Comment on Chapiro et al, page 3560
Hematopoietic development is exquisitely regulated by a spectrum of transcription factors that participate in lineage-commitment decisions and terminal differentiation. Perhaps not surprisingly, it has become apparent that mutations in many of these hematopoietic transcription factors can contribute to pathogenesis of leukemia.
Examples include fusion genes involving the components of core binding factor (eg, RUNX1-ETO and CBFB-SMMHC) and mutations resulting in loss of function of CEBPA, PU.1, and GATA-1, among others. What has been more surprising is that transforming potential of several of these alleles is remarkably sensitive to gene dosage effects. For example, approximately 20% dosage of a hypomorphic PU.1 allele is highly leukemogenic in murine models, whereas complete loss, or heterozygous dosage, of PU.1 is not.1 FIG1
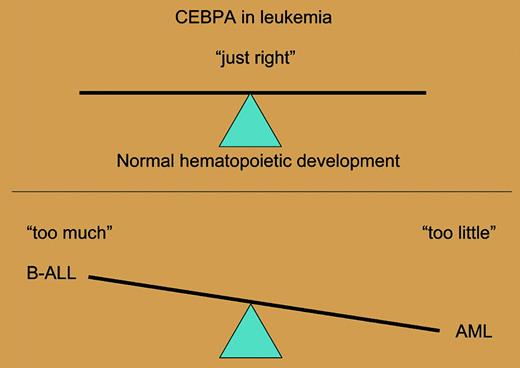
Overexpression or loss of function of CEBPA can contribute to B-ALL or AML, respectively.
Overexpression or loss of function of CEBPA can contribute to B-ALL or AML, respectively.
Against this backdrop, there are many lines of evidence indicating that loss of function of CEBPA is leukemogenic, including dominant-negative mutations in CEBPA itself and suppression of CEBPA mRNA or CEBPA protein expression by various leukemogenic fusion proteins such as RUNX1-ETO, CBFB-MYH11, and FLT3-ITD.2 Collectively, these data would suggest that CEBPA has a tumor-suppressor function in leukemogenesis.
However, in this issue of Blood, Chapiro and colleagues report a remarkable and important discovery: chromosomal translocations associated with B-cell acute lymphoblastic leukemia (B-ALL) result in overexpression of CEBPA. Thus, it appears that either loss of function of CEBPA or gain of function of CEBPA has leukemogenic potential. These observations will need to be further explored in cell culture and murine models of transformation but, at face value, they suggest that too much, or too little, transcriptional activity of key hematopoietic transcriptions factors potentiates leukemogenesis.
There are several experiments that will inform this seminal observation and important questions that are unanswered. CEBPA deficiency is associated with acute myeloid leukemia (AML), whereas CEBPA overexpression is associated with B-cell malignancies. The basis for this phenotypic delineation is not clear. At face value, this could be explained by overexpression of CEBPA from a B-cell–specific enhancer element from the IgH locus. However, it is not as simple as that, in that Graf and colleagues have shown that retroviral overexpression of CEBPA reprograms B cells into macrophages.2 It should also be noted that in the original report of mutations that result in dominant-negative activity of CEBPA in AML, there was 1 mutation that appeared to enhance transcriptional activation potential of CEPBA on reporter constructs.3
Much work remains to characterize the lineage affiliation and transforming activity of these CEBPA alleles. Previous literature has promulgated the notion that dosage of hematopoietic transcription factors is a key element in our understanding of mechanisms of leukemogenesis. However it seems clear, based on this fascinating report and other data reported in abstract form, that the dosage or expression level of CEBPA—“too much or too little”—can cause cancer; it needs to be “just right” for maintenance of normal hematopoiesis (see the figure). Furthermore, these data suggest a delicate balance between level of expression of hematopoietic transcription factors on the one hand and lineage determination and differentiation on the other. These findings also provide a compelling rationale for analysis of disease alleles as expressed from their endogenous promoters in murine models of disease. Thus, the report of Chapiro and colleagues provides data from human leukemias that serve to champion mechanistic approaches to pathophysiology of leukemogenesis based on dosage and relative levels of expression of key hematopoietic transcription factors. ▪

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal