemokine are the mean ± SEM of 5 independent experiments. Student 2-sample 2-tailed t test. N/S indicates not significant.
Migration of ZAP-70+ CLL cells toward CCL19 and CCL21, but not CXCL12, is increased when compared with ZAP-70– CLL cells. PB CLL cells were assayed in the bare filter chemotaxis assay for migration in response to CCL19, CCL21 (1 μg/mL), or CXCL12 (100 ng/mL). CLL cell migration is expressed as a migration index as described in “Patients, materials, and methods” under “Chemotaxis assay.” The concentration of the chemokines were those that induced maximal migration.18 Input and transmigrated cells were stained with anti-CD19 and anti-CD5 antibodies to determine the percentages of input CLL B cells that migrated into the lower chambers. Results for each chemokine are the mean ± SEM of 5 independent experiments. Student 2-sample 2-tailed t test. N/S indicates not significant.
Discussion
This study confirms previous findings that in CLL B cells ZAP-70 expression is associated with the presence of unmutated IgVH genes. There is, however, a proportion of patients who are ZAP-70+ with mutated IgVH genes. All these patients were shown to have between 1 and 3 additional poor prognostic markers and were more likely to be in advanced stages of the disease. Together these findings support recent indications that ZAP-70 expression is a better predictor of disease progression and need for earlier treatment.11 ZAP-70 positivity/expression appears to be identifying a subset of patients with a more progressive form of the disease.
CXCL12 treatment results in prolonged activation of ERK in ZAP-70+ but not ZAP-70– CLL B cells. (A) Lysates were collected from CLL B cells treated for varying times with CXCL12 (100 ng/mL). Western blots were probed with an anti–phospho-p44/42 ERK (P-ERK) antibody (top panel) and then with an anti-p44/42 ERK (T-ERK) antibody to demonstrate loading (bottom panel). (i) ZAP-70+ PB CLL cells. (ii) ZAP-70– PB CLL cells. (B) Densitometric quantification of P-ERK and T-ERK Western blots from 7 ZAP-70+ and 7 ZAP-70– patients was carried out and the ratio of P-ERK to T-ERK was calculated to normalize for loading. Results are presented as mean ± SEM (Student 2-sample 2-tailed t test). (C) ERK activation following addition of CXCL12 in ZAP-70+ CLL B cells is sustained for up to 2 hours.
CXCL12 treatment results in prolonged activation of ERK in ZAP-70+ but not ZAP-70– CLL B cells. (A) Lysates were collected from CLL B cells treated for varying times with CXCL12 (100 ng/mL). Western blots were probed with an anti–phospho-p44/42 ERK (P-ERK) antibody (top panel) and then with an anti-p44/42 ERK (T-ERK) antibody to demonstrate loading (bottom panel). (i) ZAP-70+ PB CLL cells. (ii) ZAP-70– PB CLL cells. (B) Densitometric quantification of P-ERK and T-ERK Western blots from 7 ZAP-70+ and 7 ZAP-70– patients was carried out and the ratio of P-ERK to T-ERK was calculated to normalize for loading. Results are presented as mean ± SEM (Student 2-sample 2-tailed t test). (C) ERK activation following addition of CXCL12 in ZAP-70+ CLL B cells is sustained for up to 2 hours.
This study demonstrates that CCR7 levels are increased on ZAP-70+ cells when compared with ZAP-70– CLL B cells and that this confers an increased ability to respond to its ligands, CCL19 and CCL21. In addition, ZAP-70 identifies CLL cells that can induce sustained activation of ERK following CXCL12 stimulation, which can promote preferential survival in these cells (Figure 6B).
Knockout mouse models have shown the crucial importance of CCL19 and CCL21 in both T- and B-lymphocyte trafficking.16,17,21 Studies in B cells have shown that antigen engagement upregulates expression of CCR7 and can facilitate the movement of these cells into the LN and localization to the B-/T-cell boundary.20 Antigen-stimulated normal B cells respond less strongly to CCL19 and CCL21 than T cells and it is proposed that this is the reason why they do not enter the central T zone but localize to the T-/B-zone boundary.20 In keeping with this, B cells transduced with a Flag-tagged retrovirus containing CCR7 were more responsive to CCL19 and CCL21 in both ex vivo chemotaxis assays and in situ hybridization studies. These cells localized to the T-cell zone and failed to localize within the B-cell follicles.20 Together these findings suggest that increases in CCR7 expression can alter B-cell responsiveness to CCL19 and CCL21 and consequently can alter the area to which the B cell localizes within the LN. These studies have confirmed previous findings that CCR7 is upregulated on the surface of circulating PB CLL B cells when compared with healthy control PB B cells.18,22,43 This suggests that all CLL lymphoid cells, regardless of their ZAP-70 status, may be better than resting normal B cells at migrating to the LN. An association between CCR7 expression and clinical staging has been observed previously, although correlations between mutational status of the IgVH gene, CD38 positivity, and CCR7 expression were not found.18,22,43,44 This study shows for the first time that there is an association between ZAP-70 positivity and enhanced CCR7 expression (Figure 2A). As increases in CCR7 expression have been documented following antigen contact, this could be reflecting either an increased level of antigen contact, which is likely to be the case for both ZAP-70+ and ZAP-70– CLL B cells, as both cell types have been shown to resemble activated B cells,45 or an increased ability to respond to antigen contact. This is particularly relevant in the ZAP-70+ CLL B cells in view of the finding that ZAP-70 can enhance signaling through the B-cell receptor (BCR).46,47
CXCL12-induced ERK activation is inhibited by anti-CXCR4 mAb and PTX but not the PI3K inhibitor wortmannin in ZAP-70+ cells, and inhibition of ERK activation in ZAP-70+ CLL cells results in partial inhibition of CXCL12-mediated survival. CLL B cells were pretreated with media (alone), anti-CXCR4 antibody, wortmannin, or PTX prior to CXCL12 treatment. (A) Anti-CXCR4 mAb and PTX inhibit ERK activation, whereas wortmannin was without effect. A representative experiment of 4 is shown. (B) CLL B cells from 3 ZAP-70+ and 3 ZAP-70– CLL patients were pretreated with PD98059 (50 μM for 1 h) prior to the addition of CXCL12 (500 ng/mL) and viability was assessed at 24 hours using annexin V/PI staining. Results are presented as mean ± SEM (Student 2-sample 2-tailed t test; *P < .05).
CXCL12-induced ERK activation is inhibited by anti-CXCR4 mAb and PTX but not the PI3K inhibitor wortmannin in ZAP-70+ cells, and inhibition of ERK activation in ZAP-70+ CLL cells results in partial inhibition of CXCL12-mediated survival. CLL B cells were pretreated with media (alone), anti-CXCR4 antibody, wortmannin, or PTX prior to CXCL12 treatment. (A) Anti-CXCR4 mAb and PTX inhibit ERK activation, whereas wortmannin was without effect. A representative experiment of 4 is shown. (B) CLL B cells from 3 ZAP-70+ and 3 ZAP-70– CLL patients were pretreated with PD98059 (50 μM for 1 h) prior to the addition of CXCL12 (500 ng/mL) and viability was assessed at 24 hours using annexin V/PI staining. Results are presented as mean ± SEM (Student 2-sample 2-tailed t test; *P < .05).
The increased CCR7 expression on the surface of ZAP-70+ cells conferred an increased ability to respond to both its ligands, CCL19 and CCL21, shown by both an increase in cellular chemotaxis and F-actin polymerization when compared with ZAP-70– cells. This may provide the ZAP-70+ cells with a selective advantage over the ZAP-70– cells, in that they may have an increased propensity to migrate into the LN through interactions with CCL19 and CCL21 on high endothelial venules (HEVs). Receptor endocytosis can occur following binding of the receptor to its respective ligand, resulting in lower surface expression.41,42 Therefore the finding that CCR7 levels are lower in LN CLL B cells than in PB CLL B cells is suggestive of it being involved in migration to the LN.18,22,43,44 Once within the LN, these cells could also be capable of migrating to the T-cell area, increasing the likelihood of contact with Th cells. Contact with CD40L+CD4+ T cells aids in CLL cell proliferation and survival through upregulation of survivin48 and through stimulating release of CCL17 and CCL22, chemokines that will attract further CD4+ Th cells,23 perpetuating the cycle. These studies have not shown that ZAP-70 directly contributes to the CLL cells' ability to respond differently to CCL19 and CCL21; however, increased CCR7 expression on the surface of ZAP-70+ cells and the subsequent increase in responsiveness to CCR7 ligands could well be a reflection of the activation status of the cell.20 Indeed, several groups have now demonstrated that ZAP-70+ cells have an enhanced ability to stimulate activation of intracellular signaling pathways following stimulation of the BCR, contributing to an enhanced cellular activation status.46,47 ZAP-70 may well play an indirect role through enhancing cellular activation mediated by BCR signaling. This could be further evidence to suggest that this disease is antigen driven.49,50
CXCL12 has previously been shown to have both a chemotactic effect and a prosurvival effect on CLL B cells, being a crucial mechanism through which stromal cells or NLCs support CLL B cells in vitro.28,42 The finding that CLL B cells proliferate at sites where stromal cells are present suggests that CXCL12 could be an important factor in CLL pathogenesis. This study confirms an increased level of CXCR4 on the surface of CLL B cells when compared with B cells from healthy volunteers.41,42 Treatment of CLL B cells with CXCL12 resulted in a rapid increase in intracellular F-actin levels, cellular chemotaxis, and ERK activation. The increase in intracellular F-actin levels, an early indicator of chemotaxis, was comparable in both ZAP-70+ and ZAP-70– CLL B cells. In keeping with this there was no significant difference in cellular chemotaxis between the 2 cell types following treatment with CXCL12. In contrast, the ERK activation profile of these 2 cell types differed. In the ZAP-70+ cells a rapid and sustained ERK activation profile (up to 2 hours) was observed, which contrasted with that seen in ZAP-70– cells. Sustained ERK activation can lead to the initiation of transcription of genes involved in both proliferation and survival.28,32,51 Previous studies have shown that sustained ERK activation (up to 2 hours) is required for the production and stabilization of intermediate early gene products, such as c-Fos, which can then control cellular survival and proliferative responses.51,52 These findings suggest that the sustained activation of ERK observed in ZAP-70+ cells could confer both survival and proliferative advantages to the CLL cell. In keeping with the finding of rapid and sustained ERK activation upon appropriate stimulation of ZAP-70+ cells, this study also shows that survival was enhanced in these cells following exposure to CXCL12. This was partially inhibited by blocking ERK activation using the MEK inhibitor PD98059, indicating that ERK does have a role in promoting survival in these cells. In ZAP-70– CLL cells, no significant increase in survival was observed following addition of CXCL12, and survival of these cells was unaffected by inhibition of ERK (Figure 6B). ERK activation in ZAP-70+ cells required ligation of CXCR4 and G-protein activation, as it was prevented in cells pretreated with a CXCR4 blocking antibody or pertussis toxin. PI3K activation, however, was not required, as shown by the inability of wortmannin to inhibit ERK activation in these cells.
In summary, this study demonstrates that ZAP-70+ cells are more responsive to signals derived from their surrounding environment. A model of the differences between a stable form of the disease with a low level of cellular turnover and a progressive form with a higher level of cellular turnover is shown in Figure 7. ZAP-70 identifies cells that may have an increased propensity to migrate to the LN and an increased ability to respond to survival signals. The consequently larger numbers of CLL cells within the LN would enable recruitment of more Th cells to further aid in their proliferation and survival. This access to a favorable niche microenvironment could potentially result in a faster accumulation of tumor load and hence a more progressive form of the disease.
Stable and progressive CLL model. In stable CLL (ZAP-70–), moderate levels of surface CCR7 expression facilitate low levels of CLL cell recruitment to the LN. Once within this environment they are stimulated to produce CCL17 and CCL21 that aid in the recruitment of CD4+CD40L+ Th cells that promote CLL proliferation and survival. In progressive CLL, ZAP-70+ CLL cells have a greater propensity to migrate to the LN, due to increased surface expression of CCR7. The higher numbers of CLL cells within this environment can recruit greater numbers of CD4+CD40L+ Th cells, leading to the formation of more proliferation centers. In addition, the ZAP-70+ CLL cells are more responsive to the survival factor CXCL12. Collectively this will result in an accumulation of CLL cells within the LN, a higher tumor burden, and cellular turnover.
Stable and progressive CLL model. In stable CLL (ZAP-70–), moderate levels of surface CCR7 expression facilitate low levels of CLL cell recruitment to the LN. Once within this environment they are stimulated to produce CCL17 and CCL21 that aid in the recruitment of CD4+CD40L+ Th cells that promote CLL proliferation and survival. In progressive CLL, ZAP-70+ CLL cells have a greater propensity to migrate to the LN, due to increased surface expression of CCR7. The higher numbers of CLL cells within this environment can recruit greater numbers of CD4+CD40L+ Th cells, leading to the formation of more proliferation centers. In addition, the ZAP-70+ CLL cells are more responsive to the survival factor CXCL12. Collectively this will result in an accumulation of CLL cells within the LN, a higher tumor burden, and cellular turnover.
Prepublished online as Blood First Edition Paper, December 6, 2005; DOI 10.1182/blood-2005-04-1718.
Supported by funding provided by the Plymouth District Leukaemia Fund (PDLF).
S.J.R. designed and performed research, analyzed data, and wrote the paper; A.G.P. designed and supervised research and wrote the paper; J.A.C. contributed to research design and wrote the paper; C.M. performed IgVH mutation analysis; M.A.C. designed IgVH mutational status study and performed IgVH mutation analysis; H.D.A. designed IgVH mutational status study and contributed to the paper; A.G. and S.M. performed the FISH analysis; D.O. directed the FISH component of the study; and J.F. and B.S.C. reviewed the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Claire Adams for technical advice and scientific discussions.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal