Abstract
The interleukin-3 receptor (IL-3R) subunits are overexpressed on acute myeloid leukemia (AML) blasts compared with normal hematopoietic cells and are thus potential targets for novel therapeutic agents. Both fluorescence-activated cell sorter (FACS) analysis and quantitative real-time reverse transcription-polymerase chain reaction (QRT-PCR) were used to quantify expression of the IL-3Rα and βc subunits on AML cells. QRT-PCR for both subunits was most predictive of killing of AML colony-forming cells (AML-CFCs) by diphtheria toxin-IL-3 fusion protein (DT388IL3). Among 19 patient samples, the relative level of the IL-3Rα was higher than the IL-3Rβc and highest in CD34+CD38-CD71- cells, enriched for candidate leukemia stem cells, compared with cell fractions depleted of such progenitors. Overall, the amount of IL-3Rβc subunit did not vary among sorted subpopulations. However, expression of both subunits varied by more than 10-fold among different AML samples for all subpopulations studied. The level of IL-3Rβc expression versus glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (set at 1000) ranged from 0.14 to 13.56 in CD34+CD38-CD71- cells from different samples; this value was correlated (r = .76, P = .05) with the ability of DT388IL3 to kill AML progenitors that engraft in β2-microglobin-deficient nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (n = 7). Thus, quantification of IL-3R subunit expression on AML blasts predicts the effectiveness IL-3R-targeted therapy in killing primitive leukemic progenitors.
Introduction
Acute myeloid leukemia (AML) is a clonal disorder of hematopoiesis characterized by the accumulation of large numbers of myeloid blast cells that fail to differentiate into functional blood cells.1 In spite of these obvious abnormalities in cell maturation there are now considerable data demonstrating a hierarchy of malignant progenitor cells in AML similar to that found in normal hematopoiesis and the existence of a small subpopulation of leukemia stem cells (LSCs) that possesses extensive proliferative capacity and the potential for self-renewal.2,3 Leukemic cells that initiate long-term hematopoiesis in stromal coculture (AML LTC-ICs) and suspension cultures and engraft in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (NOD/SCID leukemia-initiating cells [N/SL-ICs]) are typically present at a frequency of fewer than 1 in 10 000 malignant cells and express a cell-surface phenotype similar to that seen on normal multipotential long-term culture-initiating cells (LTC-ICs) and lymphomyeloid NOD/SCID mouse competitive repopulating units.4-7 Several recent studies have shown that N/SL-ICs can be phenotypically defined as CD34+, CD38-, human leukocyte antigen (HLA)-DR-, CD90-, CD71-, CD117-, and CD123+. This antigenic profile is intriguing in that it overlaps with normal stem cells (CD34+, CD38-, CD71-, HLA-DR-) but also has leukemia-specific features (CD90-, CD117-, and CD123++).4,8-16 The frequent relapses observed in patients with AML who achieve an initial remission of their leukemia suggest that LSCs might be relatively resistant to conventional chemotherapy and also might be responsible for re-establishing the malignant clone in patients with leukemia. These cells are thus obvious targets for novel therapeutic approaches in AML.
The human interleukin-3 receptor (IL-3R) is a heterodimeric structure.17 The α subunit is essential for ligand binding and confers specificity on the receptor. The common β subunit (βc), which is shared by the granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-5 receptors, is required for high-affinity ligand binding and signal transduction. The IL-3R is expressed on leukemic blasts from most patients with AML.14,18,19 The α subunit, in particular, is often expressed at much higher levels than are typically seen in normal hematopoietic cells and progenitors and is also detected on subpopulations of AML blasts enriched for N/SL-ICs.14,20 Thus, the IL-3R may be an appropriate target for cytotoxic drugs designed to selectively kill AML cells while sparing their normal hematopoietic-cell counterparts.
Fusion proteins in which the diphtheria toxin (DT) catalytic and translocation domains are genetically fused to a growth factor are a novel class of molecules which can selectively target and kill malignant cells expressing the relevant receptor.21-26 After ligand binding and receptor-mediated endocytosis, the toxin translocates to the cytosol, where it ADP-ribosylates elongation factor 2, leading to inactivation of protein synthesis and cell death.27-30 DT388IL3 consists of the truncated form of diphtheria toxin (DT388) linked to human IL-3. This fusion protein has been shown to target and kill cells which express the IL-3R, including AML blasts and progenitors.31 Previous studies have shown that this molecule will kill AML colony forming cells (AML-CFCs), long-term culture-initiating cells (AML LTC-ICs), and N/SL-ICs from many patient samples while showing little or no toxicity against analogous normal bone marrow progenitors.18 Although DT388IL3 is effective against many AML samples, only partial eradication of malignant progenitors was seen with some leukemias in spite of detectable expression of high-affinity IL-3R on target blasts. More recently, we determined that the level of IL-3R subunit expression (particularly the βc subunit) as detected by quantitative real-time reverse transcription-polymerase chain reaction (QRT-PCR) in AML blasts correlated with the effectiveness of DT388IL3 in killing AML-CFCs.32 However, in this study the relationship was not strong for a subset of patient samples and its usefulness in predicting the ability of DT388IL3 to kill N/SL-ICs was not investigated thoroughly.
The current study examined the possibility that expression of the IL-3R subunits might be heterogeneous among AML cells and progenitors. We were particularly interested in the possibility that IL-3R expression on the most primitive AML cells (ie, the candidate LSCs that initiate long-term engraftment and proliferation in immunodeficient mice), might be different than the expression detected on the majority population of leukemic cells that have limited proliferative capacity. The level of IL-3R subunit expression was measured by QRT-PCR in subpopulations of leukemic cells enriched for AML progenitors with a multiparameter fluorescence-activated cell sorter (FACS) strategy. These levels were then compared with the ability of DT388IL3 to kill N/SL-ICs from many of the same AML samples. The results demonstrate that the level of expression of the IL-3Rβc (but not the IL-3Rα) in unsorted blasts as well as subpopulations enriched for N/SL-ICs predicted the ability of DT388IL3 to kill these progenitors from most patient samples. A phase 1/2 clinical trial testing DT388IL3 in patients with relapsed or refractory AML is currently in progress. These data suggest that screening patient blast samples for expression of IL-3R subunits may allow prediction of therapeutic benefit with this agent.
Patients, materials, and methods
Blood and marrow samples
Peripheral-blood (PB) samples from 19 newly diagnosed patients with AML and bone marrow (BM) from 3 healthy allogeneic transplant donors were obtained with the approval of the Clinical Research Ethics Board of the University of British Columbia and after obtaining informed consent from patients with AML and from donors in accordance with the Declaration of Helsinki. Light-density mononuclear cells were isolated and cryopreserved as previously described.5 Diagnosis and classification of AML were based on the criteria of the French-American-British (FAB) group.33 Cytogenetic analysis was performed on diagnostic bone marrow samples of patients with AML. Thawed cells were washed twice in Iscove modified Dulbecco medium (IMDM) containing 10% fetal calf serum (FCS) before use in the experiments described below. Normal BM was enriched for CD34+ cells using an immunomagnetic separation technique (EasySep; StemCell Technologies, Vancouver, BC, Canada) prior to the FACS isolation of subpopulations described under “FACS analysis and subpopulation isolation of AML and normal cells.”
DT388IL-3 fusion protein
DT388IL3 was constructed by joining the human IL-3 cDNA to a truncated diphtheria toxin (DT388) sequence that lacks the native binding site via an intervening (G4S)2 linker. Recombinant protein was prepared, purified, stored, and tested as described previously.34 This material was found to kill IL-3R+ cell lines and AML-CFCs from primary AML samples expressing high-affinity IL-3Rs.19,31,34 The IL-3R binding affinity of this fusion toxin is similar to that of native human IL-3.22 In addition, the toxicity of DT388IL3 against IL-3R-bearing TF1/Bcl2 cells was reduced more than 120-fold by coincubation with excess human IL-3.31
Cultures of AML cells
AML cells were incubated for 24 hours at 37°C at a concentration of 1 × 106 cells/mL with or without DT388IL3. Equal fractions of the cells recovered from cultures with or without toxin were assayed in AML-CFC assay or in β2-microglobin-deficient NOD/SCID mice without regard to change in cell numbers.
AML-CFC assays were performed by plating cells at 0.2 to 1.0 × 105 cells/mL in methylcellulose-based medium (StemCell Technologies) as previously described. Cultures were scored after 14 days at 37°C in a humidified atmosphere with 5% CO2 for the presence of clusters (4-20 cells) and colonies (more than 20 cells).5
FACS analysis and subpopulation isolation of AML and normal cells
Subpopulation analysis and isolation from AML PB and CD34+-enriched normal BM cells were performed with monoclonal antibodies directly coupled to the fluorochromes fluorescein isothiocyanate (FITC), phycoerythrin (PE), and cyanin5-succinimidylester (Cy5). Anti-CD34-Cy5 and anti-CD71-FITC were kind gifts from Dr Peter Lansdorp (Terry Fox Laboratory, Vancouver, BC, Canada); Anti-CD38-PE was obtained from Becton Dickinson (San Jose, CA). Biotinylated antibodies recognizing CD123 (IL-3Rα) and CD131 (IL-3Rβc) were visualized by subsequent labeling with streptavidin-PE or -allophycocyanin (APC) (Becton Dickinson). Cells were analyzed on the basis of fluorescence intensity on a dual laser FACSaria instrument (Becton Dickinson). Gates were set to exclude nonviable propidium iodide+ (PI+) cells, and 100% of cells labeled with an irrelevant isotype control antibody. For analysis of IL-3R subunit expression the mean fluorescence intensity (MFI) and percentage of total AML blasts labeled with anti-CD123 and anti-CD131 were calculated using Cell Quest Software (Becton Dickinson).
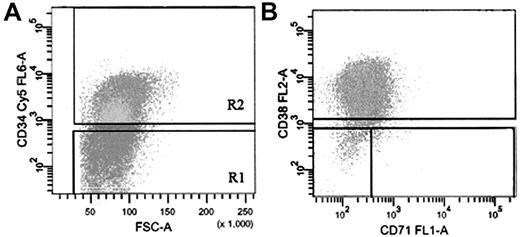
Subpopulations of AML and normal BM cells labeled with anti-CD34, anti-CD38, and (for AML cells) anti-CD71 were sorted into IMDM (StemCell Technologies) with 20% FCS in microcentrifuge tubes at 4°C. Sort windows used to isolate the specific subfractions are shown in Figure 1. Sorted fractions were washed and used for RNA isolation and QRT-PCR analysis.
QRT-PCR analysis
Total RNA was extracted from sorted cells using the Absolutely RNA, Microprep, or Nanoprep kits (Stratagene, La Jolla, CA). The RT reaction was performed in 20 μL with superscript II reverse transcriptase (Invitrogen, Burlington, ON, Canada) using random hexamer oligonucleotides (Amersham Pharmacia, Piscataway, NJ). Real-time PCR was performed using 12.5 μL 2 × SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μL of 20 pM-specific primers, 1 to 2 μL cDNA, and water to a final volume of 25 μL. Specific forward and reverse primers to produce approximately 100-bp amplicons for optimal amplification in real-time PCR of reverse-transcribed cDNA for human IL-3Rα were 5′-GACCTGTACTTGAACGTTGCC and 5′-GAAACGACACCCGATACGTGT, for human IL-3Rβc were 5′-GCAGCATGTCGGCCTTCACTA and 5′-GTCCCCGAATCCTACAGGGAA, and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were (5′-CCCATCACCATCTTCCAGGAG and 5′-CTTCTCCATGGTGGTGAAGACG. Real-time PCR and data analysis were performed on an iCycler iQ system, using iCycler iQ Real-time Detection Software (Bio-Rad, Hercules, CA). The relative quantification data of human IL-3Rα and IL-3Rβc compared with a reference gene (GAPDH) was generated on the basis of a mathematical model for relative quantification in real-time RT-PCR as described.32,35,36
FACS strategy for isolation of AML cells based on expression of CD34, CD38, and CD71. (A) Cells labeled with anti-CD34-Cy5 (R2) were sorted on the basis of their expression of CD38-PE and CD71-FITC (B). Gate R1 cells were isolated as the CD34- population. Sort gates in panels A and B were set to exclude 100% of the cells labeled with an isotype-control antibody.
FACS strategy for isolation of AML cells based on expression of CD34, CD38, and CD71. (A) Cells labeled with anti-CD34-Cy5 (R2) were sorted on the basis of their expression of CD38-PE and CD71-FITC (B). Gate R1 cells were isolated as the CD34- population. Sort gates in panels A and B were set to exclude 100% of the cells labeled with an isotype-control antibody.
Transplantation and detection of AML cells in β2-microglobulin-deficient NOD/SCID mice
β2-microglobulin-deficient NOD/SCID (β2m-/- NOD/SCID) mice37 were bred and maintained under sterile conditions in the British Columbia Cancer Research Center Joint Animal Facility according to protocols approved by the Animal Care Committee of the University of British Columbia. These animals were used based on our previous investigations demonstrating that they could allow more efficient engraftment of many AML patient samples than NOD/SCID animals.38 Mice, 8 to 10 weeks of age, received 350 cGy from a 137Cs source 16 to 24 hours before injection of AML cells. AML cells (1 to 3 × 107) previously incubated for 24 hours with or without DT388IL3 were injected via the tail vein. Femoral bone marrow aspirations were performed every 4 weeks after the injection of AML cells under isoflurane inhalation anesthesia.39 At 16 weeks, mice were killed by CO2 inhalation, and BM cells were obtained from the 4 long bones by flushing with α-MEM with 50% FCS.
Mouse BM cells were prepared for FACS analysis as previously described7 and analyzed for the expression of CD45, a human-specific panleukocyte marker, using an antibody prepared in our center from American Type Culture Collection (ATCC) (clone no. HB10508; Rockville, MD). FACS analysis was performed on a Becton Dickinson FACScan or FACSort flow cytometer. The percentage of CD45+ cells was determined after excluding 99.9% of cells labeled with the isotype control and nonviable cells. Nonspecific binding of CD45 on mouse BM cells is reliably less than 0.1% to 0.3%.7 Thus, a difference between the immunoglobulin G1-negative (IgG1-) control and the CD45 expression of the treated mice of more than 0.1% was regarded as evidence for engraftment of human cells. Values shown for engraftment of AML cells in mouse BM are the mean values (± SD) of CD45+ cells (%) obtained for all mice in the cohort that survived to the time of analysis.
Fluorescence in situ hybridization
Cells from mouse BM were cytocentrifuged onto glass microscope slides and then fixed in methanol-glacial acetic acid. The human-specific centromeric repeat probe used to detect the +8 abnormality (plasmid D8Z2; ATCC) and the yeast artificial chromosome clone containing 550 kb human DNA encompassing the breakpoint on 16p13 (CEPHy904E02 854; Max Planck Institute, Berlin, Germany) to detect the inv(16) were labeled by nick translation with digoxigenin (DIG; Boehringer Mannheim, Mannheim, Germany). Chromosomes 8 and 16 probes and slides were treated as previously described.5 For probe detection, slides were incubated with a sheep anti-DIG-FITC antibody (Boehringer Mannheim) and counter-stained with PI as previously described.5 Slides were visualized on a Zeiss Axioplan (Oberkochen, Germany) fluorescence microscope using double or triple bandpass filters (Omega Optical, Battlebore, NC) to allow simultaneous visualization of FITC and PI signals.
Statistical analysis
Mean values of IL-3R subunit expression in subpopulations of AML and normal BM cells and correlation coefficients between variables were determined in Excel (Microsoft, Redmond, WA), and the significance of differences and correlations determined using a Student t test. A P value of less than .05 was considered significant.
Results
Table 1 shows the clinical characteristics of the 19 patients with AML patients whose cells were tested in this study. Their ages at diagnosis ranged from 17 to 75 years and the FAB subtype of these leukemias was M4 (n = 10), M1 (n = 5), or M5 (n = 3). Cytogenetic analysis of the diagnostic BM specimens revealed abnormalities associated with a good prognosis [inv(16)] in 3 patients and a poor prognosis in 6 of these 19 patients according to the Medical Research Council (MRC, United Kingdom) or Southwest Oncology Group (SWOG) criteria.40,41
Patient characteristics
Patient no. . | Age, y/sex . | FAB . | Marrow cytogenetics . | WBC, × 109/L (% blasts in PB) . |
|---|---|---|---|---|
| 1 | 60/M | M5b | 47, XY, +8 | 170 (71) |
| 2 | 35/F | M1 | 46, XX | 87 (94) |
| 3 | 61/F | M4 | 44-45, XX, –3, –5, –7, –8 add(8),p23 | 24 (26) |
| 4 | 65/F | M4 | 46,XX | 205 (88) |
| 5 | 75/F | M4 | 46,XX, del(11q)(q14;q25) | 215 (90) |
| 6 | 59/M | M1 | 46, XYinv(16)(p13;q22) | 76 (90) |
| 7 | 22/F | M4 | 46, XX,t(6;9)(p23;q34) | 29 (38) |
| 8 | 59/M | M4 | 46, XY | 101 (90) |
| 9 | 64/M | M1 | 49, XY, +10, +?13, +19 | 189 (75) |
| 10 | 70/F | M4 | 46, XX | 74 (16) |
| 11 | 63/F | M1 | Unsuccessful | 295 (95) |
| 12 | 74/M | M5 | 46, XY, t(8;16)(p11;p13) | 85 (60) |
| 13 | 41/M | M4eo | 46, XY, inv(16) | 238 (90) |
| 14 | 56/M | ND | 45, X, –Y | 300 (95) |
| 15 | 53/M | M4 | 47, XY, –4,del(5)(q12;q23)add(7)(p12), –12, –17 | 73 (46) |
| 16 | 52/F | M4eo | 46, XX | 155 (45) |
| 17 | 21/M | M1 | 46, XY, add(6)(p23)?+(6;11)(p27;q23) | 320 (90) |
| 18 | 49/M | M4eo | 46, XY, inv(16)(p13;q22)idem, +15, +22 | 126 (60) |
| 19 | 17/F | M5a | 47, XX, +8,del(5)(q31;q?33) | 205 (90) |
Patient no. . | Age, y/sex . | FAB . | Marrow cytogenetics . | WBC, × 109/L (% blasts in PB) . |
|---|---|---|---|---|
| 1 | 60/M | M5b | 47, XY, +8 | 170 (71) |
| 2 | 35/F | M1 | 46, XX | 87 (94) |
| 3 | 61/F | M4 | 44-45, XX, –3, –5, –7, –8 add(8),p23 | 24 (26) |
| 4 | 65/F | M4 | 46,XX | 205 (88) |
| 5 | 75/F | M4 | 46,XX, del(11q)(q14;q25) | 215 (90) |
| 6 | 59/M | M1 | 46, XYinv(16)(p13;q22) | 76 (90) |
| 7 | 22/F | M4 | 46, XX,t(6;9)(p23;q34) | 29 (38) |
| 8 | 59/M | M4 | 46, XY | 101 (90) |
| 9 | 64/M | M1 | 49, XY, +10, +?13, +19 | 189 (75) |
| 10 | 70/F | M4 | 46, XX | 74 (16) |
| 11 | 63/F | M1 | Unsuccessful | 295 (95) |
| 12 | 74/M | M5 | 46, XY, t(8;16)(p11;p13) | 85 (60) |
| 13 | 41/M | M4eo | 46, XY, inv(16) | 238 (90) |
| 14 | 56/M | ND | 45, X, –Y | 300 (95) |
| 15 | 53/M | M4 | 47, XY, –4,del(5)(q12;q23)add(7)(p12), –12, –17 | 73 (46) |
| 16 | 52/F | M4eo | 46, XX | 155 (45) |
| 17 | 21/M | M1 | 46, XY, add(6)(p23)?+(6;11)(p27;q23) | 320 (90) |
| 18 | 49/M | M4eo | 46, XY, inv(16)(p13;q22)idem, +15, +22 | 126 (60) |
| 19 | 17/F | M5a | 47, XX, +8,del(5)(q31;q?33) | 205 (90) |
WBC indicates white blood cell; M, male; F, female; and ND, not done
Comparison of QRT-PCR and FACS for detection of IL-3R subunit expression
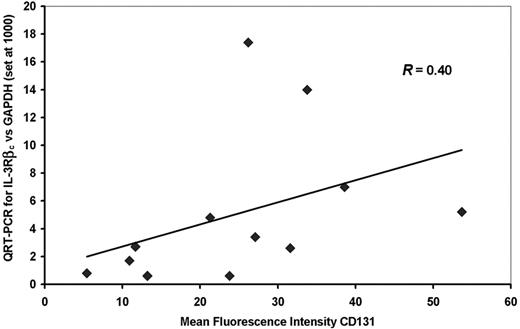
In initial experiments the levels of expression of IL-3R subunits in AML blasts as detected by QRT-PCR was compared with detection by FACS analysis for 12 of the 19 patient samples shown on Table 1 using antibodies which detect IL-3Rα (CD123) and IL-3Rβc (CD131) and primers and conditions for QRT-PCR described in “Patients, methods, and materials,” and previously.32 As shown in Figure 2 there was a weak correlation between the level of IL-3Rβc expression detected by QRT-PCR and MFI for this molecule detected by FACS for CD131 (r = 0.40). No significant correlation was seen between IL-3Rα subunit expression detected by QRT-PCR and MFI detected for CD123. In addition, there was no significant correlation between the percentages of CD123+ or CD131+ cells quantified by FACS and the MFI or the value obtained by QRT-PCR for the same molecule. MFI observed for CD131 (but not CD123) correlated significantly (r = 0.74) with the percentage of AML-CFCs killed by DT388IL3 while the percentages of CD131+ or CD123+ cells did not predict the AML-CFC killed. QRT-PCR levels for both the IL-3Rα and βc subunit expressions correlated significantly (r = 0.59 and 0.70, respectively) with AML-CFCs killed with DT388IL3 (Table 2). Because QRT-PCR values for both the IL-3Rα and βc subunits from a given AML sample predicted DT388IL3 cytotoxicity against AML-CFCs in both this and prior evaluations,32 while FACS analysis for only the IL-3Rβc subunit was predictive of the same endpoint, QRT-PCR was chosen for analysis of IL-3R expression in subsequent experiments on subpopulations of AML cells.
Correlation between IL-3R subunit quantitation on AML blasts by FACS (MFI) and QRT-PCR and AML-CFC killing by DT388IL3
. | FACS-MFI . | . | AML-CFC*kill . | |
|---|---|---|---|---|
. | CD123 . | CD131 . | . | |
| QRT-PCR | ||||
| IL-3Rα | 0.095 | – | 0.59† | |
| IL-3Rβc | – | 0.40 | 0.70† | |
| AML-CFC* kill | 0.23 | 0.74† | – | |
. | FACS-MFI . | . | AML-CFC*kill . | |
|---|---|---|---|---|
. | CD123 . | CD131 . | . | |
| QRT-PCR | ||||
| IL-3Rα | 0.095 | – | 0.59† | |
| IL-3Rβc | – | 0.40 | 0.70† | |
| AML-CFC* kill | 0.23 | 0.74† | – | |
Data expressed as correlation coefficients between the values indicated as determined for each of 12 of the 19 AML samples listed on Table 1 and according to “Patients, materials, and methods.”
–indicates not done
Percentage of AML-CFCs killed after 24 hours of exposure to 50 ng/mL DT388IL3
P < .05, Student t test
IL-3R subunit expression on subpopulations of AML and normal BM cells enriched for progenitors
Figure 1 shows a typical FACS profile of AML cells stained with antibodies against CD34, CD38, and CD71. Previous data had demonstrated that the CD34+CD38- phenotype includes N/SL-ICs from most patient samples while addition of CD71 negativity to this phenotype increases the enrichment of sorted cells for these primitive progenitors.15 These studies have also demonstrated that AML-CFCs are largely CD34+ in most patient samples but are more heterogeneous in their expression of CD388,15 (ie, most of these more mature progenitors express this antigen and thus are found in the CD34+CD38+ subpopulation, while N/SL-ICs are excluded). CD34- cells from most AML samples have little progenitor activity. Thus, PB cells from the 19 patients with AML (Table 1) were sorted into 4 subpopulations defined as CD34+CD38-CD71-, CD34+CD38-, CD34+CD38+, and CD34-. As shown in Table 3, the large proportion of AML cells from 16 of these patients were CD34+. Overall, mean frequencies of the CD34+CD38-CD71-, CD34+CD38-, and CD34+CD38+ subpopulations were 3.2% (range, 0.05%-25.3%), 5.3% (range, 0.08%-38.4%), and 40.2% (range, 0.7%-90.9%), respectively, while 27% (range, 2.1%-80.9%) of cells were CD34-.
Proportion of AML cells in FACS-isolated subpopulations
Patient no. . | CD34+CD38–CD71–, %* . | CD34+CD38–, %† . | CD34+CD38+, %‡ . | CD34–, %§ . |
|---|---|---|---|---|
| 1 | 1.5 | 1.9 | 44.5 | 12.3 |
| 2 | 17.4 | 35.3 | 10.2 | 14.8 |
| 3 | 0.9 | 2.4 | 56 | 11.8 |
| 4 | 0.3 | 0.6 | 40 | 29.6 |
| 5 | 2.9 | 3.8 | 64.9 | 8.6 |
| 6 | 0.3 | 0.4 | 62.4 | 10.8 |
| 7 | 0.4 | 0.7 | 43.1 | 26.3 |
| 8 | 0.08 | 0.1 | 17.3 | 69.3 |
| 9 | 2.6 | 4.1 | 71 | 5.8 |
| 10 | 2.7 | 2.9 | 43.4 | 21.4 |
| 11 | 0.05 | 0.08 | 0.7 | 67.1 |
| 12 | 0.1 | 0.1 | 8.7 | 43 |
| 13 | 1.6 | 1.8 | 62.6 | 6.1 |
| 14 | 3.2 | 5.3 | 45.2 | 26.1 |
| 15 | 25.3 | 38.4 | 24.3 | 19.5 |
| 16 | 0.4 | 0.6 | 52.5 | 33.2 |
| 17 | 0.6 | 0.6 | 90.9 | 2.1 |
| 18 | 0.2 | 0.3 | 23.5 | 24.3 |
| 19 | 0.7 | 0.8 | 2.1 | 80.9 |
Patient no. . | CD34+CD38–CD71–, %* . | CD34+CD38–, %† . | CD34+CD38+, %‡ . | CD34–, %§ . |
|---|---|---|---|---|
| 1 | 1.5 | 1.9 | 44.5 | 12.3 |
| 2 | 17.4 | 35.3 | 10.2 | 14.8 |
| 3 | 0.9 | 2.4 | 56 | 11.8 |
| 4 | 0.3 | 0.6 | 40 | 29.6 |
| 5 | 2.9 | 3.8 | 64.9 | 8.6 |
| 6 | 0.3 | 0.4 | 62.4 | 10.8 |
| 7 | 0.4 | 0.7 | 43.1 | 26.3 |
| 8 | 0.08 | 0.1 | 17.3 | 69.3 |
| 9 | 2.6 | 4.1 | 71 | 5.8 |
| 10 | 2.7 | 2.9 | 43.4 | 21.4 |
| 11 | 0.05 | 0.08 | 0.7 | 67.1 |
| 12 | 0.1 | 0.1 | 8.7 | 43 |
| 13 | 1.6 | 1.8 | 62.6 | 6.1 |
| 14 | 3.2 | 5.3 | 45.2 | 26.1 |
| 15 | 25.3 | 38.4 | 24.3 | 19.5 |
| 16 | 0.4 | 0.6 | 52.5 | 33.2 |
| 17 | 0.6 | 0.6 | 90.9 | 2.1 |
| 18 | 0.2 | 0.3 | 23.5 | 24.3 |
| 19 | 0.7 | 0.8 | 2.1 | 80.9 |
Mean ± SD = 3.2 ± 6.6
Mean ± SD = 5.3 ± 11.2
Mean ± SD = 40.2 ± 25.3
Mean ± SD = 27 ± 22.9
Comparison of FACS analysis and QRT-PCR for detection of IL-3Rβc. The MFI of AML cells labeled with anti-CD131 is compared with the relative amount of IL-3Rβc mRNA detected by QRT-PCR for 12 patient samples. There was a correlation between these 2 values (r = .40) that did not reach statistical significance. The diagonal line is a regression line relating the QRT-PCR to the MFI values.
Comparison of FACS analysis and QRT-PCR for detection of IL-3Rβc. The MFI of AML cells labeled with anti-CD131 is compared with the relative amount of IL-3Rβc mRNA detected by QRT-PCR for 12 patient samples. There was a correlation between these 2 values (r = .40) that did not reach statistical significance. The diagonal line is a regression line relating the QRT-PCR to the MFI values.
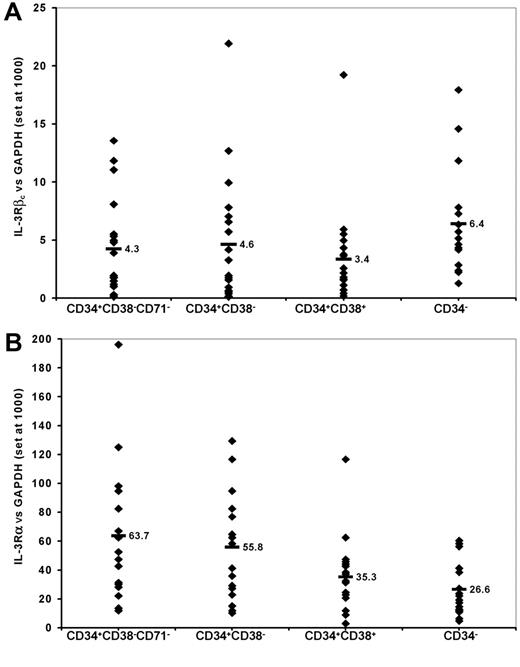
QRT-PCR was used to detect expression of the IL-3Rα and βc subunits in RNA isolated from the 4 sorted subpopulations of AML cells from these 19 patient samples. As shown in Figure 3, the median levels of expression of the α subunit were higher than that of the βc subunit for each isolated subpopulation (range, 4- to 15-fold). The level of βc expression detected among the subpopulations of cells enriched for primitive progenitors (CD34+CD38- cells or such cells lacking CD71) was not significantly different from that detected in the subpopulations depleted of such cells (CD34+CD38+ or CD34- cells). However, expression of the IL-3Rα subunit was significantly higher among CD34+CD38-CD71- and CD34+CD38- cells than among CD34+CD38+ or CD34- cells (P < .002, paired t test).
IL-3R subunit expression on AML cells as detected by QRT-PCR. RNA was extracted from AML cells sorted into the subpopulations indicated as described in “Patients, materials, and methods.” Values shown are the levels of expression of βc (A) and α (B) subunits of the IL-3R relative to the expression of GAPDH arbitrarily set at 1000 for each of 19 patient samples. Each data point represents the value for an individual patient sample. The horizontal bars and adjacent numbers indicate the mean level of expression for each of the α and βc subunits in the different subpopulations.
IL-3R subunit expression on AML cells as detected by QRT-PCR. RNA was extracted from AML cells sorted into the subpopulations indicated as described in “Patients, materials, and methods.” Values shown are the levels of expression of βc (A) and α (B) subunits of the IL-3R relative to the expression of GAPDH arbitrarily set at 1000 for each of 19 patient samples. Each data point represents the value for an individual patient sample. The horizontal bars and adjacent numbers indicate the mean level of expression for each of the α and βc subunits in the different subpopulations.
RNA also was isolated from CD34+CD38- and CD34+CD38+ cells from 3 normal BM samples. QRT-PCR demonstrated that the mean (± SD) level of expression of the IL-3R α and βc subunits (relative to GAPDH set at 1000) was 21.6 (± 8.6) and 9.0 (± 5.1), respectively, for CD34+CD38- cells, and 8.2 (± 1.8) and 26.2 (± 3.8), respectively, for CD34+CD38+ cells. These values for the α subunit were significantly (P ≤ .002) lower than those obtained for the same subpopulations of AML cells. In contrast, the βc subunit was expressed at similar (in CD34+CD38- cells) or higher (P = .003 in CD34+CD38+ cells) levels among normal than among malignant cells.
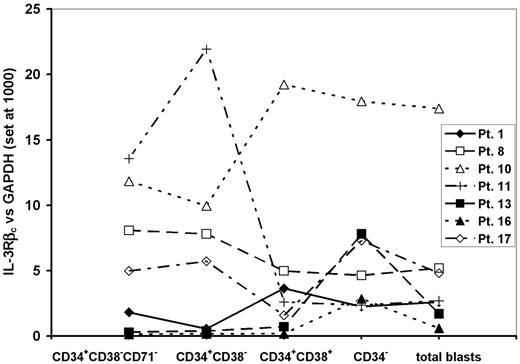
In our previous studies we had defined an expression of the IL-3Rβc subunit relative to GAPDH of 3 or less as being associated with poor killing of AML-CFCs.32 To assess the value of this measurement in predicting cytotoxicity of DT388IL3 against N/SL-ICs from different patient samples, AML blasts from 7 samples, which had levels of IL-3Rβc expression above or below that level in the sorted subpopulations were selected. Three of these AML samples had low levels of IL-3Rβc expression among CD34+CD38-CD71- AML cells (expression levels for samples 1, 16, and 13 were 1.8, 0.14, and 0.3, respectively), while the remaining 4 patient samples had higher levels of expression (expression for samples 8, 17, 10, and 11 was 8, 5, 11.8, and 13.5, respectively). Comparison of this level of expression with that seen in the other analyzed subpopulations and unsorted AML blasts from the same sample showed no consistent differences (Figure 4). Among these 7 samples expression of the IL-3Rα subunit was higher among CD34+CD38- cells compared with cells expressing CD38 or lacking CD34 as described for the entire patient group (Figure 3). However, there were no significant differences in the expression of IL-3Rα between the 3 samples with low expression and the 4 samples with high expression of IL-3Rβc in any subpopulation studied.
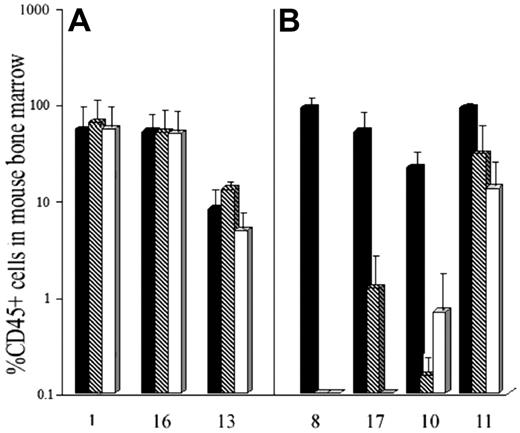
AML blasts from these same samples were incubated for 24 hours with or without DT388IL3 at 50 or 250 ng/mL and then injected into cohorts of sublethally irradiated β2m-/- NOD/SCID mice. For the 3 samples with low levels of IL-3Rβc expression, exposure to either DT388IL3 concentration did not change the level of engraftment detected in mouse bone marrow 12 weeks later indicating that N/SL-ICs from these samples had survived this treatment (Figure 5A). Fluorescence in situ hybridization (FISH) analysis of BM cells harvested from mice injected with DT388IL3-treated or -untreated AML cells from patients 1 and 13 demonstrated the presence of the +8 and inv(16) chromosome abnormalities, respectively, in a similar proportion of cells to that found in the patients'diagnostic BM sample (data not shown). In contrast, with 3 samples that expressed high levels of IL-3Rβc complete eradication of N/SL-ICs from samples 8 and 17 and near-complete eradication from sample 10 was achieved with 250 ng/mL of DT388IL3. Although sample 11 expressed the IL-3Rβc subunit at levels similar to these 3 samples, only a modest (but statistically significant [P < .05]) reduction in engraftment of AML cells was observed (Figure 5B). The percentage of reduction in AML cell engraftment achieved with prior DT388IL3 treatment of these 7 patient samples correlated most strongly with the level of IL-3Rβc expression in CD34+CD38-CD71- cells (r = 0.76) compared with the other FACS-sorted subpopulations, but a correlation was also seen with the expression of the same molecule on unsorted blast cells (r = 0.56).
Discussion
DT388IL3 is a novel fusion protein which is currently undergoing clinical evaluation as a possible new therapeutic agent for the treatment of AML. Although data from tissue culture and mouse models had suggested that this drug would be active against many human AMLs, it also suggested that some leukemias would be much more sensitive than others.18,19,31 This was particularly true when DT388IL3 was tested against AML progenitors that engraft in NOD/SCID mice (N/SL-ICs).18 Thus, for some samples DT388IL3 was highly cytotoxic when tested against AML-CFCs but much less so against N/SL-ICs, while for other samples the drug had little effect against any class of AML progenitors. Evaluation of the expression of high- and low-affinity IL-3-binding sites on AML blasts demonstrated that the presence of IL-3Rs was necessary but not always sufficient to ensure leukemic progenitor kill by the fusion protein.19,34 Previous studies had also demonstrated that AML blast populations highly enriched for or depleted of progenitor activity could be obtained by isolating cells that varied in their expression of CD34, CD38, and CD71.8,15 N/SL-ICs are largely restricted to the CD34+CD38- subpopulation of AML blasts from most patient samples and can be further enriched by excluding CD71+ cells, while CD34- cells are typically depleted of all progenitor activity.8,15,16
Comparison of the relative level of expression of the IL-3Rβc subunit between different subpopulations of AML cells from 7 patient samples tested for DT388IL3 sensitivity in β2-microglobin-deficient NOD/SCID mice. QRT-PCR was performed on FACS-sorted cells of the indicated cell-surface phenotype as described for Figure 3.
Comparison of the relative level of expression of the IL-3Rβc subunit between different subpopulations of AML cells from 7 patient samples tested for DT388IL3 sensitivity in β2-microglobin-deficient NOD/SCID mice. QRT-PCR was performed on FACS-sorted cells of the indicated cell-surface phenotype as described for Figure 3.
IL-3Rβc subunit expression detected by QRT-PCR predicts cytotoxicity of DT388IL3 against AML progenitors which engraft in β2-microglobin-deficient NOD-SCID mice. (A) AML cells from 3 patients (nos. 1, 16, and 13 from Table 1) expressing low levels of IL-3Rβc and (B) 4 patients (nos. 8, 17, 10, and 11 from Table 1) who had high levels of IL-3Rβc expression were incubated for 24 hours with or without DT388IL-3 at 50 and 250 ng/mL and then injected intravenously into sublethally irradiated mice as described in “Patients, materials, and methods.” Data are shown as the mean level of engraftment of human AML cells in cohorts of mice receiving treated or untreated cells 8 to 12 weeks previously as detected by FACS analysis of mouse BM cells for CD45+ human cells. ▪ indicates untreated cells;  , cells treated with 50 ng/mL DT388IL-3; and □, cells treated with 250 ng/mL DT388IL-3. Error bars indicate SEM level engraftment for all mice in the cohort. The level of engraftment was significantly (P < .05; Student t test) less than that obtained for the untreated control cells for the 4 patient samples treated with both doses of DT338IL3 shown in panel B.
, cells treated with 50 ng/mL DT388IL-3; and □, cells treated with 250 ng/mL DT388IL-3. Error bars indicate SEM level engraftment for all mice in the cohort. The level of engraftment was significantly (P < .05; Student t test) less than that obtained for the untreated control cells for the 4 patient samples treated with both doses of DT338IL3 shown in panel B.
IL-3Rβc subunit expression detected by QRT-PCR predicts cytotoxicity of DT388IL3 against AML progenitors which engraft in β2-microglobin-deficient NOD-SCID mice. (A) AML cells from 3 patients (nos. 1, 16, and 13 from Table 1) expressing low levels of IL-3Rβc and (B) 4 patients (nos. 8, 17, 10, and 11 from Table 1) who had high levels of IL-3Rβc expression were incubated for 24 hours with or without DT388IL-3 at 50 and 250 ng/mL and then injected intravenously into sublethally irradiated mice as described in “Patients, materials, and methods.” Data are shown as the mean level of engraftment of human AML cells in cohorts of mice receiving treated or untreated cells 8 to 12 weeks previously as detected by FACS analysis of mouse BM cells for CD45+ human cells. ▪ indicates untreated cells;  , cells treated with 50 ng/mL DT388IL-3; and □, cells treated with 250 ng/mL DT388IL-3. Error bars indicate SEM level engraftment for all mice in the cohort. The level of engraftment was significantly (P < .05; Student t test) less than that obtained for the untreated control cells for the 4 patient samples treated with both doses of DT338IL3 shown in panel B.
, cells treated with 50 ng/mL DT388IL-3; and □, cells treated with 250 ng/mL DT388IL-3. Error bars indicate SEM level engraftment for all mice in the cohort. The level of engraftment was significantly (P < .05; Student t test) less than that obtained for the untreated control cells for the 4 patient samples treated with both doses of DT338IL3 shown in panel B.
Using FACS analysis other investigators have demonstrated that the IL-3Rα subunit is typically highly expressed on AML blasts, including the CD34+CD38- subpopulation.14,20 More recently this finding has been confirmed and extended by using MFI to quantify expression of both the IL-3R subunits on AML blasts labeled with anti-CD123 and anti-CD131.42 This measurement was also shown to correlate with the percentage of AML cells undergoing apoptosis when exposed to DT388IL3. As an alternative to flow cytometry we have used QRT-PCR to show that the level of IL-3R subunit expression in AML blasts predicted the killing of AML-CFCs by DT388IL3 and its variants.32 However, with both techniques some patient samples showed relative resistance to DT388IL3 cytotoxicity in spite of high-level IL-3R expression.
In initial experiments described here, flow cytometry was compared with QRT-PCR for its ability to detect and quantify the IL-3R subunits. Both techniques provided quantitative and reproducible data. However, there was no correlation between the FACS data (either MFI or percentage of positive cells) and QRT-PCR for the IL-3Rα subunit. While a correlation was seen between MFI detected by FACS and QRT-PCR for the IL-3Rβc subunit, it was relatively weak and did not reach statistical significance (Table 2). These data indicate that RNA and protein levels for these molecules do not necessarily correspond in AML blasts. Our data differ from the recent report from Testa et al, who found that MFI detected by anti-CD123 antibodies predicted AML blast apoptosis induced by DT388IL3.42 This difference might be explained by the different endpoint in our studies (AML-CFC killing), the use of different antibodies, and the larger number of samples tested in the earlier investigation. Nevertheless, in the current study we were able to demonstrate a correlation between expression of both IL-3R subunits as detected by QRT-PCR and AML-CFC killing with DT388IL3. These data confirm those in our previous report and are also consistent with our previous conclusion that the relationship is strongest for the IL-3Rβ subunit.32 c
Since LSCs are the most relevant target population for novel antileukemic therapy such as DT388IL3 we used QRT-PCR to study the expression of IL-3R subunits on subpoulations of AML cells known to be enriched for or depleted of leukemic progenitors that engraft in immunodeficient (β2m-/- NOD/SCID) mice. Among 19 AML patient samples no consistent difference in expression of the IL-3Rβc subunit was detected between the most primitive CD34+CD38-CD71- cell fraction and any other subpopulation (Figure 3A). On the other hand, the IL-3Rα subunit was most highly expressed among cells enriched for candidate LSCs which engraft in mice (Figure 3B). This and the generally much higher level of expression of the IL-3Rα subunit compared with the βc subunit across all cell populations studied suggest that expression of the latter molecule is more likely than the former to be a limiting factor in the formation of high-affinity IL-3R-binding sites on N/SL-ICs.
Although N/SL-ICs are a small proportion of the total cells present in even the most enriched population studied here,15 and the number of AML samples studied in β2m-/- NOD/SCID mice was limited to 7, the correlation between IL-3Rβc expression in CD34+CD38-CD71- cells and DT388IL3 cytotoxicity against cells engrafting in mice was strong. In fact, a correlation between expression of this molecule in unsorted blast cells and reduced engraftment of DT388IL3-treated AML cells was also seen. This and the overall expression data shown on Figure 3 suggest that there is little difference between the expression of the IL-3Rβc among AML blasts and progenitors. On the other hand, while expression of the IL-3Rα subunit may vary considerably among different AML-cell fractions, it does not appear to limit the cytotoxicity of DT388IL3 against candidate LSCs.
In initial studies investigating the relative cytotoxicity of DT388IL3 against normal and leukemic progenitor cells, a striking lack of toxicity was seen against primitive normal multipotential progenitors, including those with lymphomyeloid potential, that are detected in NOD/SCID mice.18 This was somewhat surprising given the fact that expression of both IL-3R subunits has been demonstrated on CD34+CD38- cells from normal BM as well as cord blood and fetal liver using RT-PCR.43 We have confirmed the findings of this earlier study. However, using the QRT-PCR technique described in the current report the relative level of expression of the IL-3Rα subunit among both CD34+CD38- and CD34+CD38+ normal BM cells was found to be generally lower than that detected in the same population of AML cells. This difference may partly explain the low toxicity observed with DT388IL3 against primitive normal hematopoietic cells. Other investigators have also shown that expression of the IL-3Rα subunit is generally higher on primitive AML cells than on comparable normal cell populations.14,20 Interestingly, the expression of the βc subunit was similar among normal and malignant cells. Thus, it remains possible that the clinical experience with DT388IL3 will uncover more myelosuppressive activity for this fusion protein than has so far been suggested by the preclinical data. Alternatively, events that take place after DT388IL3 binding to the receptor may differ between normal and malignant cells. It seems feasible that enhanced association of DT388IL3 with the IL-3Rα subunits on AML cells (which must also express sufficient βc subunits to form the high-affinity binding complex) might accelerate the formation and internalization of the α/βc receptor complex,44 resulting in more efficient delivery of DT molecules to the cytoplasm of AML progenitors.
A phase 1/2 clinical trial of DT388IL3 is currently under way in patients with relapsed or refractory AML.45 Data presented here suggest that it may be possible to use quantification of target molecules on AML blasts to predict which patients are likely to benefit from therapy with this fusion protein. The use of QRT-PCR for the IL-3R subunits is currently being tested in this context. Furthermore, these results suggest that detailed evaluation of subpopulations of malignant cells with different functional properties may reveal similarities and differences in gene expression that are important for the rational development of targeted antileukemic therapy.
Prepublished online as Blood First Edition Paper, August 1, 2006; DOI 10.1182/blood-2006-04-013813.
Supported by the Canadian Institute for Health Research and National Cancer Institute of Canada (D.E.H.), National Institutes of Health (NIH) grants RO1CA76178and RO1CA90263, and Leukemia and Lymphoma Society grant 6006-05 (A.E.F.).
The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal