Abstract
To identify new cytogenetic abnormalities associated with leukemogenesis or disease outcome, T-cell acute lymphoblastic leukemia (T-ALL) patient samples were analyzed by means of the array-comparative genome hybridization technique (array-CGH). Here, we report the identification of a new recurrent and cryptic deletion on chromosome 11 (del(11)(p12p13)) in about 4% (6/138) of pediatric T-ALL patients. Detailed molecular-cytogenetic analysis revealed that this deletion activates the LMO2 oncogene in 4 of 6 del(11)(p12p13)-positive T-ALL patients, in the same manner as in patients with an LMO2 translocation (9/138). The LMO2 activation mechanism of this deletion is loss of a negative regulatory region upstream of LMO2, causing activation of the proximal LMO2 promoter. LMO2 rearrangements, including this del(11)(p12p13) and t(11;14) (p13;q11) or t(7;11)(q35;p13), were found in the absence of other recurrent cytogenetic abnormalities involving HOX11L2, HOX11, CALM-AF10, TAL1, MLL, or MYC. LMO2 abnormalities represent about 9% (13/138) of pediatric T-ALL cases and are more frequent in pediatric T-ALL than appreciated until now.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a high-risk malignancy of thymocytes, and accounts for 10% to 15% of pediatric ALL cases. T-ALL often presents with a high tumor-mass that is accompanied by a rapid progression of disease. About 30% of T-ALL cases relapse within the first years during or following treatment and eventually die.1
Genetic analyses of T-ALL have elucidated an enormous heterogeneity in genetic abnormalities including chromosomal translocations, deletions, amplifications, and mutations.2 These abnormalities result in the aberrant expression of transcription factors such as the basic helix-loop-helix (bHLH) genes MYC, TAL1(SCL), TAL2, LYL1, or bHLHB1; genes involved in transcriptional regulation such as the cysteine-rich LIM-domain-only genes LMO1 or LMO2; or the Krüppel-like zinc-finger gene BCL11B. Abnormalities can also affect genes that are involved in embryonic development such as the homeodomain genes HOX11/TLX1 and HOX11L2/TLX3; members of the HOXA cluster; as well as signaling molecules such as the tyrosine kinase ABL13-6 (reviewed in Raimondi7 and Rubnitz and Look8 ). Other translocations lead to the formation of specific fusion products and include CALM-AF109 or MLL rearrangements. Mutational mechanisms may also enhance gene activity as, for example, activating mutations in the NOTCH1 gene were recently identified in about 50% of human T-ALLs.10
LMO2 encodes a protein that participates in the transcription factor complex, which includes E2A, TAL1, GATA1, and LDB1 in erythroid cells.11,12 Within this transcription complex, LMO2 mediates the protein-protein interactions by recruiting LDB1, whereas TAL1, GATA1, and E2A regulate the binding to specific DNA target sites.13 This complex regulates the expression of several genes in various cellular backgrounds including C-KIT,14 EKLF,15 and RALDH.16 In normal T-cell development, LMO2 is expressed in immature CD4/CD8 double-negative thymocytes, and is down-regulated during T-cell maturation.17,18 In various mouse models, ectopic expression of LMO2 leads to clonal expansion of T cells, eventually leading to T-ALL development. LMO2-mediated leukemogenesis seems restricted to the T cell, as transgenic mice with constitutive expression of LMO2 in all tissues develop malignancies involving the T-lineage only.19-22 The long latency of leukemia in these mice suggests that T-ALL development requires secondary mutations in addition to the activation of LMO2.22,23
LMO2-driven oncogenesis in humans was suggested by its frequent involvement in the chromosomal translocations t(11;14)(p13;q11) and t(7;11)(q35;p13) in T-ALL, in which the TCRA/D or TCRB locus is fused to LMO2.24,25 Direct proof came from the retroviral IL2Rgc gene therapy trial for X-linked severe combined immunodeficiency (X-SCID) patients, in which 2 patients developed T-ALL after retroviral insertions in the LMO2 gene.26,27
In this study, we report the identification of a new recurrent and cryptic deletion (del(11)(p12p13)) in about 4% (6/138) of pediatric T-ALL patients. Detailed molecular-cytogenetic analysis revealed that this deletion activates the LMO2 oncogene in 4 of 6 of these del(11)(p12p13)-positive T-ALL patients, mainly through deletion of negative regulatory sequences upstream of LMO2. The relation to other recurrent cytogenetic abnormalities, the immunophenotypic characteristics, and the clinical outcome of this new cryptic abnormality in pediatric T-ALL are discussed.
Materials and methods
Patient samples
Viably frozen diagnostic bone marrow or peripheral blood samples from 64 pediatric T-ALL patients and clinical and immunophenotypic data were provided by the Dutch Childhood Oncology Group (DCOG). Patients were considered positive if more than 25% of total cells were positive for a specific immunophenotypic marker. A second pediatric T-ALL cohort (n = 74) was obtained from the German Co-operative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL). The patients' parents or their legal guardians provided informed consent to use leftover material for research purposes in accordance with the rules of the review board of Erasmus MC and the Declaration of Helsinki. Leukemic cells were isolated and enriched from these samples as previously described.28 All resulting samples contained 90% or more leukemic cells, as determined morphologically by May-Grünwald-Giemsa (Merck, Darmstadt, Germany)-stained cytospins. Viably frozen T-ALL cells were used for DNA and RNA extraction, and a minimum of 5 × 106 leukemic cells was lysed in Trizol reagent (Gibco BRL, Life Technologies, Breda, The Netherlands) and stored at -80°C. A total of 25 × 103 leukemic cells was used for cytospin slides for fluorescence in situ hybridization (FISH) and stored at -20°C. For the preparation of metaphase slides, a minimum of 5 × 106 leukemic cells was cultured for 72 hours in serum-free medium (JRH Biosciences, Lenexa, KS) in the presence of IL7 (10 ng/mL) and IL2 (10 ng/mL), and harvested according to standard cytogenetic techniques.
Genomic DNA isolation, RNA extraction, and cDNA synthesis
Genomic DNA and total cellular RNA were isolated according to the manufacturer's protocol, with minor modifications. An additional phenolchloroform extraction was performed and the DNA was precipitated with isopropanol along with 1 μL (20 μg/mL) glycogen (Roche, Almere, The Netherlands). After precipitation, RNA pellets were dissolved in 20 μL RNAse-free TE-buffer (10 mM Tris-HCl, 1 mM EDTA, pH = 8.0). The RNA concentration was quantified spectrophotometrically. Following a denaturation step of 5 minutes at 70°C, 1 μg RNA was reverse transcribed to single-stranded cDNA using a mix of random hexamers (2.5 μM) and oligodT primers (20 nM). The RT reaction was performed in a total volume of 25 μL containing 0.2 mM of each dNTP (Amersham Pharmacia BioTech, Piscataway, NJ), 200 U Moloney murine leukemia virus reverse transcriptase (M-MLV RT; Promega, Madison, WI), and 25 U RNAsin (Promega). Conditions for the RT reaction were 37°C for 30 minutes, 42°C for 15 minutes, and 94°C for 5 minutes. The cDNA was diluted to a final concentration of 8 ng/μL and stored at -80°C.
BAC array-comparative genomic hybridization (BAC array-CGH)
Bacterial artificial chromosome (BAC) array-CGH analysis was performed using a dye-swap experimental design to minimize false-positive results. Patient genomic DNA (2 μg) and male/female reference DNA (2 μg; Promega) were fragmented by sonification (VibraCell Model VC130; Sonics & Materials, Newtown, CT). DNA fragmentation was verified by agarose gel electrophoresis. Individual reference and experimental samples were then purified using the QIAQuick polymerase chain reaction (PCR) clean-up kit (Qiagen, Valencia, CA). Labeling reactions with Cy5-dUTP and Cy3-dUTP (PerkinElmer, Wellesley, MA) were performed with 5 μg purified restricted DNA using the Bioprime labeling kit (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. The patient and reference DNA were combined, denatured, and applied to the 1-Mb GenomeChip V1.2 Human BAC arrays (2632 BAC clones spotted on a single array; Spectral Genomics, Houston, TX) according to the manufacturer's protocol. Hybridization and washing procedures were performed as recommended, and the slides were scanned on a GenePix 4000B Microarray Scanner (Axon Instruments, Union City, CA). Cy3 and Cy5 fluorescent intensities at each DNA spot were quantified using GenePix Pro 4.0 Microarray Image Analysis Software, and the data were subsequently imported into SpectralWare software (Spectral Genomics). Using this software, background intensities were subtracted and initial fluorescence ratios were log2 transformed. The ratios for each clone were subsequently plotted into chromosome-ideograms. At this stage, known large-scale copy number polymorphisms were not considered disease related.29
Oligo array-CGH
Oligo array-CGH analysis was performed, as previously described,30 on the human genome CGH Microarray 44A (Agilent Technologies, Palo Alto, CA), which consists of approximately 40 000 60-mer oligonucleotide probes that span both coding and noncoding sequences with an average spatial resolution of approximately 35 Kb. Briefly, 10 μg genomic reference or patient DNA was digested with AluI (20 U) and RsaI (20 U) (Invitrogen) overnight at 37°C. Verification of DNA fragmentation, purification, and Cy-3 or Cy-5 labeling was as described under “BAC array-comparative genomic hybridization (BAC array-CGH).” Experimental and reference targets for each hybridization were pooled and mixed with 50 μg human Cot-1 DNA (Invitrogen), 100 μg yeast tRNA (Invitrogen), and 1x hybridization control targets (SP310; Operon Technologies, Alameda, CA) in a final volume of 500 μL in situ hybridization buffer (Agilent Technologies). The hybridization mixtures were denatured at 95°C for 3 minutes, preincubated at 37°C for 30 minutes, and hybridized to the array in a microarray hybridization chamber (Agilent Technologies) for 14 to 18 hours at 65°C in a rotating oven (Robbins Scientific, Mountain View, CA). The array slides were washed in 0.5x SSC/0.005% Triton X-102 at room temperature for 5 minutes, followed by 5 minutes at 37°C in 0.1x SSC/0.005% Triton X-102. Slides were dried and scanned using a 2565AA DNA microarray scanner (Agilent Technologies). Microarray images were analyzed using feature extraction software (version 8.1; Agilent Technologies), and the data were subsequently imported into array-CGH analytics software v3.1.28 (Agilent Technologies).
FISH procedure
BACs were obtained from BAC/PAC Resource Center (Children's Hospital, Oakland, CA). BAC DNA was isolated using DNA MiniPrep plasmid kit (Promega) and labeled with biotin-16-dUTP/digoxigenin-11-dUTP (Roche) by nick translation.31 BAC clones RP11-646J21, RP11-98C11, and RP11-603J2 were used for the characterization of the telomeric breakpoints of the del(11)(p12p13), whereas the centromeric breakpoints were localized using RP11-36H11, RP11-769M16, and RP11-465C16. LMO2 translocations were identified using a split-FISH procedure with the LMO2 flanking BAC clones RP11-646J21 and RP11-98C11. FISH analysis was performed on freshly prepared interphase and metaphase slides from methanol/acetic acid cell suspensions or cytospins stored at -20°C. Slides were pretreated by an RNase and pepsin treatment, fixed with acid-free formaldehyde, and denatured at 72°C. Probes were denatured (4 minutes at 72°C in 70% formamide/2x SSC) in the presence of a 100-fold excess of human Cot-1 DNA (Gibco BRL). Following preannealing at 37°C for 30 minutes, biotinylated probes were hybridized overnight at 37°C, and visualized by fluorescein avidin (Vector Laboratories, Burlingame, VT) and biotinylated anti-avidin D sandwich detection (affinity purified; Vector Laboratories). The digoxigenin hybridization signal was detected using anti-digoxigenin-rhodamine (Boehringer Mannheim, Mannheim, Germany) and donkey anti-sheep Texas Red (Jackson ImmunoResearch Laboratories, Westgrove, PA). Cells were counterstained with DAPI/Vectashield mounting medium (Vector Laboratories). Fluorescence signals were visualized on a Zeiss Axioplan II fluorescence microscope (Zeiss, Sliedrecht, The Netherlands) equipped with double and triple bandpass filters for simultaneous visualization of rhodamine-TR/FITC/DAPI, as well as a Plan-Apochromat 100×/1.40 NA oil iris objective lens. Images were captured by using PSI MacProbe 4.3 software (Applied Imaging, Newcastle-Upon-Tyne, United Kingdom). TAL1, HOX11L2, HOX11, and MLL chromosomal rearrangements or the SIL-TAL1 deletion was determined using FISH kits (DakoCytomation, Glostrup, Denmark),32 hybridized, and scored as described by the manufacturer. The CALM-AF10 chromosomal rearrangement was detected using FISH as previously described.32
Genomic and cDNA PCR
The genomic breakpoint in T-ALL patient 1950 was determined by long-range PCR using forward primer 5′-GATGCCTTCCCTCATGTA-3′ (intron 1 RAG2) and reverse primer 5′-CGCAGTGCCTAGAACAGT-3′ (intron 1 LMO2). PCR reactions were performed using 200 ng genomic DNA (200 ng/μL), 10 pmol primers, 10 nmol dNTPs, 4 mM MgCl2, 1.25 U ampliTaq gold (Applied Biosystems, Foster City, CA) in 10 × PCR buffer II (Applied Biosystems) in a total volume of 50 μL. After the initial denaturation at 94°C for 10 minutes, PCR was performed for 15 cycles of 95°C for 15 seconds, 60°C for 1 minute, and 68°C for 3 minutes followed by 15 cycles consisting of 95°C for 15 seconds, 60°C for 1 minute, and 68°C for 3 minutes (+10 s/cycle).
To identify the RAG2-LMO2 fusion gene in T-ALL patient 1950, cDNA PCR (RT-PCR) was performed in the presence of forward primer 5′-GTGGGCAGTCAGTGAATC-3′ (exon 1 RAG2) and reverse primer 5′-TGCAAGTTCAGGTTGAAA-3′ (exon 2 LMO2) in a total volume of 50 μL. Following initial denaturation at 95°C for 10 minutes, reactions were amplified for 39 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Ligation-mediated PCR
Ligation-mediated PCR (LM-PCR) was performed as previously described.33 Briefly, 1 μg patient (2846) and control (2720) DNA was digested with blunt-end restriction enzymes (EcoRV, DraI, PvuII, and StuI), and 50 μM of an adaptor was ligated to both ends of the restriction fragments. The ligation products were subjected to 2 rounds of PCR with nested adaptor-specific primers AP1 (5′-GCT AGA TAC GAC TCA GTA TAG-3′) and AP2 (5′-TAT AGG CGC ACG AAC G-3′) and nested LMO2 intron 1-specific primers LMO2F (5′-CAG CCA CAT GGG TAG AAC-3′) and LMO2F nested (5′-TGG CAT TAG GGT ATG GAA-3′). The band that differed in size from the expected band in the control patient, lacking the del(11)(p12p13), was excised from the gel and purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) and subjected to direct nucleotide sequencing.
Quantitative real-time RT-PCR (RQ-PCR)
Expression levels of LMO2, TAL1, HOX11, and HOX11L2 transcripts and the CALM-AF10 fusion product were quantified relative to the expression level of the endogenous housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using real-time RT-PCR in an ABI 7700 sequence detection system (Applied Biosystems) as described previously.28,34 The expression levels relative to the GAPDH housekeeping gene were calculated following the equation: relative expression level as percentage of GAPDH expression = 2-ΔCt × 100%, whereby ΔCt = Cttarget - CtGAPDH.
Primer and probe combinations were designed using Oligo 6.22 software (Molecular Biology Insight, Cascade, CO) and were purchased from Eurogentec (Seraing, Belgium). Primers and probe had melting temperatures of 65°C to 66.5°C and 73°C to 75°C, respectively, and performed with 95% efficiency or higher as determined from slopes of standard curves. This allows direct normalization of the target reaction to GAPDH expression levels at the Ct level.34 Primers and probe for the detection of the housekeeping gene GAPDH have been described previously.28 For the detection of total LMO2 transcripts derived from the LMO2 distal promoter (upstream of exon 1), RAG2-LMO2 fusion transcripts and transcripts derived from the LMO2 proximal promoter (exon 3), the forward primer 5′-TTG GGG ACC GCT ACT T-3′, and reverse primer 5′-ATG TCC TGT TCG CAC ACT-3′ were used in combination with the probe 5′-(FAM)-AAG CTC TGC CGG AGA GAC TAT CT-3′. For the detection of distal LMO2 transcripts and/or RAG2-LMO2 fusion transcripts, forward primer 5′-TCA ACC TGA ACT TGC AGT AG-3′ and reverse primer 5′-TCT CTC GGG AAG GTC TAT TT-3′ were used in combination with the probe 5′-(FAM)-AAC CAG AGA CAG AGG GAA GCT G-3′. For CALM-AF10, 5′ and 3′ CALM-AF10 fusion transcripts were detected in separate reactions using the CALM-AF10 forward primer 5′-TTA ACT GGG GGA TCT AAC TG-3′ in combination with the 5′ fusion transcript reverse primer 5′-GCT GCT TTG CTT TCT CTT C-3′ or the 3′ fusion transcript reverse primer 5′-CCC TCT GAC CCT CTA GCT TC-3′, both in combination with the common CALM-AF10 probe 5′-(FAM)-CTT GGA ATG CGG CAA CAA TG-(TAMRA)-3′. For detection of HOX11 expression levels, the forward primer 5′-CTC ACT GGC CTC ACC TT-3′ and reverse primer 5′-CTG TGC CAG GCT CTT CT-3′ were used in combination with the probe 5′-(FAM)-CCT TCA CAC GCC TGC AGA TC-(TAMRA)-3′. For detection of HOX11L2 expression levels, forward primer 5′-TCT GCG AGC TGG AAA A-3′ and reverse primer 5′-GAT GGA GTC GTT GAG GC-3′ were used in combination with probe 5′-(FAM)-CCA AAA CCG GAG GAC CAA GT-(TAMRA)-3′. For the detection of TAL1 transcripts, the forward primer 5′-TGC CTT CCC TAT GTT CAC-3′ and reverse primer 5′-AAG ATA CGC CGC ACAAC-3′ were used in combination with probe 5′-(FAM)-CCT TCC CCC TAT GAG ATG GAG A-(TAMRA)-3′. The SIL-TAL1 primers (ENF601, ENR664) and probe (ENP641) for the detection of a SIL-TAL1 deletion were used as recommended by the Europe Against Cancer Program.32
Statistical analysis
Kaplan-Meier curves were constructed in SPSS 11.0 software (SPSS, Chicago, IL) in a stratified analysis pairwise over strata, and P values were determined using the log-rank test. An event was defined as a relapse or nonresponse after induction therapy. The Mann-Whitney U test was used to analyze differences in LMO2 expression levels between subgroups. Data were considered statistically significant for P values less than .05.
Results
New recurrent deletion del(11)(p12p13) in pediatric T-ALL
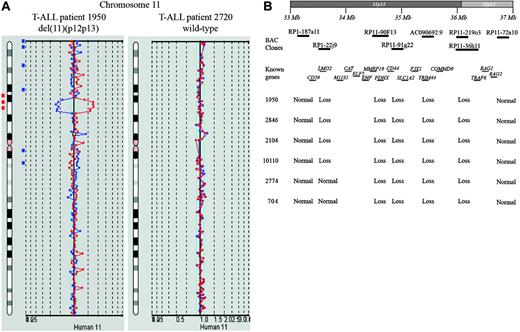
To identify new chromosomal abnormalities in pediatric T-ALL related to outcome and/or leukemogenesis, BAC array-CGH analysis was performed on a selected cohort of 30 of 64 clinically and karyotypically well-defined diagnostic T-ALL patient samples treated according to DCOG protocols. A recurrent loss of genomic material at chromosomal band 11p12-11p13 was found in 2 of 30 pediatric T-ALL cases (Figure 1A). Analysis of this pediatric T-ALL cohort and a second independent cohort (n = 74) treated according to the COALL97 protocol using FISH confirmed the presence of the del(11)(p12p13) in both positive patients, but also revealed 4 additional patients with this same deletion (data not shown). BAC array-CGH analysis of these additional positive cases confirmed the presence of this del(11)(p12p13) (Figure 1B). This deletion is therefore present in about 4% (6/138) of pediatric T-ALL patients. In 4 patients (1950, 2846, 2104, and 10 110), the deleted region was flanked by the BAC clones RP1-187A11 (11p13) and RP11-72A10 (11p12), and comprised the clones RP1-22J9, RP11-90F13, RP11-91G22, AC090692.9, RP11-219O3, and RP11-36H11. In the 2 remaining cases (2774 and 704), the deleted area was smaller and flanked by the clones RP1-22J9 and RP11-72A10 (Figure 1B).
New recurrent deletion, del(11)(p12p13), in pediatric T-ALL. (A) Chromosome 11 ideogram and corresponding BAC array-CGH plot of test DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patients 1950 (left panel) and 2720 (right panel). (B) Overview of BAC array-CGH results for the 11p12-11p13 region for the 4 DCOG and the 2 COALL T-ALL patients with del(11)(p12p13). The BAC clones present on the DNA array and located on chromosome bands 11p12-11p13 are shown. Specific genes located in this region are indicated. Depicted genome positions are based on the UCSC Genome Browser at http://genome.ucsc.edu/.
New recurrent deletion, del(11)(p12p13), in pediatric T-ALL. (A) Chromosome 11 ideogram and corresponding BAC array-CGH plot of test DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patients 1950 (left panel) and 2720 (right panel). (B) Overview of BAC array-CGH results for the 11p12-11p13 region for the 4 DCOG and the 2 COALL T-ALL patients with del(11)(p12p13). The BAC clones present on the DNA array and located on chromosome bands 11p12-11p13 are shown. Specific genes located in this region are indicated. Depicted genome positions are based on the UCSC Genome Browser at http://genome.ucsc.edu/.
The resolution of the BAC array-CGH system as used for our analysis is approximately 1 Mb. To determine the exact telomeric and centromeric breakpoints for this del(11)(p12p13) in pediatric T-ALL, we used the oligo array-CGH system of Agilent Technologies with a resolution of approximately 35 Kb. In agreement with the BAC array-CGH data, the oligo array-CGH analysis showed identical genomic losses at 11p12-11p13 for these 6 patients albeit at higher resolution (Figure 2A-G). Detailed analysis of the telomeric breakpoints indicated that both LMO2 probes located in intron 2 hybridized in a 1:1 ratio in patients 1950 (Figure 2B), 2846, 10 110, 2774, and 704 (Figure 2G), indicating that this part of LMO2 was retained. In all of these cases, both probes situated in M11S1 were deleted (Figure 2B,G). For patient 2104, the LMO2 intron 2 probes were lost, whereas the telomeric part of LMO2 (exon 6) was retained (Figure 2E), indicating that the genomic breakpoint is probably located downstream of LMO2 intron 2. At the centromeric breakpoint, the hybridization signals of both RAG1 and RAG2 probes were altered and indicated that one copy of both RAG1 and RAG2 genes was lost in patients 1950, 2846, and 10 110, whereas they had retained the LOC119710 locus (Figure 2C,G). For patients 2104 and 704, the centromeric breakpoint seemed to be situated in the FLJ14213 gene (Figure 2F-G). For patient 2774, both FLJ14213 probes were lost, whereas the TRAF6 probes hybridized normally.
FISH analysis confirms array-CGH data
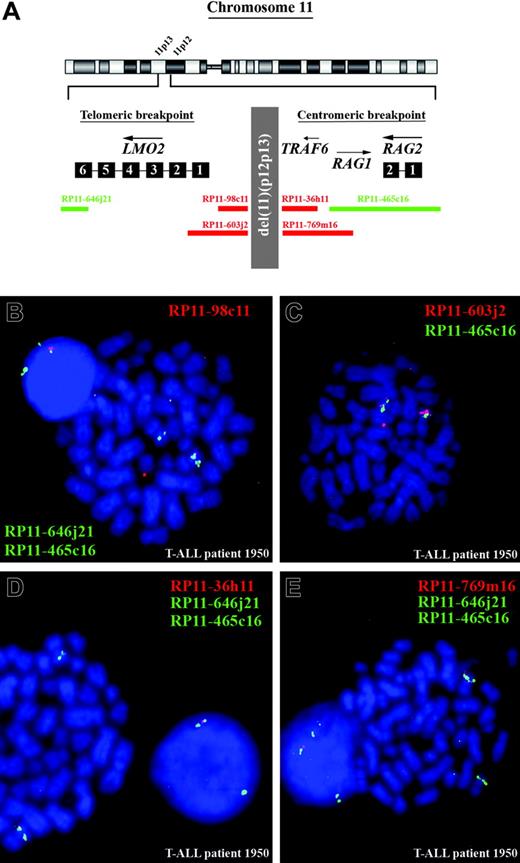
To confirm the BAC and oligo array-CGH data and to further characterize the exact breakpoints of this del(11)(p12p13), metaphase and interphase cells of the positive T-ALL cases were analyzed by FISH. In Figure 3A, the genomic positions of the FISH probes are visualized. For patient 1950, FISH analysis with RP11-465C16, which covers both RAG genes, RP11-646J21, which covers the telomeric part of LMO2, and RP11-98C11, which is located directly centromeric of LMO2, confirmed heterozygous loss of a region directly upstream of LMO2 (Figure 3B). The RP11-603J2 probe that includes part of the LMO2 locus was partly retained in the mutant allele (Figure 3C), indicating that the telomeric breakpoint of the del(11)(p12p13) was situated in a 9-kb region surrounding exon 1 of LMO2. Similar analysis in this patient of the centromeric breakpoint indicated that both RP11-36H11 (Figure 3D) and RP11-769M16 (Figure 3E) were deleted, whereas at least part of RP11-465C16 was retained. This confirms that the telomeric breakpoint of the del(11)(p12p13) in this patient was located in or just flanking the RAG genes.
FISH analysis for the 5 other cases with del(11)(p12p13) (Table 1) confirmed that this deletion also targeted LMO2 in patients 2846, 2104, and 10 110. However, for patients 2774 and 704 (Table 1) both RP11-603J2 and RP11-98C11 probes showed a normal hybridization pattern, suggesting that in these cases the break had occurred upstream of LMO2 between the LMO2 and M11S1 genes.
FISH analysis in 6 pediatric T-ALL patients with del(11)(p12p13)
. | No. of hybridization signals . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Telomeric breakpoint . | . | . | Centromeric breakpoint . | . | . | |||||
| Patient ID . | 646j21 . | 603j2 . | 98c11 . | 36h11 . | 769m16 . | 465c16 . | |||||
| 1 950 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 846 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 104 | 2 | 1 | 1 | 2* | 2* | 2 | |||||
| 10 110 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 774 | 2 | 2 | 2 | 2* | 2* | 2 | |||||
| 704 | 2 | 2 | 2 | 2* | 2* | 2 | |||||
. | No. of hybridization signals . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Telomeric breakpoint . | . | . | Centromeric breakpoint . | . | . | |||||
| Patient ID . | 646j21 . | 603j2 . | 98c11 . | 36h11 . | 769m16 . | 465c16 . | |||||
| 1 950 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 846 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 104 | 2 | 1 | 1 | 2* | 2* | 2 | |||||
| 10 110 | 2 | 2* | 1 | 1 | 1 | 2 | |||||
| 2 774 | 2 | 2 | 2 | 2* | 2* | 2 | |||||
| 704 | 2 | 2 | 2 | 2* | 2* | 2 | |||||
Intensity difference between the hybridization signal on the wild-type and the mutated allele
A single patient from the DCOG T-ALL cohort had a classical t(11;14)(p13;q11) by conventional cytogenetics. To determine the exact frequency of classical t(11;14)(p13;q11) or the t(7;11)(q34; p13) translocations involving LMO2, both the DCOG and the COALL cohorts (n = 138) were analyzed by FISH using LMO2 flanking BAC clones (data not shown). In total, 9 cases were identified that contained a translocation involving LMO2 (9/138, 6.5%), including the patient also positive by conventional cytogenetics. Including these LMO2-translocated patients, the frequency of LMO2 rearrangements (t(11;14)(p13;q11), t(7;11)(q34;p13), or del(11)(p12p13)) was 9.4% (13/138) in total.
Molecular characterization of deletion, del(11)(p12p13), in 6 pediatric T-ALL patients. Chromosome 11 ideogram and corresponding oligo array-CGH plot of test DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patient 1950 (A) and patient 2104 (D). Hybridization signals in the absence of amplifications or deletions scatter around the “zero” line, indicating equal hybridization for patient and reference DNA. Hybridization signals around the -2X or +2X lines represent loss of the corresponding region in the patient DNA. Detailed analysis of the telomeric breakpoints in patients 1950 (B) and 2104 (E) and the centromeric breakpoints in patients 1950 (C) and 2104 (F) of the deletion, del(11)(p12p13). (G) Overview of oligo array-CGH results in the potential breakpoint regions for 4 DCOG and the 2 COALL T-ALL patients with del(11)(p12p13). The 60-mer oligos present on the DNA array and located in the telomeric and centromeric breakpoint regions, as well as the specific genes located in this region with their transcription direction, are shown. N indicates normal; L, loss; and U, noninformative.
Molecular characterization of deletion, del(11)(p12p13), in 6 pediatric T-ALL patients. Chromosome 11 ideogram and corresponding oligo array-CGH plot of test DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patient 1950 (A) and patient 2104 (D). Hybridization signals in the absence of amplifications or deletions scatter around the “zero” line, indicating equal hybridization for patient and reference DNA. Hybridization signals around the -2X or +2X lines represent loss of the corresponding region in the patient DNA. Detailed analysis of the telomeric breakpoints in patients 1950 (B) and 2104 (E) and the centromeric breakpoints in patients 1950 (C) and 2104 (F) of the deletion, del(11)(p12p13). (G) Overview of oligo array-CGH results in the potential breakpoint regions for 4 DCOG and the 2 COALL T-ALL patients with del(11)(p12p13). The 60-mer oligos present on the DNA array and located in the telomeric and centromeric breakpoint regions, as well as the specific genes located in this region with their transcription direction, are shown. N indicates normal; L, loss; and U, noninformative.
Molecular characterization of del(11)(p12p13)
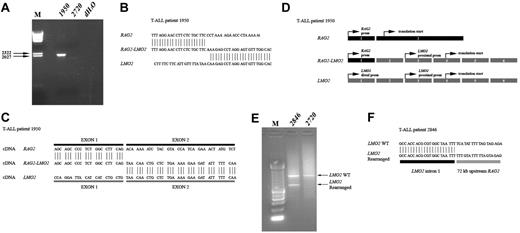
We next characterized the genomic breakpoint of the del(11)(p12p13) in T-ALL patient 1950 in more detail. Long-range PCR analysis on genomic DNA using primers situated in intron 1 of RAG2 and intron 1 of LMO2 revealed a specific band of approximately 2000 bp (Figure 4A) for patient 1950 that was not present in a del(11)(p12p13)-negative control (2720). Sequence analysis showed the exact positions of the genomic breakpoints in both intron regions (Figure 4B). It was expected that this deletion gives rise to a fusion of exon 1 of RAG2 to exon 2 of LMO2, which was confirmed at the mRNA level (Figure 4C-D). Subsequent RT-PCR failed to detect RAG2-LMO2 fusion products in any of the remaining del(11)(p12p13)-positive T-ALL patients. Therefore, we performed ligation-mediated PCR (LM-PCR) in order to determine additional genomic breakpoints. In patient 2846, LM-PCR with an LMO2 intron 1-specific primer revealed an aberrant PCR product in addition to the expected wild-type band (Figure 4E). Sequence analysis showed that in this case LMO2 intron 1 sequences were fused to a region located 72-kb upstream of RAG2 (Figure 4F).
Del(11)(p12p13) correlates with a mature immunophenotype and high LMO2 expression in T-ALL
Immunophenotypic analysis of LMO2-rearranged cases revealed that patients with the del(11)(p12p13) did not express CD34, CD33, or CD1 but expressed mCD3 (Table 2). None of these cases expressed TCRγδ, whereas 2 patients expressed TCRαβ (1950 and 2104) and 2 patients were CD4/CD8 double positive (1950 and 2846) in agreement with an immunophenotypic mature developmental stage. The LMO2-translocated patients were immunophenotypically more immature. Two of 3 cases expressed CD1, but none expressed mCD3 and/or the TCR. All 3 cases were CD4/CD8 double positive, consistent with an early cortical developmental stage.
Immunophenotypic characteristics of LMO2-rearranged pediatric T-ALL patients
. | . | Positive cells, % . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID . | LMO2 rearrangement . | CD34 . | CD33 . | CD1 . | CD4 . | CD8 . | cytCD3 . | mCD3 . | TCRαβ . | |||||||
| 1950 | del(11)(p12p13) | 0 | 0 | 0 | 85 | 91 | 90 | 80 | Pos | |||||||
| 2846 | del(11)(p12p13) | 0 | 10 | 0 | 48 | 57 | 75 | 26 | Neg | |||||||
| 2104 | del(11)(p12p13) | 0 | 0 | 0 | 6 | 1 | 98 | 45 | Pos | |||||||
| 2698 | t(11;14)(p13;q11) | 1 | 8 | 0 | 42 | 69 | 93 | 3 | Neg | |||||||
| 2789 | t(11;14)(p13;q11) | 1 | 12 | 25 | 59 | 73 | 85 | 12 | Neg | |||||||
| 2735 | t(11;14)(p13;q11) | 4 | 1 | 71 | 90 | 94 | 82 | 8 | Neg | |||||||
. | . | Positive cells, % . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID . | LMO2 rearrangement . | CD34 . | CD33 . | CD1 . | CD4 . | CD8 . | cytCD3 . | mCD3 . | TCRαβ . | |||||||
| 1950 | del(11)(p12p13) | 0 | 0 | 0 | 85 | 91 | 90 | 80 | Pos | |||||||
| 2846 | del(11)(p12p13) | 0 | 10 | 0 | 48 | 57 | 75 | 26 | Neg | |||||||
| 2104 | del(11)(p12p13) | 0 | 0 | 0 | 6 | 1 | 98 | 45 | Pos | |||||||
| 2698 | t(11;14)(p13;q11) | 1 | 8 | 0 | 42 | 69 | 93 | 3 | Neg | |||||||
| 2789 | t(11;14)(p13;q11) | 1 | 12 | 25 | 59 | 73 | 85 | 12 | Neg | |||||||
| 2735 | t(11;14)(p13;q11) | 4 | 1 | 71 | 90 | 94 | 82 | 8 | Neg | |||||||
cytCD3 indicates cytoplasmic CD3; mCD3, membrane CD3; Pos, positive; and Neg, negative
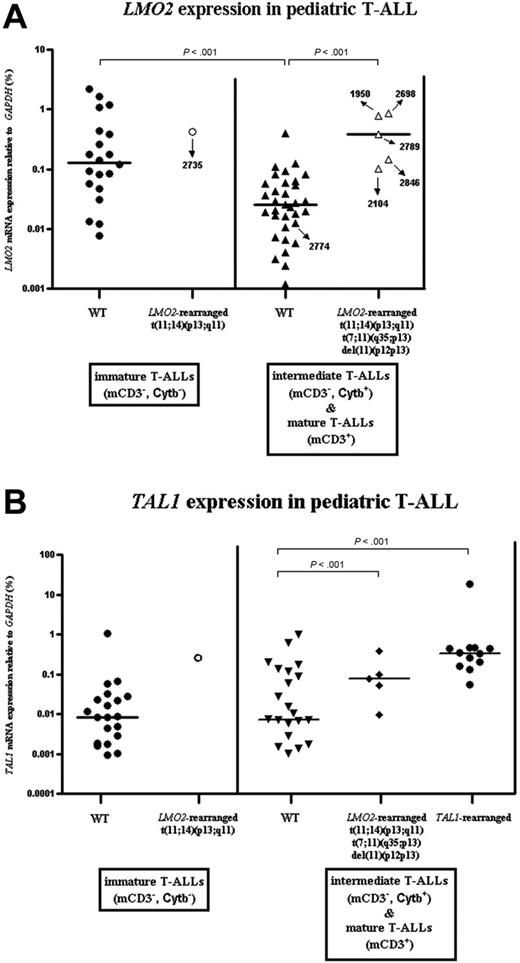
LMO2 mRNA expression levels of LMO2-rearranged versus nonrearranged cases were measured using RQ-PCR on 59 DCOG T-ALL patient samples for which immunophenotypic data were available. Since LMO2 is highly expressed in T-ALL samples with an immature immunophenotype,33 we divided T-ALL samples into 2 categories: the first category included the immature double-negative cases (CD4-/CD8-, mCD3-, Cytβ-), whereas the second category comprised more mature cases with evidence for TCRβ rearrangements (Cytβ+) and/or TCR/CD3 expression.35 For LMO2 nonrearranged cases (WT), LMO2 expression was significantly higher in the immature T-ALL cases compared with the immunophenotypically more advanced patients (Figure 5A, Mann-Whitney, P < .001). LMO2-rearranged cases had significantly higher LMO2 levels compared with the LMO2 nonrearranged T-ALL patients with a comparable immunophenotype (P < .001). LMO2 expression was low for patient 2774 (Figure 5A), which was in line with the observation that the deletion breakpoints did not affect the LMO2 gene.
FISH analysis confirms the presence of del(11)(p12p13) in T-ALL patient 1950. (A) Chromosome ideogram and overview of the genomic position of the BAC clones used for FISH analysis, located in the telomeric and centromeric breakpoint regions. (B) Dual-color FISH analysis on metaphase spreads of patient 1950 using RP11-465C16 (green), RP11-646J21 (green), and RP11-98C11 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse. The extrachromosomal red signal represents background. (C) Dual-color FISH analysis on metaphase spreads of the same patient using RP11-465C16 (green) and RP11-603J2 (red). The intensity of the red signal is lower compared with the wild-type allele of chromosome 11, suggesting that only part of RP11-603J2 is deleted. (D) Dual-color FISH analysis on metaphase spreads using RP11-465C16 (green), RP11-646J21 (green), and RP11-36H11 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse. (E) Dual-color FISH analysis on metaphase spreads using RP11-465C16 (green), RP11-646J21 (green), and RP11-769M16 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse.
FISH analysis confirms the presence of del(11)(p12p13) in T-ALL patient 1950. (A) Chromosome ideogram and overview of the genomic position of the BAC clones used for FISH analysis, located in the telomeric and centromeric breakpoint regions. (B) Dual-color FISH analysis on metaphase spreads of patient 1950 using RP11-465C16 (green), RP11-646J21 (green), and RP11-98C11 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse. The extrachromosomal red signal represents background. (C) Dual-color FISH analysis on metaphase spreads of the same patient using RP11-465C16 (green) and RP11-603J2 (red). The intensity of the red signal is lower compared with the wild-type allele of chromosome 11, suggesting that only part of RP11-603J2 is deleted. (D) Dual-color FISH analysis on metaphase spreads using RP11-465C16 (green), RP11-646J21 (green), and RP11-36H11 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse. (E) Dual-color FISH analysis on metaphase spreads using RP11-465C16 (green), RP11-646J21 (green), and RP11-769M16 (red). The wild-type allele of chromosome 11 shows 2 green and 1 red signal, whereas on the mutated allele the red signal is lost and both green signals fuse.
LMO2 rearrangements in relation to other oncogenic events in pediatric T-ALL
In order to determine the relation between LMO2 rearrangements and other recurrent cytogenetic abnormalities in pediatric T-ALL, we screened all 13 LMO2-rearranged T-ALL patients for abnormalities at the TAL1, HOX11L2, HOX11, CALM-AF10, MLL, and cMYC loci using FISH analysis and RQ-PCR (data not shown). None of the LMO2-positive cases showed rearrangements of any of these loci. Nevertheless, in the del(11)(p12p13)-positive patient 2774 without involvement of LMO2, a SIL-TAL1 interstitial deletion was identified. This indicates that del(11)(p12p13)-positive T-ALL with elevated LMO2 levels together with LMO2-translocated T-ALL samples reflect a separate cytogenetic subgroup without detectable TAL1, HOX11L2, HOX11, CALM-AF10, MLL, and cMYC abnormalities.
We further determined TAL1 expression levels by RQ-PCR (Figure 5B). These analyses showed that for intermediate and mature T-ALL patients, TAL1 is significantly more highly expressed in both LMO2-rearranged (Figure 5B, Mann-Whitney, P < .001) and TAL1-rearranged (Figure 5B, Mann-Whitney, P < .001) cases, compared with non-LMO2/TAL1-rearranged samples.
LMO2 activation induced by enhanced activity of the LMO2 proximal promoter
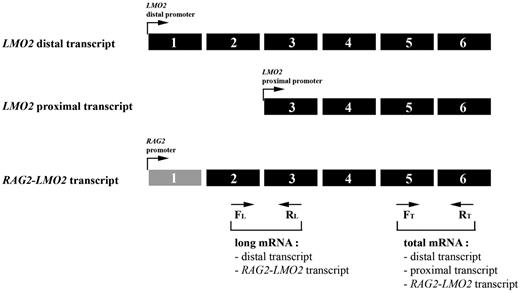
In patient 1950, the del(11)(p12p13) resulted in a RAG2-LMO2 gene fusion in which the distal LMO2 promoter is replaced by the RAG2 promoter (Figure 4C). However, a comparable fusion product was not detected in any of the remaining 3 del(11)(p12p13)-positive patients with elevated LMO2 levels, suggesting that RAG2-LMO2 fusion products were either expressed at very low levels or that other genomic regions were fused to LMO2, as found for patient 2846 (Figure 4F). We hypothesized that LMO2 rearrangements due to the del(11)(p12p13) could result in the loss of a negative regulatory domain upstream of LMO2, with subsequent activation of the proximal promoter (exon 3), a situation comparable with LMO2-translocated patients.36,37 To elucidate which kind of LMO2 transcripts are predominantly expressed, we developed a double RQ-PCR: one RQ-PCR can quantify LMO2 transcripts derived from the distal LMO2 promoter as well as RAG2-LMO2 fusion products. The second RQ-PCR quantifies total LMO2 transcripts derived from both the distal and proximal LMO2 promoters as well as RAG2-LMO2 fusion products (Figure 6 and Table 3). These analyses revealed that LMO2 transcripts derived from the distal promoter or RAG2-LMO2 fusion products in del(11)(p12p13)-positive patients (1950 and 2846) represent only 5.5% to 9.3% of total LMO2 transcripts (Figure 6 and Table 3). For both LMO2-translocated patients (2789 and 2735) also 8.5% to 10% of total LMO2 transcripts originate from the distal promoter.
Activation of the proximal promoter of LMO2 in LMO2-rearranged T-ALL patients
. | Total LMO2 mRNA . | . | Long LMO2 mRNA . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|
| Patient ID, by mutation . | dCt . | Expression level . | dCt . | Expression level . | Long transcripts, % . | Proximal transcripts, % . | ||
| Del(11)(p12p13) | ||||||||
| 1950 | 6.14 | 0.58577 | 8.52 | 0.05421 | 9.3 | 90.7 | ||
| 2846 | 7.24 | 0.19499 | 10.15 | 0.01062 | 5.4 | 94.6 | ||
| LMO2 translocation | ||||||||
| 2789 | 6.81 | 0.29974 | 9.11 | 0.03005 | 10.0 | 90.0 | ||
| 2735 | 6.96 | 0.25799 | 9.43 | 0.02182 | 8.5 | 91.5 | ||
. | Total LMO2 mRNA . | . | Long LMO2 mRNA . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|
| Patient ID, by mutation . | dCt . | Expression level . | dCt . | Expression level . | Long transcripts, % . | Proximal transcripts, % . | ||
| Del(11)(p12p13) | ||||||||
| 1950 | 6.14 | 0.58577 | 8.52 | 0.05421 | 9.3 | 90.7 | ||
| 2846 | 7.24 | 0.19499 | 10.15 | 0.01062 | 5.4 | 94.6 | ||
| LMO2 translocation | ||||||||
| 2789 | 6.81 | 0.29974 | 9.11 | 0.03005 | 10.0 | 90.0 | ||
| 2735 | 6.96 | 0.25799 | 9.43 | 0.02182 | 8.5 | 91.5 | ||
dCt indicates the expression of indicated promoter transcripts relative to the housekeeping gene GAPDH
Molecular characterization of del(11)(p12p13) in T-ALL patients 1950 and 2846. (A) Long-range PCR analysis on genomic DNA of patient 1950 using primers situated in intron 1 of RAG2 and intron 1 of LMO2 revealed a specific band of approximately 2000 bp. Patient 2720 served as a negative control. (B) Sequence analysis confirmed the exact position of the genomic breakpoint. (C) PCR analysis on cDNA of this patient revealed a RAG2-LMO2 fusion gene, in which exon 1 of RAG2 was fused to exon 2 of LMO2. (D) Gene (exon) structure of both RAG2 and LMO2 shows that the translation initiation sites are situated in exon 2 and exon 4, respectively. As a consequence, translation of the RAG2-LMO2 fusion gene will also be initiated in exon 4. (E) LM-PCR analysis on HincII-digested genomic DNA from patient 2846 using an LMO2 intron 1-specific primer revealed an aberrant PCR product of approximately 600 bp. The expected wild-type product is approximately 1000 bp and is visible in both patients 2846 and 2720, who served as a negative control. (F) Sequence analysis confirmed that in patient 2846 the LMO2 intron 1 sequences are fused to a genomic region upstream of RAG2. prom indicates promoter.
Molecular characterization of del(11)(p12p13) in T-ALL patients 1950 and 2846. (A) Long-range PCR analysis on genomic DNA of patient 1950 using primers situated in intron 1 of RAG2 and intron 1 of LMO2 revealed a specific band of approximately 2000 bp. Patient 2720 served as a negative control. (B) Sequence analysis confirmed the exact position of the genomic breakpoint. (C) PCR analysis on cDNA of this patient revealed a RAG2-LMO2 fusion gene, in which exon 1 of RAG2 was fused to exon 2 of LMO2. (D) Gene (exon) structure of both RAG2 and LMO2 shows that the translation initiation sites are situated in exon 2 and exon 4, respectively. As a consequence, translation of the RAG2-LMO2 fusion gene will also be initiated in exon 4. (E) LM-PCR analysis on HincII-digested genomic DNA from patient 2846 using an LMO2 intron 1-specific primer revealed an aberrant PCR product of approximately 600 bp. The expected wild-type product is approximately 1000 bp and is visible in both patients 2846 and 2720, who served as a negative control. (F) Sequence analysis confirmed that in patient 2846 the LMO2 intron 1 sequences are fused to a genomic region upstream of RAG2. prom indicates promoter.
Clinical relevance of LMO2 rearrangements in pediatric T-ALL
To study the prognostic relevance of LMO2 rearrangements in pediatric T-ALL, Kaplan-Meier disease-free-survival (DFS) curves were created for LMO2-rearranged cases versus LMO2 wild-type cases. In a stratified analysis of the combined DCOG and COALL cohorts (n = 138), LMO2 rearrangements had borderline significance for poor outcome (log-rank, P = .03).
Discussion
LMO2 has been identified as an oncogene in T-ALL due to its involvement in the translocation t(11;14)(p13;q11) or t(7;11)(q35; p13), in which the TCR-LMO2 fusion results in a constitutive activation of the LMO2 gene.15,16 However, high LMO2 expression levels have also been reported in the absence of translocations,32,38 suggesting that alternative mechanisms may exist in T-ALL, resulting in the activation of LMO2.
Using the genome-wide array-CGH technique for the detection of genomic amplification and/or deletion areas, we identified a new recurrent deletion in pediatric T-ALL cases (ie, the del(11)(p12p13)). Screening pediatric T-ALL samples showed that this deletion is present in about 4% of pediatric T-ALL patients (6/138 cases). The genomic breakpoints are located in intron 1 of RAG2 and intron 1 of LMO2 for patient 1950, placing LMO2 under the control of the RAG2 promoter. As expected, a RAG2-LMO2 fusion product could be cloned. Since exon 1 of RAG2 does not contain a translation initiation-site and the translation initiation-site of LMO2 is located in exon 3, this fusion product will produce normal LMO2 protein. However, RAG2-LMO2 fusions could not be identified in any of the remaining del(11)(p12p13)-positive T-ALL patients, suggesting that the localization of genomic breakpoints in these deletions is heterogeneous. This was demonstrated by the identification of the genomic breakpoint in patient 2846, in whom the deletion caused fusion of a genomic region approximately 72 Kb upstream of the RAG2 gene with LMO2 intron 1 sequences. Although the exact genomic breakpoints of both other del(11)(p12p13)-positive cases (2104 and 10 110) remain to be identified, oligo array-CGH, FISH, and RQ-PCR analyses predict involvement of LMO2 in these cases in the same manner as in patients 1950 and 2846.
Recently, it has been proposed that deletion of negative regulatory sequences, located approximately 3000 bp upstream of exon 1 of LMO2, could contribute to ectopic LMO2 expression in T-cell leukemia.37 Of interest, this negative regulatory element was consistently removed in 4 del(11)(p12p13)-positive T-ALL cases that target LMO2, and may therefore provide a mechanism for the enhanced LMO2 activation in pediatric T-ALL. However, based upon the RAG2-LMO2 fusion product that was identified in patient 1950, promoter substitution could also contribute to elevated levels of LMO2 expression. Our RQ-PCR data supported only marginal contribution of the distal LMO2 promoter from the remaining wild-type allele or RAG2-LMO2 fusion products to the total LMO2 mRNA levels in del(11)(p12p13)-positive patients. Also, 2 LMO2-translocated patients as analyzed by RQ-PCR demonstrated low distal LMO2 promoter activity, confirming enhanced proximal promoter activity due to the loss of this negative regulatory domain.36,37
LMO2 and TAL1 expression in pediatric T-ALL. Relative expression levels of LMO2 (A) and TAL1 (B) as percentage of GAPDH expression levels for 59 pediatric T-ALL patients (DCOG cohort). Patients were divided into 2 maturation stages according to their cytoplasmatic TCRβ (Cytβ) and membrane CD3 (mCD3) expression. Median expression levels are indicated by horizontal bars.
LMO2 and TAL1 expression in pediatric T-ALL. Relative expression levels of LMO2 (A) and TAL1 (B) as percentage of GAPDH expression levels for 59 pediatric T-ALL patients (DCOG cohort). Patients were divided into 2 maturation stages according to their cytoplasmatic TCRβ (Cytβ) and membrane CD3 (mCD3) expression. Median expression levels are indicated by horizontal bars.
Array-CGH and FISH data indicated that the deletion area for both del(11)(p12p13)-positive patients 2774 and 704 is smaller compared with the other 4 del(11)(p12p13)-positive patients. For both patients, the deletion seems to be located upstream of the negative regulatory region of LMO2 as patient 2774 does not have elevated LMO2 expression levels. These 2 cases may support the hypothesis that the minimally deleted region on chromosome 11 further contains a tumor-suppressor gene that could contribute to the pathogenesis of T-ALL.
LMO2-rearranged cases of the DCOG cohort including those with the del(11)(p12p13) as well as the 3 patients with a t(11; 14)(p13;q11) expressed significantly higher levels of LMO2 than LMO2 nonrearranged T-ALL samples with a comparable immunophenotypic development stage (ie, the cortical or mature T-cell developmental stage). The expression was comparable with immature T-ALL patients who highly express LMO2 as a consequence of their immature developmental stage.17,18 Nevertheless, a number of immunophenotypically more advanced T-ALL patients demonstrated high LMO2 expression levels in the absence of currently known LMO2 rearrangements, yet other alternative mechanisms leading to LMO2 activation in pediatric T-ALL may exist.
LMO2-rearranged pediatric T-ALL samples with the del(11)(p12p13) may have a maturation stage that is more advanced compared with LMO2-translocated patients. Two of 3 cases with the del(11)(p12p13) involving the LMO2 gene were TCRαβ positive. In contrast, none of the LMO2-translocated patients expressed CD3 and/or TCRαβ, but 2 of these patients expressed CD1 conform an early cortical developmental stage. Whether this reflects true differences in maturation stage between patients with the del(11)(p12p13) and the LMO2-translocated cases needs to be validated in a larger panel of patients. Similar variations in cortical and mature T-cell developmental stages were also observed for TAL1-rearranged T-ALL patients in the same cohort.32 Deregulation of LMO2 or TAL1 may lead to a similar T-cell developmental arrest. TAL1 and LMO2 act in the same pentameric transcription complex, and deregulation of either or both genes may lead to the (in)activation of identical target genes.
The frequency of LMO2 rearrangements in both cohorts combined is about 9%, and includes 4 patients with LMO2 rearrangements due to the del(11)(p12p13) and 9 cases with a t(11;14)(p13; q11) or the t(7;11)(q35;p13). These LMO2 abnormalities were shown to be independent of other recurrent cytogenetic abnormalities including TAL1, HOX11L2, HOX11, CALM-AF10, MLL, or cMYC.32 Patient 2774, who was del(11)(p12p13) positive but lacked LMO2 activation, had an interstitial SIL-TAL1 deletion.
Since LMO2 and TAL1 are frequently coexpressed in mature T-ALL cases and since no TAL1 deletions and/or translocations were observed in the LMO2-rearranged cases, we determined TAL1 mRNA expression in the 59 T-ALL samples for which LMO2 expression data were available. These analyses confirmed that TAL1 is significantly more highly expressed in both LMO2- and TAL1-rearranged T-ALL cases, compared with non-LMO2/TAL1-rearranged samples. These data further suggest that for del(11)(p12p13)-positive patients, alternative mechanisms of TAL1 activation besides TAL1 deletions and translocations may exist in T-ALL or that TAL1 may be a direct target gene for LMO2-driven transcription.
Elevated LMO2 expression by activation of the LMO2 proximal promoter. Relative expression of long and total mRNA transcript levels of LMO2 as measured by the RQ-PCR strategy. Long transcripts including the RAG2-LMO2 fusion transcript can be measured by the exon 2/3 primer combination, whereas the total amount of LMO2 transcript was measured using an exon 5/6 primer combination. Expression of proximal promoter transcripts is calculated by subtracting the long-transcript expression from the total expression. FL indicates forward primer long mRNA transcript; RL, reverse primer long mRNA transcript; FT, forward primer total mRNA transcript; and RT, reverse primer total mRNA transcript.
Elevated LMO2 expression by activation of the LMO2 proximal promoter. Relative expression of long and total mRNA transcript levels of LMO2 as measured by the RQ-PCR strategy. Long transcripts including the RAG2-LMO2 fusion transcript can be measured by the exon 2/3 primer combination, whereas the total amount of LMO2 transcript was measured using an exon 5/6 primer combination. Expression of proximal promoter transcripts is calculated by subtracting the long-transcript expression from the total expression. FL indicates forward primer long mRNA transcript; RL, reverse primer long mRNA transcript; FT, forward primer total mRNA transcript; and RT, reverse primer total mRNA transcript.
The presence of LMO2 rearrangements predicted for poor outcome in a stratified analysis of both the DCOG and COALL pediatric T-ALL cohorts. This prognostic significance has to be looked at cautiously due to the low number of patients, and a larger panel of T-ALL patients is needed to validate these findings. The presence of LMO2 translocations did not predict for poor outcome in a previous study.39
In conclusion, we report the identification of a new cryptic cytogenetic abnormality (ie, the del(11)(p12p13)) in 6 pediatric T-ALL patients targeting the LMO2 gene in 4 cases. For del(11)(p12p13)-positive patients involving LMO2, the proximal LMO2 promoter is highly activated due to the deletion of negative regulatory sequences upstream of LMO2. Abnormalities involving LMO2, including the del(11)(p12p13), the t(7;11)(q35;p13), and the t(11;14)(p13;q11), are more common in pediatric T-ALL (9%) as appreciated until now. LMO2 abnormalities are independent from other recurrent cytogenetic abnormalities as frequently present in T-ALL.
Prepublished online as Blood First Edition Paper, July 27, 2006; DOI 10.1182/blood-2006-04-019927.
Supported by the Ter Meulen Fund, the Royal Netherlands Academy of Arts and Sciences, and the Foundation “De Drie Lichten.” P.V.V. is financed by the Sophia Foundation for Medical Research (SSWO-440). C.L. was funded in part by a transnational research grant from the Leukemia and Lymphoma Society.
The authors declare no competing financial interests.
P.V.V. designed and performed research and wrote the paper; C.L. collaborated on the array-CGH study; E.R.v.W. collected and made available patient samples; T.H., M.P., and J.B.-G. performed and designed FISH analysis; M.v.G. collected immunophenotypic data and performed expression analysis; A.J.P.V. and W.A.K. supervised the DCOG trials; J.P.P.M., H.B.B., and R.P. wrote the grant, designed research, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We are greatly indebted to Dr A. W. Langerak for comments and critical discussion. We also thank Dr W. Dorlijn and R. Finch of Agilent Technologies, and Dr W. van Workum and A. Wijfjes of ServiceXS (Leiden, The Netherlands) for technical support in the oligo array-CGH analysis.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal