CD94/NKG2C+ natural killer (NK) cells are increased in healthy individuals infected with human cytomegalovirus (HCMV), suggesting that HCMV infection may shape the NK cell receptor repertoire. To address this question, we analyzed the distribution of NK cell subsets in peripheral blood lymphocytes (PBLs) cocultured with HCMV-infected fibroblasts. A substantial increase of NK cells was detected by day 10 in samples from a group of HCMV+ donors, and CD94/NKG2C+ cells outnumbered the CD94/NKG2A+ subset. Fibroblast infection was required to induce the preferential expansion of CD94/NKG2C+ NK cells that was comparable with allogeneic or autologous fibroblasts, and different virus strains. A CD94-specific monoclonal antibody (mAb) abrogated the effect, supporting an involvement of the lectinlike receptor. Purified CD56+ populations stimulated with HCMV-infected cells did not proliferate, but the expansion of the CD94/NKG2C+ subset was detected in the presence of interleukin-15 (IL-15). Experiments with HCMV deletion mutants indicated that the response of CD94/NKG2C+ NK cells was independent of the UL16, UL18, and UL40 HCMV genes, but was impaired when cells were infected with a mutant lacking the US2-11 gene region. Taken together the data support that the interaction of CD94/NKG2C with HCMV-infected fibroblasts, concomitant to the inhibition of human leukocyte antigen (HLA) class I expression, promotes an outgrowth of CD94/NKG2C+ NK cells.
Introduction
Human cytomegalovirus (HCMV) infection generally follows a subclinical course, but may lead to severe disorders in immunocompromised individuals and is a main cause of infectious congenital diseases. HCMV remains latent in immunocompetent hosts, undergoing occasional reactivation.1 Studies in murine models revealed that an effective defense against CMV requires the participation of natural killer (NK) and T cells.2,3 Detection of antibodies and CD8+ T lymphocytes specific for HCMV antigens allow an assessment of the adaptive immune response to the pathogen.4,5 To escape from CD8+ T cells, HCMV inhibits the expression of human leukocyte antigen (HLA) class I molecules and interferes with antigen presentation using a set of glycoproteins (US2, US3, US6, US10, and US11) whose genes are clustered within the unique short (US) region of the virus genome.6-8 The loss of HLA class I molecules in HCMV-infected cells impairs the engagement of inhibitory receptors and prompts the activation of NK cell effector functions; reciprocally, the virus has developed several strategies to evade NK-mediated recognition.9
The nature of receptor-ligand interactions involved in the NK cell response to CMV-infected cells is incompletely understood. In strains of mice expressing the Ly49H receptor, NK cell functions are triggered upon recognition of the m157 mouse CMV (MCMV) glycoprotein, becoming essential to control replication;10,11 by contrast, human activating NK cell receptors (NKRs) specific for HCMV molecules have not been identified. The involvement of activating killer immunoglobulin (Ig)-like receptors (KIRs) and natural cytotoxicity receptors (NCRs; ie, NKp46, NKp30, and NKp44) in the response to HCMV is uncertain. The interaction of the pp65 HCMV tegument protein with NKp30 has been reported to inhibit rather than to activate NK cell functions.12 The ability of the UL16 HCMV molecule to interfere with the surface expression of NKG2D ligands,13-15 and the evidence that similar evasion mechanisms operate in MCMV infection,16,17 support an important role for this killer lectinlike receptor (KLR) in the antiviral defense.18 Recently, the UL141 HCMV molecule has been shown to inhibit the expression in infected cells of CD155, a ligand for the DNAM-1 stimulating receptor.19
HCMV may also escape NK-mediated surveillance by keeping inhibitory receptors for HLA class I molecules engaged. The viral UL18 molecule binds with high affinity to the ILT2 (CD85j) inhibitory receptor,20,21 though its role in immune evasion has not been precisely elucidated.9 On the other hand, HLA-E appears constitutively resistant to the action of US2 and US11,22 and it also becomes refractory to the action of US6 when bound to a peptide from the leader sequence of the HCMV UL40 protein.23,24 Thus, the HLA class Ib molecule may be preserved in infected cells and interfere with the NK cell response by engaging the inhibitory CD94/NKG2A KLR.25
We recently reported26 that healthy HCMV-seropositive individuals displayed increased proportions of NK and T cells that expressed the triggering CD94/NKG2C KLR, which binds HLA-E with a lower affinity than CD94/NKG2A.27,28 Increased numbers of circulating CD85j+ T lymphocytes were also significantly associated to HCMV infection. Taken together, the data supported that the challenge exerted by HCMV on the innate immune system might shape the NKR repertoire.
Signaling by NCRs and KIRs may control the proliferation and/or survival of NK cells;29,30 we recently reported that CD94/NKG2C+ NK- and T-cell subsets divided in response to stimulation with an HLA class I-deficient tumor cell line transfected with HLA-E.31 On that basis, we hypothesized that CD94/NKG2C-mediated recognition of virus-infected cells might promote the expansion of NKG2C+ cell subsets, as described for Ly49H+ cells in MCMV-infected mice.32 To address this issue, we analyzed the NKR repertoire in peripheral blood lymphocytes (PBLs) cocultured with HCMV-infected fibroblasts; little information is available on the NK cell response in this system, widely used to study cytotoxic T lymphocytes (CTLs) specific for viral antigens. Our results indicate that stimulation of PBLs from HCMV+ donors with virus-infected fibroblasts elicited a preferential expansion of CD94/NKG2C+ NK cells, and that a cognate interaction of the activating KLRs with HCMV-infected cells may contribute to drive the proliferation of this lymphocyte subset.
Materials and methods
Subjects
Heparinized blood samples were obtained from 15 healthy adult individuals (aged 23-51 years); written informed consent was obtained, and the study protocol was approved by the Comite de Etica e Investigacion-Institut Municipal d'Assistencia Sanitaria (CEIC-IMAS). Standard clinical diagnostic tests were used to analyze serum samples for circulating IgG antibodies against CMV (Abbott Laboratories, Abbott Park, IL); 4 donors were seronegative (HCMV-) and 11 were seropositive (HCMV+).
Cell lines
MRC-5 fetal human lung fibroblasts and human foreskin fibroblast (HFF) cell lines were obtained from the American Type Culture Collection (Manassas, VA) and grown in Dulbecco modified essential medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, penicillin, and streptomycin (referred to as complete medium). Primary fibroblast lines were established from skin biopsies obtained from 2 subjects following standard procedures; briefly, tissue samples were minced, treated with 0.2% collagenase A (Roche Diagnostics, Mannheim, Germany), washed, and grown in complete medium.
Cell lines were screened and shown to be negative for Mycoplasma contamination by polymerase chain reaction (PCR) using primers as described.33 Fibroblast lines used were between passages 8 and 15 (HFF), 20 and 30 (MRC-5 cells), and 4 and 6 (primary fibroblast lines).
HCMV preparations and infection of fibroblasts
Stocks of HCMV strains AD169, Towne, and Toledo and HB5-derived mutants were prepared, and titers of infectious virus determined by standard procedures on HFF or MRC-5 cells. Viral preparations were screened for Mycoplasma as indicated. The Towne strain of HCMV was purified as described34 with slight modifications. Ultraviolet (UV)-inactivation of virus was performed using a 30 W Phillips lamp (model G13; Phillips, Eindhoven, The Netherlands) and confirmed by monitoring expression of the IE1 protein on MRC-5 cells by indirect immunofluorescence. Recombinant HCMV genomes were generated in Escherichia coli. The AD169-derived HCMV bacterial artificial chromosome plasmid (BACmid) HB5,35 which lacks the US2-6 gene region, was used to generate all HCMV mutants described in the study.
The HB5-ΔUS2-11 mutant was constructed by homologous recombination, as described,36 with some modifications. Briefly, 5′:TCACACATACCTTTGTGCATACGGTTTATATATGACCATCCACGCTTATAACGAACCTACGATTTATTCAACAAAGCCACG and 3′:TGCTATAAGACAGCCTTACAGCTTTTGAGTCTAGACAGGGTAACAGCCTTCCCTTGTAAGGCCAGTGTTACAACCAATTAACC PCR primers were used to amplify the kanamycin gene, adding sequences homologous to regions upstream of US7 and downstream of US11, respectively. The 5′ primer corresponds to nucleotide (nt) 197237 to 197296 of the coding strand, and the 3′ primer to nt 201356 to 201297 of the antisense strand of AD169 (GenBank no. NC_001347). The PCR product was transformed into E coli carrying the HB5 BACmid and expressing the Red recombinase system from the plasmid pkD46.37 Recombinant clones were selected at 43°C under chloramphenicol and kanamycin selection, and correct mutagenesis was confirmed by restriction digest analysis, PCR, and Southern blot analysis of isolated BACmid DNA. Reconstitution of the virus mutant was carried out by transfecting 5 μg BACmid into MRC-5 cells using superfect (Quiagen, Hilden, Germany). Human CMV recombinants HB5-ΔUL14-20 and HB5-ΔUL40-42 have been previously described.36
Human fibroblasts in 24- or 96-well plates at 70% confluence were mock or HCMV infected (multiplicity of infection [MOI] = 1). After a 2-hour adsorption period, the inoculum was removed and cells were washed with phosphate-buffered saline (PBS) and incubated in complete medium; in some experiments infection was carried out at a different MOI (0.01-5).
Lymphocyte cultures
PBLs were obtained by centrifugation of heparinized blood on Ficoll-Hypaque (Lymphoprep; Axis-Shield PoC AS, Oslo, Norway). PBLs were incubated in 24-well (2 × 106/well) or 96-well (2 × 105/well) plates in complete medium in the presence of mock- or HCMV-infected fibroblasts; in some experiments PBLs were stimulated with free HCMV (0.05 infectious particles/cell). PBLs were added to infected fibroblasts immediately after the 2-hour adsorption period; in time-course experiments coculture was delayed 24 to 72 hours after infection. All cultures were supplemented with IL-2 (10 U/mL) (Proleukin [human recombinant interleukin 2]; Chiron, Emeryville, CA) at day 3, and in some experiments with IL-15 (10 U/mL; Peprotech Ec, London, United Kingdom). Cells were incubated for 10 to 12 days, fed every 3 days, and eventually split. Some experiments were set up in a Transwell permeable support (0.4 μm; Corning Inc, NY); infected fibroblasts were cultured with and without PBLs, and the phenotype of PBLs incubated separately in the upper chamber was analyzed. CD94/NKG2C+ NK cell clones were derived as previously described.31
Antibodies, immunofluorescence, and flow cytometry analysis
Monoclonal antibodies (mAbs) specific for CD56, CD94, NKG2A, KIR, ILT2, NCR, and NKG2D have been previously detailed.26 HP-1F7 anti-HLA class I was generated in our laboratory;38 3D12 anti-HLA-E was provided by Dr D. Geraghty (Fred Hutchinson Cancer Research Center, Seattle, WA); anti-NKG2C (MAB1381) and anti-NKG2C-PE were from R&D Systems (Minneapolis, MN).
For immunofluorescence staining, cells were pretreated with human aggregated Ig (10 μg/mL) to block FcR, and subsequently labeled with the different mAb and analyzed by flow cytometry (FACScan; Becton Dickinson, Mountain View, CA), as described.26 In experiments where blocking mAbs (CD94 or CD56) were used, harvested cells were stained with anti-CD3-PerCP, anti-NKG2A-FITC, or anti-NKG2C-PE. In some experiments, cells were sorted (FACSvantage; Becton Dickinson) after labeling with CD56-PE and NKG2C-PE.
Detection of HCMV IE1 was performed by indirect immunofluorescence with mAb 8130 (Chemicon, Temecula, CA) and an FITC-labeled anti-mouse IgG (Sigma, St Louis, MO).
Proliferation and cytotoxicity assays
As described,31 PBLs were resuspended in RPMI-1640 (107/mL) and incubated for 10 minutes at 37°C with the intracellular fluorescent dye 5(6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) (2 μM). After 2 washes, CFSE-labeled PBLs were cultured on mock- and HCMV-infected MRC-5. Fluorescence-activated cell-sorting (FACS) analysis was performed at 7 days, after staining cells with anti-NKG2C or anti-NKG2A mAbs by indirect immunofluorescence with PE-tagged F(ab′)2 rabbit anti-mouse Ig (Dakopatts, Glostrup, Denmark). The proportions of dividing cells within each subset were calculated as described.39
CD94/NKG2C+ NK cell clones were tested in a 4-hour 51Cr-release assay against mock- and HCMV (Towne)-infected (24 or 72 hours) autologous or HFF fibroblasts,40 at different effector-target ratios. Cells were treated with Trypsin-EDTA (Invitrogen Gibco, Grand Island, NY) labeled with 51Cr and used in standard cytotoxicity assays.41 In parallel samples, effector cells were preincubated with CD94- or CD56-specific mAbs for 30 minutes before adding targets. All assays were set up in triplicate and specific lysis was calculated as described.41
Statistical analysis
Statistical analysis was performed by the Mann-Whitney U test, using the SPSS 9.0 software (SPSS, Chicago, IL). Results were considered significant at the 2-sided P level of .05.
Results
Preferential expansion of NKG2C+ NK cells in PBLs stimulated with HCMV-infected fibroblasts
We previously reported that increased proportions of CD94/NKG2C+ NK cells and ILT2+ (CD85j+) T lymphocytes were detectable in PBLs from HCMV+ donors, indicating that the viral infection might shape the NK cell receptor repertoire.26 To address whether HCMV could promote the expansion of CD94/NKG2C+ lymphocytes, PBLs from HCMV+ donors were cocultured either with mock- or HCMV (Towne)-infected MRC-5 fibroblasts, in the presence of an exogenous supply of IL-2 (10 U/mL); at different time-points the proportions and phenotype of NK cells were comparatively assessed. In vitro stimulation of PBLs with HCMV-infected fibroblasts has been extensively used to analyze CTLs specific for viral antigens, but NK cells have not been systematically studied in this system.
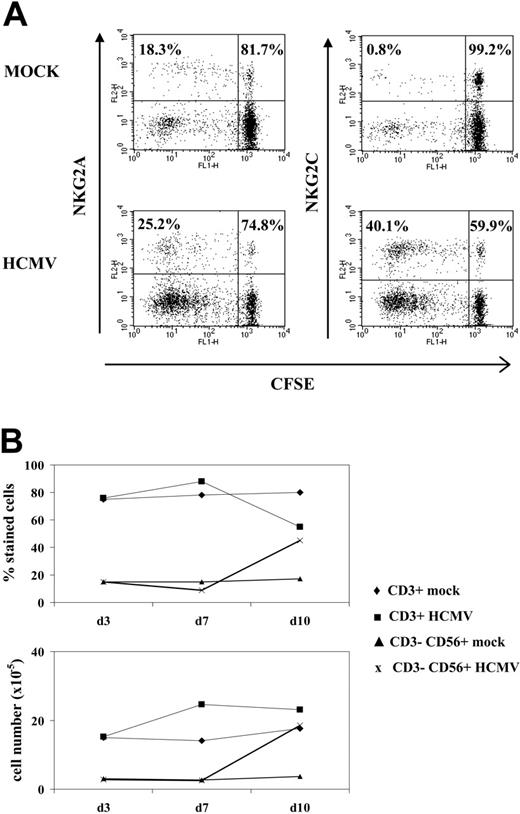
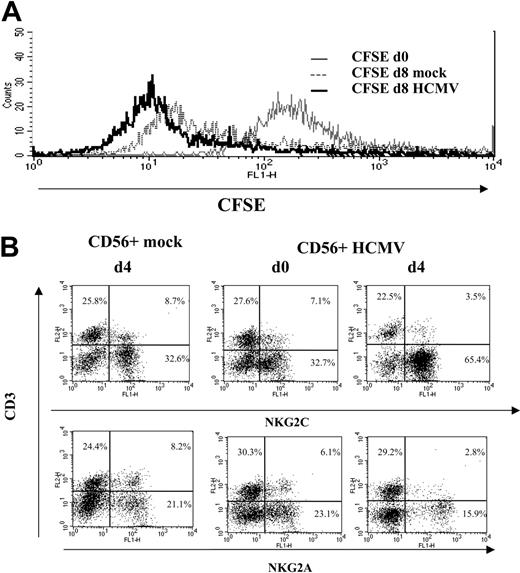
A proliferative response of PBLs to HCMV-infected cells was observed by phase-contrast microscopy. Cell recovery after 10 to 12 days, referred to the input, was 1.5- to 5-fold in cultures with HCMV-infected fibroblasts and 0.8- to 1.5-fold with mock-infected cells in different experiments (n = 25). Studies carried out with CFSE-labeled PBLs also showed that cell proliferation stimulated by HCMV-infected MRC-5 cells exceeded that induced by mock-infected cultures, allowing a phenotypic analysis of the dividing cells (Figure 1A). Compared with NKG2A+ lymphocytes, NKG2C+ cells mainly divided upon stimulation by HCMV-infected MRC-5 cells; yet, only a fraction proliferated.
Analysis at different time-points of PBLs cocultured with HCMV-infected fibroblasts indicated that T cells were predominant during the first week (Figure 1B), but a shift in the distribution of lymphocyte populations was observed by days 10 to 12. Remarkably, NK cells substantially increased (Figure 1B), becoming up to 35% to 80% of the population in different experiments (n = 25), and NKG2C+ lymphocytes outnumbered the NKG2A+ subset (Figure 2A); the proportions of cells coexpressing both KLRs were negligible (data not shown). The NKG2C+ cell recovery in cultures with HCMV-infected cells was 5- to 25-fold that obtained with mock-infected fibroblasts; the NKG2C+/NKG2A+ ratios were 0.3 to 1.3 and 2.2 to 6.7 in mock- and HCMV-infected cultures, respectively. Together with a majority of NKG2C+ NK cells, small proportions of CD3+ NKG2C+ and NKG2A+ T lymphocytes were also identified (Figure 2A). By contrast, the phenotype of PBLs from HCMV-seronegative donors (n = 4) was comparable upon incubation with virus- or mock-infected MRC-5 cells (Figure 2A). It is of note that the late outgrowth of NKG2C+ cells was not perceived in samples from a group of HCMV+ donors (5 of 11), in which T cells remained the predominant population all over the culture. Compared with responders (Rs), fresh PBLs from the nonresponder (NR) group displayed significantly lower proportions of NKG2C+ cells (NR = 2.2± 1.5 vs R = 10.1 ± 7.3; P = .03) and reduced NKG2C/NKG2A ratios (NR = 0.3 ± 0.3 vs R = 1.7 ± 1.1; P = .02).
NK cell proliferation in response to HCMV-infected fibroblasts. (A) CFSE-labeled PBLs from an HCMV+ donor were cocultured with mock- and Towne-infected (MOI = 1) MRC-5 cells as described in “Materials and methods.” Flow cytometry analysis was carried out at day 7, after staining cells with anti-NKG2C or -NKG2A mAbs; numbers correspond to the proportions of dividing and nondividing cells in NKG2C+ and NKG2A+ populations, calculated as described in “Materials and methods.” The experiment is representative of 6 performed. (B) PBLs were stimulated with Towne-infected fibroblasts as described in panel A, and flow cytometry analysis was carried out at different time points with anti-CD3-PerCP and anti-CD56-PE, counting the numbers of recovered cells. The percentages and the calculated numbers of NK- and T-cell populations are represented.
NK cell proliferation in response to HCMV-infected fibroblasts. (A) CFSE-labeled PBLs from an HCMV+ donor were cocultured with mock- and Towne-infected (MOI = 1) MRC-5 cells as described in “Materials and methods.” Flow cytometry analysis was carried out at day 7, after staining cells with anti-NKG2C or -NKG2A mAbs; numbers correspond to the proportions of dividing and nondividing cells in NKG2C+ and NKG2A+ populations, calculated as described in “Materials and methods.” The experiment is representative of 6 performed. (B) PBLs were stimulated with Towne-infected fibroblasts as described in panel A, and flow cytometry analysis was carried out at different time points with anti-CD3-PerCP and anti-CD56-PE, counting the numbers of recovered cells. The percentages and the calculated numbers of NK- and T-cell populations are represented.
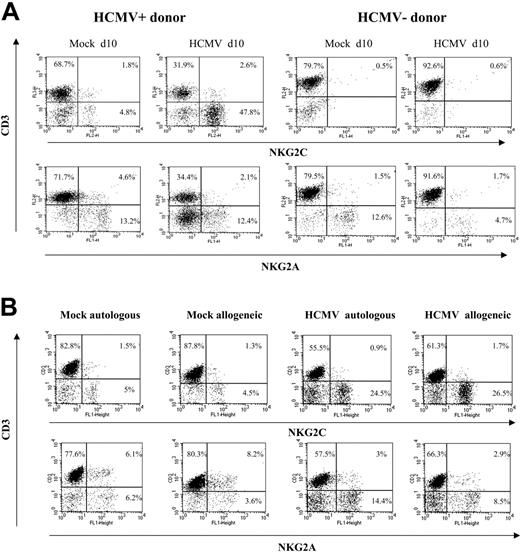
The expansion of NKG2C+ cells was detectable when PBLs were cocultured with HCMV-infected HFFs (data not shown) or autologous skin fibroblasts (Figure 2B). Together with the limited NK cell proliferation in response to mock-infected MRC-5 cells, these results ruled out alloreactivity as the main stimulus responsible for driving the proliferation of NKG2C+ NK cells, and strongly supported a central role for the virus. The effect was comparable upon infection with different HCMV strains (ie, AD169 and Toledo), as well as with a purified Towne preparation, thus excluding a role for cellular products present in crude virus stocks (not shown); subsequent experiments were performed using either Towne or AD169, as specified in every case.
Preferential expansion of CD94/NKG2C+ NK cells upon stimulation with HCMV-infected autologous or allogeneic fibroblasts. (A) PBLs from HCMV+ and HCMV- donors were cocultured with mock- and AD169-infected (MOI = 1) MRC-5 cells. Two-color analysis was carried out at day 10 with an anti-CD3 mAb combined to anti-NKG2C or -NKG2A mAbs. The results are representative of the patterns of response observed in 6 of 11 HCMV+ individuals, and in 4 HCMV- donors. (B) PBLs from an HCMV+ donor were cocultured with mock- and AD169-infected allogeneic (MRC-5) or autologous fibroblasts, and samples were analyzed at day 10, as described in panel A. Similar results were obtained in 6 different experiments with PBLs from 2 different individuals.
Preferential expansion of CD94/NKG2C+ NK cells upon stimulation with HCMV-infected autologous or allogeneic fibroblasts. (A) PBLs from HCMV+ and HCMV- donors were cocultured with mock- and AD169-infected (MOI = 1) MRC-5 cells. Two-color analysis was carried out at day 10 with an anti-CD3 mAb combined to anti-NKG2C or -NKG2A mAbs. The results are representative of the patterns of response observed in 6 of 11 HCMV+ individuals, and in 4 HCMV- donors. (B) PBLs from an HCMV+ donor were cocultured with mock- and AD169-infected allogeneic (MRC-5) or autologous fibroblasts, and samples were analyzed at day 10, as described in panel A. Similar results were obtained in 6 different experiments with PBLs from 2 different individuals.
Distribution of NKRs and NCRs in PBLs stimulated with HCMV-infected fibroblasts
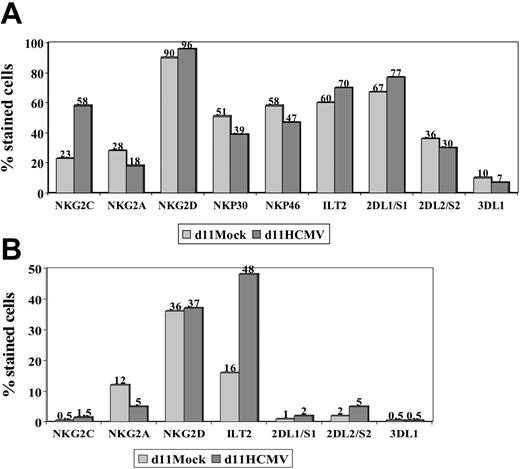
The expression of other NKRs (ie, KIR2D, KIR3D, NKG2D, ILT2) and NCRs (ie, NKp46 and NKp30) was studied in NK and T cells stimulated with HCMV-infected fibroblasts, as previously done in PBLs from HCMV+ donors. Expression of NKG2D, NKp46, and NKp30 was similar in mock- and HCMV-infected cultures (Figure 3), except for a slight reduction of NKp46 and NKp30 in the latter; it is of note that NKG2C+ NK cells were reported to express lower levels of these receptors than the NKG2A+ subset.26 Some differences in the distribution of KIRs were also noticed, but did not follow a reproducible pattern in samples from different donors. By contrast, the proportions of ILT2+ T lymphocytes were found to be significantly increased (P = .04) upon stimulation with HCMV-infected fibroblasts (mean ± SD = 51 ± 47; range, 30-82; n = 6) compared with mock-infected cultures (mean ± SD = 18 ± 12; range, 8-40).
Fibroblast infection is essential to promote the expansion of NKG2C+ cells
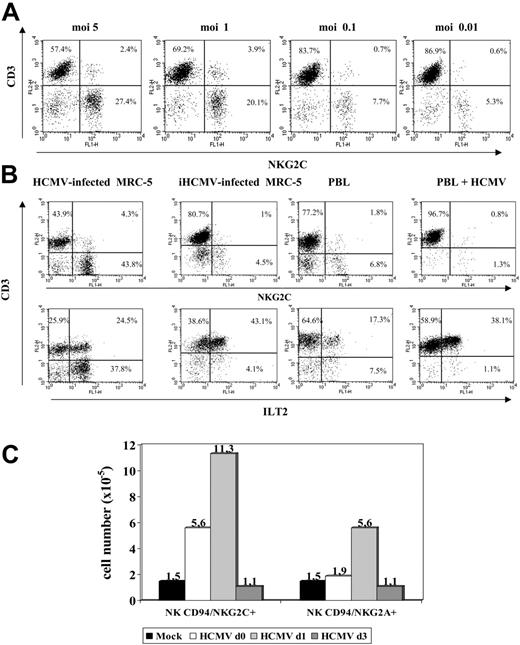
An additional set of experiments was carried out to define the conditions required for HCMV to promote the expansion of NKG2C+ cells that was dependent on the infectious dose used (Figure 4A). Treatment of MRC-5 with UV-inactivated HCMV (Figure 4B) abolished the effect. On the other hand, incubation of PBLs with the virus in the absence of fibroblasts induced a proliferative response of T lymphocytes, including the ILT2+ subset, but did not expand NK cells (Figure 4B). Infected fibroblasts, either alone or with PBLs, did not stimulate NK cell proliferation in PBL samples cultured separately in a transwell system (not shown). Taken together, these findings indicate that a direct interaction between PBLs and HCMV-infected fibroblasts is required to induce the proliferation of NKG2C+ cells.
NKR and NCR distribution in PBLs stimulated with HCMV-infected fibroblasts. PBLs from an HCMV+ donor were cocultured with mock- and Towne-infected (MOI = 1) MRC-5 cells. Cells harvested at day 11 were stained by indirect immunofluorescence with mAbs specific for different NKRs or NCRs, followed by labeling with anti-CD56-PE and -CD3-PerCP mAbs. Three-color flow cytometry analysis was carried out gating on CD3-CD56+ (A) and CD3+ (B) populations. The data are representative of 4 different experiments.
NKR and NCR distribution in PBLs stimulated with HCMV-infected fibroblasts. PBLs from an HCMV+ donor were cocultured with mock- and Towne-infected (MOI = 1) MRC-5 cells. Cells harvested at day 11 were stained by indirect immunofluorescence with mAbs specific for different NKRs or NCRs, followed by labeling with anti-CD56-PE and -CD3-PerCP mAbs. Three-color flow cytometry analysis was carried out gating on CD3-CD56+ (A) and CD3+ (B) populations. The data are representative of 4 different experiments.
Expansion of NKG2C+ NK cells is dependent on the HCMV infection of fibroblasts and on the time of their interaction. (A) The distribution of NKG2C+ cells in response to HCMV-infected MRC-5 cells at different MOI was analyzed. The percentage of NKG2C at MOI of 0.01 was comparable to mock-infected samples (not shown). (B) PBLs from HCMV+ donors were cocultured with mock- or Towne-infected MRC-5 fibroblasts; in parallel, PBLs were incubated with the virus alone (HCMV) or fibroblasts infected with UV-inactivated Towne (iHCMV). Two-color flow cytometry analysis was performed at day 10 with anti-CD3, -NKG2C, and -ILT2 mAbs. Data are representative of 5 different experiments. (C) PBLs were incubated with Towne-infected MRC-5 cells at different time points after infection (days 0, 1, and 3). At day 10, cells were harvested, counted, and analyzed by flow cytometry; the numbers of NKG2C+ and NKG2A+ cells recovered are shown. A similar pattern of response was observed in 3 different experiments.
Expansion of NKG2C+ NK cells is dependent on the HCMV infection of fibroblasts and on the time of their interaction. (A) The distribution of NKG2C+ cells in response to HCMV-infected MRC-5 cells at different MOI was analyzed. The percentage of NKG2C at MOI of 0.01 was comparable to mock-infected samples (not shown). (B) PBLs from HCMV+ donors were cocultured with mock- or Towne-infected MRC-5 fibroblasts; in parallel, PBLs were incubated with the virus alone (HCMV) or fibroblasts infected with UV-inactivated Towne (iHCMV). Two-color flow cytometry analysis was performed at day 10 with anti-CD3, -NKG2C, and -ILT2 mAbs. Data are representative of 5 different experiments. (C) PBLs were incubated with Towne-infected MRC-5 cells at different time points after infection (days 0, 1, and 3). At day 10, cells were harvested, counted, and analyzed by flow cytometry; the numbers of NKG2C+ and NKG2A+ cells recovered are shown. A similar pattern of response was observed in 3 different experiments.
Time-course experiments were carried out culturing PBLs with MRC-5 cells at different stages after HCMV infection. As shown in Figure 4C, the expansion of NKG2C+ cells was optimal upon stimulation of PBLs at day 1, but was reduced when the coculture was delayed to day 3 after infection, revealing the existence of an optimal temporal window for the cellular interactions required to stimulate the NKG2C+ subset.
The expansion of NKG2C+ NK cells involves the CD94/NKG2C receptor and is enhanced by IL-15
Depletion of NKG2C+ lymphocytes by cell sorting abolished the effect, pointing out that the responding cells constitutively express the activating KLR (not shown); positive selection of NKG2C+ cells was not considered reliable due to the potential agonistic/antagonistic effects mediated by the receptor-specific mAb. Thus, to further dissect the process, CD56+ and CD56- populations were sorted and incubated with HCMV-infected fibroblasts. Remarkably, the CD56+ population, which included both NKG2C+ and NKG2A+ subsets, did not divide despite the presence of exogenous IL-2 (10 U/mL); in fact, IL-2Rα (CD25) expression remained negative after 48 to 72 hours (not shown). By contrast, cell proliferation was observed among the CD56- population, and the proportions of CD56+ NKG2C+ cells increased, becoming detectable (not shown). These findings indicated that additional signals, missing in the CD56+ population, were required to stimulate the proliferation of NK cells in response to HCMV-infected fibroblasts. Such complementary stimuli, provided by cells contained within the CD56- fraction, likely accounted for the expansion of residual NKG2C+ CD56+ cells (< 1%-2%) after sorting.
IL-15 plays a primordial role in regulating NK cell proliferation and differentiation, contributing to the accumulation of NK cells in CMV-infected mice.42 Purified CD56+ populations, containing NK and T cells, were cultured with either HCMV- or mock-infected MRC-5 fibroblasts, and replicate samples were supplemented with IL-15 at different time points. Experiments with CFSE-labeled cells indicated that, in both cases, CD56+ cells divided by day 8 in response to IL-15 (Figure 5A). Yet, the cytokine promoted a preferential expansion of the NKG2C+ subset in response to HCMV-infected MRC-5 when cultures were supplemented by day 4 (Figure 5B) but not earlier (day 0). Similar results were obtained with sorted CD3-CD56+ NK cell populations (data not shown). Altogether, these observations supported that the interaction of NKG2C+ cells with HCMV-infected fibroblasts enhances their responsiveness to IL-15. Further studies are required to dissect the influence of the cytokine network in the response of NKG2C+ cells to HCMV.
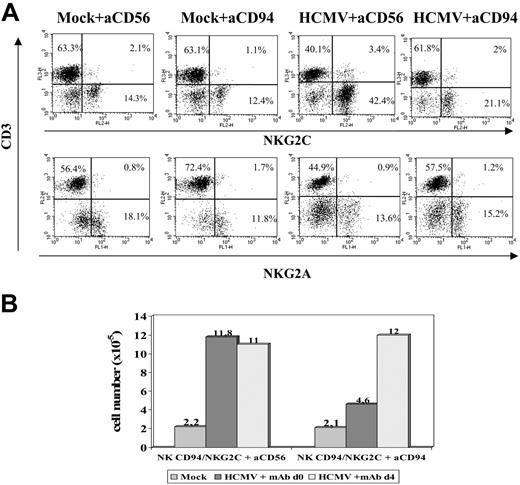
To approach whether the CD94/NKG2C receptor participated in the response to HCMV-infected cells, experiments were carried out in the presence of an F(ab′)2 anti-CD94 mAb (HP-3B1). Compared with the control (anti-CD56), the HP-3B1 mAb hampered the expansion of NKG2C+ cells when added early (days 0-1) after stimulating PBLs with HCMV-infected fibroblasts (Figure 6A). Interestingly, the inhibitory effect vanished if the mAb supply was delayed to days 3 to 4 (Figure 6B), when infected fibroblasts cocultured with PBLs were no longer detectable (not shown), further arguing against a nonspecific effect of the reagent. These results were also consistent with the existence of a temporal window in which cell interactions with infected fibroblasts, required to drive the expansion of NKG2C+ cells, take place and likely involve the CD94/NKG2C receptor.
IL-15 promotes the preferential expansion of NKG2C+ cells in CD56+ populations stimulated with HCMV-infected cells. CD56+ populations sorted from PBLs of HCMV+ donors were cultured with mock- or HCMV (AD169)-infected MRC-5 cells, infected 24 hours before, in the presence of IL-2 (10 U/mL). (A) Proliferation of CFSE-labeled CD56+ cells cultured for 8 days with mock- or HCMV-infected cells supplemented with IL-15 at day 4. (B) Distribution of NKG2A+ and NKG2C+ subsets in CD56+ lymphocytes cocultured for 8 days with mock- or HCMV-infected cells in the presence of IL-15, added at days 1 or 4. The data are representative of 6 experiments performed.
IL-15 promotes the preferential expansion of NKG2C+ cells in CD56+ populations stimulated with HCMV-infected cells. CD56+ populations sorted from PBLs of HCMV+ donors were cultured with mock- or HCMV (AD169)-infected MRC-5 cells, infected 24 hours before, in the presence of IL-2 (10 U/mL). (A) Proliferation of CFSE-labeled CD56+ cells cultured for 8 days with mock- or HCMV-infected cells supplemented with IL-15 at day 4. (B) Distribution of NKG2A+ and NKG2C+ subsets in CD56+ lymphocytes cocultured for 8 days with mock- or HCMV-infected cells in the presence of IL-15, added at days 1 or 4. The data are representative of 6 experiments performed.
By contrast, the cytolytic activity of NKG2C+ NK clones in response to HCMV-infected MRC-5 or autologous fibroblasts was not consistently inhibited by the anti-CD94 mAb, thus indicating that the KLRs did not play a dominant role in triggering NK effector functions against virus-infected fibroblasts (data not shown).
Response of CD94/NKG2C+ cells to fibroblasts infected with HCMV deletion mutants
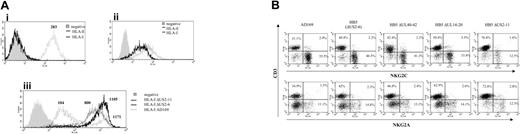
The CD94/NKG2C receptor specifically interacts with HLA-E bound to peptides derived from the leader sequences of other class I molecules, though with a lower affinity than NKG2A.27,28 It has been proposed that HLA-E may be selectively preserved during HCMV infection, impairing the response of CD94/NKG2A+ NK cells.23,24 Thus, the HLA class Ib molecule was considered a first candidate ligand for CD94/NKG2C in HCMV-infected cells. To address whether UL40 was required for the expansion of CD94/NKG2C+ cells, a targeted deletion mutant of HCMV missing the UL40-42 genes36 was studied. The HB5ΔUL40-42 mutant was constructed from a HCMV BACmid pHB5 that lacks the US2-US6 gene region. Thus, HB5 and HB5ΔUL40-42 viruses retain only US11 to inhibit HLA class I expression. In these experiments, PBLs were stimulated with MRC-5 cells infected with either AD169, HB5, HB5ΔUL40-42, or another HB5-derived targeted deletion mutant (HB5ΔUL14-20) missing the UL16 and UL18 genes.36 In line with a previous report,23 surface HLA-E was undetectable by flow cytometry in both mock- and HCMV-infected fibroblasts (Figure 7A). Compared with the marked inhibition of class I expression exerted by AD169 in MRC-5 cells, infection with either HB5, HB5ΔUL40-42, or HB5ΔUL14-20 had only a modest effect, consistent with the exclusive use of US11 to inhibit class I molecules (Figure 7Aiii). Albeit reaching a slightly lower yield compared with AD169, the late outgrowth of NKG2C+ NK cells was observed in every case, indicating that it was independent of UL40, and excluding as well a critical involvement of UL18 or UL16 (Figure 7B).
Anti-CD94 mAb blocks the expansion of NKG2C+ NK cells stimulated by HCMV-infected fibroblasts. (A) PBLs from an HCMV+ donor were cultured with mock- and Towne-infected (MOI = 1) MRC-5 cells. Either F(ab′)2 anti-CD94 or -CD56 mAbs (10 μg/mL) were added at day 0. Flow cytometry analysis with anti-CD3, -NKG2C, and -NKG2A mAbs was carried out at day 10. (B) The differential effects in the numbers of recovered cells in the presence of the anti-CD94 mAb added at day 0 or day 4 are compared. The data are representative of 5 experiments performed.
Anti-CD94 mAb blocks the expansion of NKG2C+ NK cells stimulated by HCMV-infected fibroblasts. (A) PBLs from an HCMV+ donor were cultured with mock- and Towne-infected (MOI = 1) MRC-5 cells. Either F(ab′)2 anti-CD94 or -CD56 mAbs (10 μg/mL) were added at day 0. Flow cytometry analysis with anti-CD3, -NKG2C, and -NKG2A mAbs was carried out at day 10. (B) The differential effects in the numbers of recovered cells in the presence of the anti-CD94 mAb added at day 0 or day 4 are compared. The data are representative of 5 experiments performed.
By contrast, the expansion of NKG2C+ cells was impaired in response to another HCMV mutant that lacked all major histocompatibility complex (MHC) I down-regulating genes (ie, US2 through US11 [HB5ΔUS2-11]) (Figure 7B) which, as expected, did not alter the expression of HLA class I in infected fibroblasts (Figure 7A). Similar results were obtained with another independently derived HB5ΔUS2-US11 mutant (A.A., unpublished data, January 2004).
These results suggested that normal levels of HLA class I may dampen the stimulation of CD94/NKG2C+ cells, presumably engaging the inhibitory receptors (ie, KIRs and/or ILT2) expressed by this cell subset. Further studies are required to determine whether HLA-E in HCMV-infected fibroblasts participates in engaging CD94/NKG2C and inducing the response.
Discussion
The detection of increased proportions of CD94/NKG2C+ cells in PBLs from healthy HCMV+ donors suggested that the viral infection might shape the NKR repertoire.26 In the present report, we provide direct in vitro evidence supporting that the CD94/NKG2C+ NK cell subset preferentially expands upon interaction with HCMV-infected fibroblasts, and that the KLR itself may participate in driving the proliferation. The requirements for HCMV infection to promote this effect have been defined, and its complexity partially dissected.
To assess the influence of HCMV on the NKR repertoire, PBLs were stimulated with infected fibroblasts, an experimental system conventionally employed to study virus-specific CTLs.44 T lymphocytes were predominant during the first week, but a substantial increase of NK cells was detected by days 10 to 12. NKG2C+ cells systematically outnumbered the NKG2A+ subset and a minor proportion of NKG2C+ T lymphocytes was also identified. The proliferation of NKG2C+ cells was confirmed by 2-color analysis of CFSE-labeled samples. This pattern of response observed in PBL samples from a group of HCMV+ donors, but not in HCMV- individuals, was similar regardless of the origin of fibroblasts (allogeneic vs autologous) and the virus strains tested (ie, Toledo, Towne, and AD169). Fibroblast infection was essential for the expansion of NKG2C+ NK cells, as it was dependent on the infectious dose and was not substantiated with UV-inactivated HCMV, nor when PBLs were incubated with the virus alone. The unresponsiveness of PBLs from HCMV-seronegative donors is presumably due to the absence of virus-specific memory T lymphocytes and the low/undetectable proportions of CD94/NKG2C+ cells.26
Anti-CD94 mAb F(ab′)2 markedly inhibited the response of NKG2C+ cells, supporting that signaling by the KLRs was required. An antagonistic effect of NKG2C-specific mAbs would lend stronger support to this possibility; yet, such experiments were not feasible due to the difficulties of generating F(ab′)2 fragments from the commercially available unconjugated anti-NKG2C mAb (IgG2b). Nevertheless, purified CD56+ populations, containing NKG2C+ and NKG2A+ cells, did not proliferate in response to HCMV-infected fibroblasts despite the presence of exogenous IL-2, pointing out the requirement of additional signals. The outgrowth of NKG2C+ cells became evident when the CD56+ subset was stimulated with HCMV-infected cells in the presence of exogenous IL-15, added at day 4 after the culture onset. IL-15 has been reported to be required for the accumulation of NK cells in MCMV infection.42,45 Altogether the observations are consistent with the view that signaling by the CD94/NKG2C receptor, upon interaction with infected fibroblasts, enhances the responsiveness to IL-15 and promotes the proliferation of the corresponding subset. DAP12 signaling has been shown to enhance NK cell responsiveness to IL-15.53 (communicated by A.R. French and W.M. Yokoyama; 21st International NK cell Workshop, Kawai, November 2005). The molecular basis for this effect deserves further attention, and the contribution of other signals in the expansion of NKG2C+ cells is not excluded. More extensive studies are required to dissect how HCMV-induced changes in the cytokine network may contribute to the expansion of NKG2C+ cells and the role of endogenous IL-15.
Expansion of NKG2C+ NK cells in response to fibroblasts infected with HCMV deletion mutants. (Ai) The expression of total class I molecules (HLA-I) and HLA-E was analyzed by flow cytometry in MRC-5 fibroblasts. The expression of HLA-E was also undetectable in AD169-infected MRC-5 cells (not shown). (Aii) Staining of an HLA-E+ cell line (.221-AEH) is shown for comparison. (Aiii) Expression of HLA-I in MRC-5 cells infected for 72 hours with either AD169, HB5 (ΔUS2-6), or HB5-ΔUS2-11 mutants; HLA expression in cells infected with HB5-ΔUS40-42 or HB5-ΔUS14-20 was comparable to HB5 (not shown). Production of type I IFN upon HCMV infection up-regulates HLA class I expression.43 In this experiment, the presence of a fraction of bystander noninfected cells among AD169-treated cells provides a suitable internal control. Numbers correspond to the mean fluorescence intensity (MFI) of the histograms. (B) PBLs from an HCMV+ donor were cultured in parallel with MRC-5 cells infected (MOI = 1) with the wild-type AD169 HCMV strain or different deletion mutants generated from the HB5 BACmid clone (ΔUS2-6). Two-color flow cytometry analysis was performed by day 10. Data are representative of 6 different experiments. The proportion of NKG2C+ cells in mock-infected cultures (not shown) was 10.5%.
Expansion of NKG2C+ NK cells in response to fibroblasts infected with HCMV deletion mutants. (Ai) The expression of total class I molecules (HLA-I) and HLA-E was analyzed by flow cytometry in MRC-5 fibroblasts. The expression of HLA-E was also undetectable in AD169-infected MRC-5 cells (not shown). (Aii) Staining of an HLA-E+ cell line (.221-AEH) is shown for comparison. (Aiii) Expression of HLA-I in MRC-5 cells infected for 72 hours with either AD169, HB5 (ΔUS2-6), or HB5-ΔUS2-11 mutants; HLA expression in cells infected with HB5-ΔUS40-42 or HB5-ΔUS14-20 was comparable to HB5 (not shown). Production of type I IFN upon HCMV infection up-regulates HLA class I expression.43 In this experiment, the presence of a fraction of bystander noninfected cells among AD169-treated cells provides a suitable internal control. Numbers correspond to the mean fluorescence intensity (MFI) of the histograms. (B) PBLs from an HCMV+ donor were cultured in parallel with MRC-5 cells infected (MOI = 1) with the wild-type AD169 HCMV strain or different deletion mutants generated from the HB5 BACmid clone (ΔUS2-6). Two-color flow cytometry analysis was performed by day 10. Data are representative of 6 different experiments. The proportion of NKG2C+ cells in mock-infected cultures (not shown) was 10.5%.
The nature of the putative ligand(s) expressed by HCMV-infected fibroblasts responsible for engaging CD94/NKG2C is a key open question. The observed phenomenon was reminiscent of the activation of NKG2C+ NK and T cells in response to a transfectant of the 721.221 HLA class I-deficient cell line overexpressing HLA-E+ (.221-AEH).31 This class Ib molecule was reported to be refractory to the action of US6 when bound to a peptide from the leader sequence of the HCMV UL40 protein, and appeared constitutively resistant to the action of US2 and US11.22-24 On that basis it was proposed that surface HLA-E expression may be preserved in infected cells protecting them against CD94/NKG2A+ NK cells; a CD8+ T cell subset that specifically recognizes HLA-E via the TCR might counteract this evasion mechanism.46,47 In another report,48 down-regulation of MHC class I by the HCMV US2-11 gene region was reported to dominate UL40-mediated effects in infected fibroblasts attacked by NK cells.
The analysis of HCMV deletion mutants provided valuable information. First, the response to HB5ΔUL40-42 unequivocally demonstrated that UL40 was not required to promote the expansion of NKG2C+ cells. Moreover, cells infected with the HB5ΔUL14-20 elicited a response of NKG2C+ cells comparable with that observed with wild-type HCMV, also excluding a critical role of UL16 and UL18. This is relevant considering that most NKG2C+ cells express the ILT2 (CD85j) receptor, which binds with high affinity to UL18. Moreover, 19 viral genes, present in the UL/b′ region of HCMV strain Toledo but missing in AD169 and Towne,49 are obviously dispensable for the expansion of CD94/NKG2C cells.
HCMV mutants derived from AD169 (HB5) lack the US2-US6 genes, and inhibit HLA class Ia expression using only US11, which was reported to preserve the constitutive expression of HLA-E.22 The interaction of CD94/NKG2C with HLA-E, concomitant to the inhibition of HLA class Ia expression, might contribute to drive the proliferation of NKG2C+ cells. In our hands, surface expression of HLA-E was virtually undetectable by flow cytometry in mock- and HCMV-infected fibroblasts using a bona fide specific mAb (3D12). Tomasec et al23 reported similar difficulties with detecting the class Ib molecule in fibroblasts using a mAb cross-reactive with HLA-E and HLA-C. Thus, if HLA-E bound to class I-derived nonamers is indeed the ligand responsible for engaging CD94/NKG2C in infected fibroblasts, it should be effective despite its limiting expression levels and low affinity for the activating KLRs. This is conceivable if signaling by inhibitory receptors is prevented by a reduction of HLA class Ia expression, even minimal as in cells infected with the HB5-derived mutants. The inability to expand NKG2C+ cells upon stimulation with cells infected with HB5ΔUS2-11, which fully preserved HLA class I molecules, supported this view; yet, a direct role for other gene(s) in the US7-11 region is not excluded. Alternatively, it cannot be ruled out that infected fibroblasts may display HLA-E/peptide complexes with high affinity for CD94/NKG2C or a viral ligand capable of directly engaging the receptor.
CD94 mAb did not consistently block cytotoxicity of NKG2C+ NK clones against HCMV-infected fibroblasts, questioning a dominant role of the KLRs in triggering this NK effector function. Nevertheless, these observations do not rule out a relevant participation of CD94/NKG2C+ cells in the response to HCMV-infected cells. First, this NK subset is not susceptible to HLA-E-mediated inhibition. Moreover, the KLRs may act in concert with other NKRs (ie, NKG2D, DNAM-1) and/or NCRs; in this regard, it has been reported that the NK response to some tumor cells could not be inhibited by blocking individual receptor-ligand pairs.50
Experiments with CFSE-labeled PBLs indicated that only a fraction of NKG2C+ cells proliferated. Moreover, the expansion was not perceived in PBLs from a group of HCMV+ donors that displayed lower proportions of NKG2C+ cells. Thus, it is conceivable that this NK cell subset may be heterogeneous in terms of its replication capacity, and that the proliferating “progenitor” pool responsible for the expansion may be variably represented in different individuals. Lower proportions of this subset, together with the intrinsic complexity of the coculture system where T lymphocytes also respond, may render undetectable the proliferation of NKG2C+ cells accounting for the “nonresponder” phenotype. The possibility that other signals contributing to the expansion of NKG2C+ cells may be defective in these samples, and/or that regulatory mechanisms interfere with their response, is not ruled out. Preliminary experiments indicate that the expansion of NKG2C+ cells may be perceived by stimulating CD56+ populations from “nonresponders” with HCMV-infected cells in the presence of IL-15.
Together with the skewed distribution of NKG2C+ cells, an expansion of ILT2+ T lymphocytes was observed in PBLs from HCMV+ donors stimulated with infected fibroblasts. ILT2+ T cells were also significantly increased in fresh PBLs from HCMV+ individuals,26 and in transplantation patients undergoing HCMV infections.51
The impact of HCMV infection on the CD94/NKG2C+ subset is reminiscent of the oligoclonal expansion of CTLs specific for viral antigens, that display an effector-memory phenotype and tend to substantially increase in elderly individuals.5,52 The basis for the variability in the numbers of NKG2C+ cells observed in HCMV+ donors is uncertain, but likely reflects the challenge exerted by the virus on the innate immune system. The frequency of reactivation episodes, the extension of latent infection, and/or genetic differences in HCMV clinical isolates may be relevant in this context. The analysis of NKG2C+ cells becomes a potentially useful novel parameter to monitor the host-pathogen relationship.
Prepublished online as Blood First Edition Paper, December 29, 2005; DOI 10.1182/blood-2005-09-3682.
Supported by grants from Ministerio de Educación y Ciencia (MEC; SAF2004-07632 and SAF2002-00270), European Community (QLRT-2001-01112 to H.H. and M.L.-B.). M.G. is recipient of a fellowship from Instituto de Salud Carlos III (ISCIII), Ministry of Health. A.A. is a fellow from the Ramón y Cajal program (MEC).
A.A. and M.L.-B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Dr Oscar Fornas for advice in flow cytometry analysis, to Esther Menoyo for collaborating in obtaining blood samples, to Dr Carlos Vilches for critical reading of the manuscript, and to Drs A. Moretta and D. Pende for kindly supplying reagents.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal