Abstract
The NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia (T-ALL) has recently been identified as a possible target for imatinib and related tyrosine kinase inhibitors, but exact data regarding the prognostic impact and frequency of the several putative NUP214-ABL1 mRNA transcripts are still missing. We investigated 279 adult patients with T-ALL treated within the framework of the GMALL 5/93 and 6/99 therapy trials for NUP214-ABL1 by using a novel multiplex real-time, quantitative polymerase chain reaction (PCR). Eleven (3.9%) patients were NUP214-ABL1 positive, and 5 different transcripts were observed; 8 patients had a thymic immunophenotype, 1 had an early T-cell immunophenotype, and 2 had a mature T-cell immunophenotype. NUP214-ABL1-positive and -negative patients did not differ significantly in their major clinical features. In contrast to previous reports suggesting an adverse clinical course for NUP214-ABL1-positive patients, no significant difference in overall survival was observed. Based on the results, we have established and tested a novel PCR method for simplified detection of the NUP214-ABL1 fusion gene.
Introduction
Various dysregulated tyrosine kinases found in a number of hematologic diseases are sensitive to specific tyrosine kinase inhibitors such as imatinib.1 These include the BCR-ABL1 fusion gene in chronic myeloid leukemia and a subset of acute lymphoblastic leukemia and the PDGFRB and PDGFRA fusion genes rarely found in myeloid malignancies.1,2 Because this therapeutic option offers high efficacy with comparatively low toxic side effects, efforts have been made to identify new target structures for imatinib. A novel fusion gene, resulting from episomal fusion of the ABL1 gene to the neighboring NUP214 gene, has recently been identified.3 The T-cell acute lymphoblastic leukemia (T-ALL) cell line ALL-SIL was found to carry this fusion gene and was experimentally susceptible to imatinib treatment.3 The NUP214-ABL1 fusion gene could thus serve as a target of imatinib treatment in this subset of T-ALL patients, as recently suggested.4
In this context, the prognostic impact and exact frequency of NUP214-ABL1 in adult patients with T-ALL are of interest. Moreover, because several exons of the NUP214 gene could potentially be fused in-frame to the ABL1 gene, the chromosomal breakpoint region within NUP214 is yet to be well defined. Therefore, we investigated a large number of adult patients with T-ALL treated within the framework of the German Multicenter Adult ALL (GMALL) therapy trials.
Patients, materials, and methods
No experiments on humans or animals were performed. Patients were treated within the German Multicenter Adult ALL (GMALL) therapy studies, and these therapy studies have been approved by the ethics committees of the involved study clinics. Patients provided written informed consent for participation in accordance with the Declaration of Helsinki.
Patient material
Bone marrow (n = 213) and peripheral-blood (n = 66) samples were obtained for diagnostic purposes within the framework of the GMALL therapy studies between May 1993 and May 2003. One hundred forty-two patients were enrolled in the 5/93 study and 137 patients in the subsequent 6/99 study. Details about the therapy have been reported.5,6 All samples were taken at the time of primary diagnosis and had a high blast count, as revealed by fluorescence-activated cell sorter (FACS) analysis. Genetic investigations were performed retrospectively on archived material.
Cell lines
Oligonucleotides
All oligonucleotides were obtained from metabion (Martinsried, Germany) and were high-performance liquid chromatography (HPLC) purified.
Immunophenotyping
Immunophenotyping was performed as previously described in detail.10,11 Samples were immunologically classified as pre-T cell (cyCD3+, CD7+, CD5+/-, CD2-, CD1a-, CD4-, CD8-/+, sCD3- or cyCD3+, CD7+, CD5-, CD2+, CD1a-, CD4-, CD8-, sCD3-), thymic T cell (cyCD3+, CD7+, CD5+, CD2+, CD1a+, CD4+/-, CD8+/-, sCD3+/-), or mature T cell (cyCD3+, CD7+, CD5+, CD2+, CD1a-, CD4+/-, CD8-/+, sCD3+).
RNA isolation and reverse transcription
RNA was isolated using the TRIzol method (Invitrogen, Karlsruhe, Germany) or the PureScript method (Biozym Diagnostik, Hessisch-Oldendorf, Germany). One microgram total RNA was reverse transcribed using the Ready-to-Go RT-PCR Beads (Amersham Biosciences, Freiburg, Germany) in a 33-μL total volume. The quality of the cDNA was confirmed using the ABL1 gene as a control in a separate real-time PCR reaction performed under conditions previously recommended by the Europe Against Cancer (EAC) initiative.12
Real-time quantitative PCR for NUP214-ABL1
Real-time quantitative PCR was performed on an RG3000 cycler (Corbett Research, Wasserburg am Inn, Germany) using TaqMan chemistry. Each sample was examined in duplicate. The following primers (5′-> 3′) were used: NUP-2 ACCCGGAGATGATCCCAACAAAAT, NUP-6 CGTTTTATGAGTCAGATCATCCTGTC, NUP-10 CTATACAAACCAAGTGGAAATCACCAT, NUP-13 CCTCAGTCTTGCCCTCACCATC, NUP-20 CACCAAATCCTTGCCCAAAGTACC, NUP-22 CTTCACCTGGTGTGATGGGAACTT, NUP-26 CGGTCAGTTATCATCTGGTGACAA, NUP-29 TGGGTTCAGCTTTTGCCAAGCT, NUP-31 GGCTTTGGATCCACAGCTACCT, NUP-34 CTGGTTTTGGATCAGGCACAGGA, ABL3-R CCATTTTTGGTTTGGGCTTCACACC. The TaqMan probe was ABL1-3-FAM FAM-TCCCCATTGTGATTATAGCCTAAGACCCGGAGCT-BHQ1. All primers and the probe were adjusted to a concentration of 10 μM. An equimolar mix of the 10 diluted NUP primers was prepared. The final reaction mix (25 μL) contained (final concentrations in parentheses) 3.5 μL NUP primer mix (140 nM each primer), 0.5 μL ABL3-R primer (200 nM), 1.0 μL ABL1-3-FAM probe (400 nM), and 3 μL cDNA reaction mix. The Absolute QPCR Mix (ABGene, Hamburg, Germany) containing the hot start enzyme Thermo-Start DNA polymerase was used with the following cycler program: 15 minutes at 95°C, 55 cycles (15 seconds at 95°C, 60 seconds at 60°C), 10 minutes at 25°C. For simplified multiplex, real-time quantitative PCR, only the 5 last-mentioned NUP primers were included, and the concentrations were 200 nM for each primer. Reproducible sensitivity13 of the PCR was assessed using serial dilutions of cell lines and was found to be 10-3 for the multiplex PCR.
Conventional (qualitative) PCR
All samples positive by real-time PCR were subsequently analyzed by conventional PCR to identify the exact transcript. The primers described were used in separate PCR reactions performed with the HotStarTaq Mastermix PCR kit (Qiagen, Hilden, Germany) and the following cycler program: 15 minutes at 95°C, 35 cycles of 20 seconds at 95°C, 20 seconds at 58°C, 20 seconds 72°C. Standard techniques were used to sequence purified PCR products on an ABI sequencer with the respective PCR primers as sequencing primers.
Real-time quantitative PCR for TLX1 (HOX11) and TLX3 (HOX11L2)
Aberrant TLX1 or TLX3 expression was detected by real-time PCR in a TaqMan format using the following primers and probes for TLX1 and TLX3, respectively (5′-3′): HOX11-F GATGGAGAGTAACCGCAGATACAC, HOX11-R TGCGCGGCTTCTTCTTCTT, HOX11-FAM FAM-AGGACAGGTTCACAGGTCACCCCTATCAGA-BHQ1, HOX11L2-F CAAGACCTGGTTCCAAAACCG, HOX11L2-R AGGCTGGATGGAGTCGTTGA, and HOX11L2-FAM FAM-CAGCTGCAACACGACGCCTTCCAA-BHQ1. PCR conditions were as described. RNA from the cell lines ALL-SIL and HPB-ALL were used as positive controls.
FISH
Cells fixed for fluorescence in situ hybridization (FISH) with probes for ABL1 and NUP214 were available in 4 patients (Table 1).3 In all 4 patients, an amplification of the ABL1/NUP214 gene was present (proportion of abnormal cells, 1.74%-64.6%; median spot counts/cell, 5-10).
Features of NUP214-ABL1-positive patients
Patient . | Sex . | Age, y . | Material . | Immunophenotype . | Transcript . | Cytogenetics . | HOX11 . | WBC count, × 109/L . | Clinical course . |
|---|---|---|---|---|---|---|---|---|---|
| 3 | M | 18 | BM | Thymic* | nup31a2 | 46,XY FISH: positive | TLX1–TLX3+ | 122.0 | CCR d979; alive d998 |
| 4 | M | 20 | PB | Thymic | nup31a3 | 46,XY,del(6)(q15q21)5; 46,XY10 | TLX1–TLX3– | 103.8 | CCR d768; alive d793 |
| 5 | M | 39 | BM | Thymic† | nup31a2 | 46,XY | TLX1+TLX3– | 158.6 | CCR d1457; alive d1469 |
| 6 | M | 34 | PB | Thymic | nup31a2 | 46,XY10 /45,XY,–141FISH:positive | TLX1+TLX3– | 15.6 | CCR d1105; alive d1133 |
| 7 | M | 28 | BM | Thymic | nup31a2 | NA; FISH: positive | TLX1+TLX3– | 60.2 | Death d86 (in CR) |
| 8 | M | 23 | BM | Thymic | nup29a2 | 46,XY | TLX1+TLX3– | 181.4 | CCR d1791; alive d1832 |
| 9 | M | 18 | BM | Thymic | nup29a3 | 46,XY | TLX1–TLX3– | 151.0 | CCRd2118; alive d2156 |
| 10 | F | 34 | BM | Mature | nup34a2 | 46,XX | TLX1+TLX3+ | 20.7 | Death d554 (in CR) |
| 11 | M | 31 | BM | Mature | nup34a2 | NA | NA | 1.8 | Relapse d496; death d936 |
| 12 | M | 18 | BM | Early T‡ | nup31a3 | 46,XY; FISH: positive | TLX1–TLX3+ | 150.0 | Relapse d778; alive d1014 |
| 13 | F | 27 | PB | Thymic | nup28a2 | NA | TLX1+TLX3– | 66.0 | Relapse d107; death d209 |
Patient . | Sex . | Age, y . | Material . | Immunophenotype . | Transcript . | Cytogenetics . | HOX11 . | WBC count, × 109/L . | Clinical course . |
|---|---|---|---|---|---|---|---|---|---|
| 3 | M | 18 | BM | Thymic* | nup31a2 | 46,XY FISH: positive | TLX1–TLX3+ | 122.0 | CCR d979; alive d998 |
| 4 | M | 20 | PB | Thymic | nup31a3 | 46,XY,del(6)(q15q21)5; 46,XY10 | TLX1–TLX3– | 103.8 | CCR d768; alive d793 |
| 5 | M | 39 | BM | Thymic† | nup31a2 | 46,XY | TLX1+TLX3– | 158.6 | CCR d1457; alive d1469 |
| 6 | M | 34 | PB | Thymic | nup31a2 | 46,XY10 /45,XY,–141FISH:positive | TLX1+TLX3– | 15.6 | CCR d1105; alive d1133 |
| 7 | M | 28 | BM | Thymic | nup31a2 | NA; FISH: positive | TLX1+TLX3– | 60.2 | Death d86 (in CR) |
| 8 | M | 23 | BM | Thymic | nup29a2 | 46,XY | TLX1+TLX3– | 181.4 | CCR d1791; alive d1832 |
| 9 | M | 18 | BM | Thymic | nup29a3 | 46,XY | TLX1–TLX3– | 151.0 | CCRd2118; alive d2156 |
| 10 | F | 34 | BM | Mature | nup34a2 | 46,XX | TLX1+TLX3+ | 20.7 | Death d554 (in CR) |
| 11 | M | 31 | BM | Mature | nup34a2 | NA | NA | 1.8 | Relapse d496; death d936 |
| 12 | M | 18 | BM | Early T‡ | nup31a3 | 46,XY; FISH: positive | TLX1–TLX3+ | 150.0 | Relapse d778; alive d1014 |
| 13 | F | 27 | PB | Thymic | nup28a2 | NA | TLX1+TLX3– | 66.0 | Relapse d107; death d209 |
Genetic, immunologic, and clinical features of the 11 NUP214-ABL1–positive patients. Numbering corresponds to that in Figure 2B (Nos. 1 and 2: cell lines BE-13 and ALL-SIL, respectively).
HOX11: aberrant expression of TLX1 (HOX11) and TLX3 (HOX11L2) genes; WBC count, peripheral leukocyte count per microliter; transcript, NUP214-ABL1 fusion transcript. One patient was not evaluable for HOX11 gene expression because of sparse material, and cytogenetic data were not available (NA) for 3 patients. FISH data were available for 4 patients, all showing an ABL1 and an NUP214 amplification
With CD56/CD16 (NK-like) coexpression
Without CD2 expression
With CD13 coexpression
Results
Altogether 279 adult T-ALL samples were analyzed, as described. Eleven (3.9%) samples were found to be NUP214-ABL1 positive by real-time quantitative PCR (Table 1).
Immunologic and genetic characteristics of the NUP214-ABL1-positive patients
Eight of these patients had a thymic immunophenotype, 1 had an early T-cell immunophenotype, and 2 had a mature T-cell immunophenotype. No particular immunophenotypic features distinguished NUP214-ABL1-positive patients from NUP214-ABL1-negative ones. Cytogenetic data, available for 8 patients, mostly showed a normal karyotype. Eight of 10 NUP214-ABL1-positive patients had evidence of an aberrant expression of HOX11 genes (TLX1 or TLX3; Table 1) compared with 41.5% (115 of 277 evaluable) in the negative group.
Probability of overall survival of NUP214-ABL1-positive and -negative patients. No significant differences were observed in overall survival between NUP214-ABL1-positive (dashed line) and -negative patients (solid line). Table 1 lists the detailed clinical characteristics of the 11 NUP214-ABL1-positive patients.
Probability of overall survival of NUP214-ABL1-positive and -negative patients. No significant differences were observed in overall survival between NUP214-ABL1-positive (dashed line) and -negative patients (solid line). Table 1 lists the detailed clinical characteristics of the 11 NUP214-ABL1-positive patients.
Clinical features of the NUP214-ABL1-positive patients
There were no significant differences in mean age (26.0 years vs 32.3 years; n = 268), sex (82% vs 75.5% male), mean leukocyte count at diagnosis (82.383/μL vs 86.081/μL; n = 258), CNS involvement (0% vs 9.8%; n = 235), therapy response (CR after induction I, 100% vs 75.9%), and, in particular, overall survival (Figure 1) between NUP214-ABL1-positive and -negative patients.
Discussion
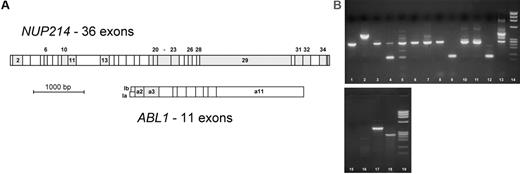
Several hypothetical in-frame fusion genes are possible with NUP214 (Figure 2A), and the breakpoint region in the NUP214 gene is not yet well characterized. Thus, forward primers were constructed in 10 different NUP214 exons to allow amplification of all possible NUP214-ABL1 in-frame transcripts. The ABL1 SH3 regulatory domain is encoded by ABL1 exons 2 and 3, and its fusion to another gene turns ABL1 into an oncogene. All experimentally observed ABL1 fusion genes (eg, BCR-ABL1) show a fusion of ABL1 exon 2 or occasionally exon 3 to the partner gene. Thus the reverse primer and probe in the real-time PCR were located in ABL1 exon 3. A real-time PCR format was chosen for detection because it is less susceptible than conventional multiplex PCR to misinterpretations caused by artifact bands given that it implies a sequence-specific labeled TaqMan probe.
Graux et al3 reported 4 adult patients with T-ALL with the NUP214-ABL1 fusion gene. Three died within the first year, and one had an early relapse in month 8 after diagnosis. No treatment data were given for these patients or a comparable NUP214-ABL1-negative group. The authors concluded that this fusion gene was “indicative of a rather aggressive course of the disease.” This assumption is not supported by the findings we obtained in a much larger and uniformly treated patient population. Overall survival did not differ significantly between the 2 groups (Figure 1). The major clinical features were comparable in NUP214-ABL1-positive and -negative patients (Table 1). Ballerini et al14 speculated that the presence or absence of NUP214-ABL1 in T-ALL might explain the previously reported heterogeneous clinical courses of TLX3-positive patients. The 3 TLX3- and NUP214-ABL1-positive patients in our series, however, had very different clinical courses (Table 1). We observed only 5 several possible NUP214-ABL1 transcripts (Figure 2B; Table 1). Taken together with the results of Graux et al,3 it appears that a limited number of all theoretically possible transcripts can actually be found in adult T-ALL. We therefore simplified the detection method and established a novel multiplex PCR in a real-time format.
Before the discovery of NUP214-ABL1, an involvement of NUP214 in acute leukemia was already known from AML. Here the carboxyterminal part of the gene is occasionally fused to the chromatin- and DNA-binding DEK gene on 6p23,15 resulting in a DEK-NUP214 (DEK-CAN) hybrid gene.16 The exact mode of leukemogenicity of DEK-NUP214 is unknown, but it is thought to interfere with nucleocytoplasmic transport processes.17 It is unknown whether NUP214-ABL1 also interferes with these transport processes.
A recently published case report described a NUP214-ABL1-positive patient who showed no response to imatinib treatment.18 It remains an open question whether most NUP214-ABL1-positive patients could benefit from imatinib treatment. It must also be kept in mind that processes other than NUP214-ABL1 fusion may lead to amplification of the ABL1 gene in T-ALL and to potential imatinib susceptibility.19,20
NUP214 and ABL1 exons. (A) In most ABL1 fusion genes (eg, BCR-ABL1), ABL1 exon 2 is fused to the partner gene, but fusion genes with ABL1 exon 3 have occasionally been described (both exons are in the same reading frame). NUP214 exons that could be joined in-frame with either ABL1 exon 2 or 3 are shaded. PCR primers were located in ABL1 exon 3 and in the 3′ part of NUP214 exons 2, 6, 10, 13, 20, 22, 26, 29, 31, and 34. (B) Agarose gel of NUP214-ABL1-positive cases. Lane 1, cell line BE-13 (transcript nup34a2); lane 2, cell line ALL-SIL (transcript nup32a2); lanes 3-13, patient samples as described in Table 1; lanes 14 and 19, ϕX174 size standard; lanes 15-18, multiplex PCR with H2O (lane 15), NUP214-ABL1-negative cDNA (lane 16), and positive samples (lanes 17, 18).
NUP214 and ABL1 exons. (A) In most ABL1 fusion genes (eg, BCR-ABL1), ABL1 exon 2 is fused to the partner gene, but fusion genes with ABL1 exon 3 have occasionally been described (both exons are in the same reading frame). NUP214 exons that could be joined in-frame with either ABL1 exon 2 or 3 are shaded. PCR primers were located in ABL1 exon 3 and in the 3′ part of NUP214 exons 2, 6, 10, 13, 20, 22, 26, 29, 31, and 34. (B) Agarose gel of NUP214-ABL1-positive cases. Lane 1, cell line BE-13 (transcript nup34a2); lane 2, cell line ALL-SIL (transcript nup32a2); lanes 3-13, patient samples as described in Table 1; lanes 14 and 19, ϕX174 size standard; lanes 15-18, multiplex PCR with H2O (lane 15), NUP214-ABL1-negative cDNA (lane 16), and positive samples (lanes 17, 18).
Appendix
Members of the GMALL study group are as follows: Aachen: Uniklinik, Med. Klinik IV (R. Osieka, R. Dada); Augsburg: Zentralklinikum (G. Schlimok); Bad Saarow: Humaine Klinikum (H. Fuss, A. West); Berlin: Charité Campus Benjamin Franklin (E. Thiel, I. W. Blau); Berlin: Vivantes-Klinikum (K.-P. Hellriegel, M. Hackenthal); Berlin: Charité Campus Virchow (R. Arnold, T. Terwey); Berlin: Charité Campus Buch (W.-D. Ludwig, R. Ratei); Berlin: Charité Campus Mitte (K. Possinger, D. Kühnhardt); Bochum: Knappschaftskrankenhaus (W. Schmiegel, G. Massenkeil); Bonn: Universitätsklinik (I. Schmidt-Wolf); Bremen: Klinikum Mitte (B. Hertenstein, S. Kaun); Bremen: Evangelische Diakonissenanstalt (K. H. Pflüger, Chr. Diekmann); Chemnitz: Klinikum (M. Haenel, G. Geiβler); Dessau: Städtisches Klinikum (A. Florschütz, E. Schwalbe); Dortmund: St. Johannes-Hospital (H.-J. Pielken, V. Hagen); Dresden: Klinikum Carl Gustav Carus (G. Ehninger, R. Naumann); Duisburg: St. Johannes-Hospital (C. Aul, A. Giagounidis); Duisburg: Johanniter-Krankenhaus (W. Lange, S. Kuhlemann); Düsseldorf: Universitätsklinik (R. Haas); Erlangen: Universität Erlangen-Nürnberg (J. R. Kalden, G. Helm); Eschweiler: St-Antonius-Hospital (R. Fuchs, F. Schlegel); Essen: Universitätsklinikum (U. Dührsen, A. Hüttmann); Essen: Universitätsklinikum der GHS (S. Seeber, M. R. Nowrousian); Essen-Werden: Kliniken Essen Süd (W. Heit, F.-K. Baur); Frankfurt: Universitätsklinikum (D. Hoelzer, N. Gökbuget); Freiburg: Universitätsklinikum (R. Mertelsmann, M. Lübbert); Gieβen: Universitätsklinikum (H. Pralle, M. Dörner); Göttingen: Universitätsklinikum (L. Trümper, F. Griesinger); Greifswald: Ernst-Moritz-Arndt-Universität (G. Dölken, B. Kallinich); Gütersloh: Städtisches Krankenhaus (C. Gropp, P. Düwel); Hagen: Katholisches Krankenhaus (H. Eimermacher, W. Lindemann); Halle/Saale: Martin-Luther-Univ. Halle-Wittenberg (H.-J. Schmoll, H.-H. Wolf); Halle/Saale: Städtisches Krankenhaus Martha-Maria (W. Schütte, U. Haak); Hamburg: Asklepios Klinik St. Georg (N. Schmitz, J. Rutjes); Hamburg: Universitätsklinikum Hamburg-Eppendorf (C. Bokemeyer, M. de Wit); Hamm: Evangelisches Krankenhaus (L. Balleisen, E. Lange); Hannover: Medizinische Hochschule (A. Ganser, H. Diedrich); Heidelberg: Universitätsklinikum (A. Ho, U. Mahlknecht); Homburg/Saar: Med. Universitätsklinik und Poliklinik (M. Pfreundschuh, B. Gleissner); Idar-Oberstein: Klinik für Knochenmarktransplantation (A. A. Fauser, Kraut); Jena: Klinikum d. Friedrich-Schiller-Universität (K. Höffken, H.-J. Fricke); Karlsruhe: Städt. Klinikum (M. Bentz, S. Wilhelm); Kassel: Klinikum (M. Wolf, B. Ritter); Kiel: Universitätsklinikum, Campus Kiel (M. Kneba, S. Irmer); Köln: Universitätsklinik (M. Hallek, P. Staib); Krefeld: Klinikum (T. Frieling, M. Planker); Leverkusen: Klinikum (N. Niederle, M. Kreβ); Lübeck: Universitätsklinikum, Campus Lübeck (T. Wagner, S. Peters); Magdeburg: Otto-von-Guericke-Universität (A. Franke, Messner); Mainz: Universitätskliniken (C. Huber, J. Beck); Mannheim: Klinikum (R. Hehlmann, K. Adam); Marburg: Klinikum Lahnberge (A. Neubauer, N. Hagner); Minden: Klinikum (H. Bodenstein, H. Lampe); München: Krankenhaus München-Schwabing (Chr. Nerl, Th. Lipp); München: Klinikum Rechts der Isar (C. Peschel, F. Schneller); Münster: Universitätsklinik (W. E. Berdel, M. Stelljes); Neubrandenburg: Dietrich-Bonhoeffer-Klinikum (H. Rühle, N. Grobe); Nürnberg: Klinikum Nord (M. Wilhelm, K. Schäfer-Eckart); Offenburg: Klinikum (F. Hirsch, I. Dresel); Oldenburg: Klinikum (C.-H. Köhne, B. Metzner); Passau: Klinikum (T. Südhoff); Potsdam: Klinikum Ernst von Bergmann (G. Maschmeyer, A. Gerhardt); Regensburg: Universitätsklinikum (R. Andreesen, A. Reichle); Regensburg: Krankenhaus d. Barmherzigen Brüder (E. Kreuser, J. Schenk); Rostock: Universität (M. Freund, C. Junghanss); Saarbrücken: Caritasklinik St. Theresia (J. Preiβ, A. Matzdorff); Sande: Nordwest-Krankenhaus (H. Mayet, F. K. Natt); Schwerin: Helios Klinikum (D. Hähling, Chr. Schult); Stralsund: Klinikum der Hansestadt (T. H. Ittel, U. Gerecke); Stuttgart: Robert Bosch-Krankenhaus (W. Aulitzky, L. Leimer); Stuttgart: Klinikum (H.-G. Mergenthaler, J. Schleicher); Trier: Krankenhaus d. Barmherzigen Brüder (C. B. Kölbel, H. Kirchen); Tübingen: Universitätsklinikum (L. Kanz, M. Schmalzing); Ulm: Medizinische Universitätsklinik (H. Döhner, M. Schmid); Wiesbaden: Dr.-Horst-Schmidt-Kliniken (N. Frickhofen, H.-G. Fuhr); Wuppertal: Helios Klinikum (A. Raghavachar); Würzburg: Universitätsklinikum (H. Einsele, M. Topp); Zwickau: Städt. Klinikum Heinrich Braun (U. Kreibich, W. Zschille).
Prepublished online as Blood First Edition Paper, July 27, 2006; DOI 10.1182/blood-2006-04-014514.
A complete list of the members of the German Multicenter Adult ALL (GMALL) study group appears in the “Appendix.”
Supported by grant 10-1988-Bu1 (to T.B. and S.S.) and 70-2657-Ho2 (to D.H.) from the Deutsche Krebshilfe, and grant NGFN-01 GR0105-6.2 from the Bundesministerium für Bildung und Forschung (R.R.).
The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank all clinic and cytogenetic laboratory staffs and all patients involved in the GMALL study group for their support. All authors thank Prof. E. Thiel (Charité CBF Berlin) for invaluable support. We are particularly indebted to M. Molkentin, C. Seide, B. Komischke, R. Lippoldt, and D. Gröger for their skillful technical assistance. We thank Dr J. Weirowski for critically reading the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal