Abstract
The existence and roles of a class of abundant regulatory RNA molecules have recently come into sharp focus. Micro-RNAs (miRNAs) are small (approximately 22 bases), non–protein-coding RNAs that recognize target sequences of imperfect complementarity in cognate mRNAs and either destabilize them or inhibit protein translation. Although mechanisms of miRNA biogenesis have been elucidated in some detail, there is limited appreciation of their biological functions. Reported examples typically focus on miRNA regulation of a single tissue-restricted transcript, often one encoding a transcription factor, that controls a specific aspect of development, cell differentiation, or physiology. However, computational algorithms predict up to hundreds of putative targets for individual miRNAs, single transcripts may be regulated by multiple miRNAs, and miRNAs may either eliminate target gene expression or serve to finetune transcript and protein levels. Theoretical considerations and early experimental results hence suggest diverse roles for miRNAs as a class. One appealing possibility, that miRNAs eliminate low-level expression of unwanted genes and hence refine unilineage gene expression, may be especially amenable to evaluation in models of hematopoiesis. This review summarizes current understanding of miRNA mechanisms, outlines some of the important outstanding questions, and describes studies that attempt to define miRNA functions in hematopoiesis.
miRNAs, the sounds of (gene) silence
MicroRNAs (miRNAs) are small, often phylogenetically conserved, non–protein-coding RNAs that mediate posttranscriptional gene repression by inhibiting protein translation or by destabilizing target transcripts. miRNAs recognize target sites, most commonly found in the 3′-untranslated regions (UTRs) of cognate mRNAs, through imperfect base-pairing, with 1 or more mismatches in sequence complementarity. miRNAs are believed to fine-regulate a diverse array of biological processes, and a convergence of genetic, biochemical, and structural studies has led to rapid growth in understanding of their synthesis and molecular mechanisms. In contrast, the precise biological functions of these approximately 22-base RNA products are less clear. To date, only a handful of studies provide definitive evidence of a role for a specific miRNA in vertebrate biology, although such roles are probably pervasive. The limits on current appreciation simply reflect the infancy of the field, and it is safe to predict a vast growth in functional studies that will refine understanding of the roles of individual miRNAs.
The history and general features of miRNAs have been reviewed extensively in a number of different contexts.1-3 Here I review the biogenesis and mechanisms of miRNAs briefly before exploring the current appreciation of the role of miRNAs in cell differentiation, with emphasis on hematopoiesis. Many of the general and biochemical insights derive from experiments conducted in worms and flies, and these studies are discussed when appropriate. Although investigation in invertebrate species also provides many functional insights,4 this review will focus on miRNA functions in vertebrate animals.
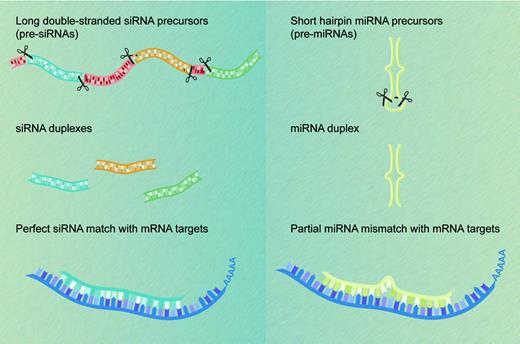
The first miRNAs, lin-4 and let-7, were each identified, some 7 years apart, on the basis of their mutant phenotypes in forward genetic screens in the worm Caenorhabditis elegans. Both entities control developmental timing by repressing translation of their respective targets, the protein-coding mRNA products of the lin-14 and lin-41 genes.5,6 Although posttranscriptional gene silencing by small regulatory RNAs was initially regarded as a biological curiosity, perhaps restricted to certain lower species, we now know that many multicellular organisms express hundreds of miRNAs that vary in abundance across cell and tissue types.7-10 miRNAs are found in multicellular plant and animal species but not in unicellular organisms such as yeast. By definition, miRNAs correspond to cloned small RNAs that, like the lin-4 and let-7 products, are processed from stem-loop precursors transcribed from genes other than the ones they regulate. Endogenous small RNAs that do not meet these criteria and are known collectively as small-interfering RNAs (siRNAs) derive from long double-stranded RNA (dsRNA) precursors that in some species may be transcribed from repeat elements, heterochromatin, or transposons.3 The 2 classes of short regulatory RNAs differ mainly in their origins and not in their functions,11 which may be grouped under the single moniker of RNA interference (RNAi), although siRNA sequences are usually perfectly complementary to those of their RNA targets (Figure 1).
Salient concepts in miRNA function
In zebrafish development, where miRNA expression dynamics are the best characterized to date, most miRNAs appear relatively late in embryogenesis and reveal tissue-specific distributions and requirements.12-14 In certain mammalian organs, including bone marrow, a handful of tissue-specific miRNAs tend to dominate the miRNA expression profile,10,15 and in a limited analysis of mouse miRNAs, expression profiles differed between fetal and adult forms of the same tissue.16 Taken together with the original identification of worm miRNAs as regulators of developmental timing, such observations have led to the notion that a principal function of miRNAs is to control cell differentiation and development. Indeed, mouse embryonic stem cells with an engineered inability to synthesize miRNAs are viable but fail to differentiate, and zebrafish embryos that are unable to produce mature miRNAs develop normally at first but display specific morphogenetic defects during gastrulation and organogenesis.13,17 Specific and prominent miRNA functions are illustrated in part by the roles of miR-196 in vertebrate limb development,18 of the miR-1 cluster in cardiomyocyte differentiation,19 and of miR-181 in maintaining a balance between circulating T and B lymphocytes.15 However, miRNAs are expressed widely in developing and adult tissues and their functions are likely to be widespread, diverse, and ranging from seminal to subtle depending on the context.
miRNAs and siRNAs: differences in biogenesis and properties. siRNAs (left) derive from long endogenous dsRNA molecules that form either long hairpins or bimolecular duplexes. Processing of these dsRNA precursors can generate many different siRNAs from both strands. In contrast, processing of the shorter hairpin structures known as pre-miRNAs (right) produces a single miRNA molecule from one arm of the hairpin precursor. siRNAs recognize their target transcripts with perfect sequence complementarity (left), whereas miRNAs typically have a limited number of mismatches with their mRNA target sequences (right). Both classes of small regulatory RNA molecules cause posttranscriptional silencing of protein-coding genes.
miRNAs and siRNAs: differences in biogenesis and properties. siRNAs (left) derive from long endogenous dsRNA molecules that form either long hairpins or bimolecular duplexes. Processing of these dsRNA precursors can generate many different siRNAs from both strands. In contrast, processing of the shorter hairpin structures known as pre-miRNAs (right) produces a single miRNA molecule from one arm of the hairpin precursor. siRNAs recognize their target transcripts with perfect sequence complementarity (left), whereas miRNAs typically have a limited number of mismatches with their mRNA target sequences (right). Both classes of small regulatory RNA molecules cause posttranscriptional silencing of protein-coding genes.
In the best-studied examples in worms and mammals, miRNAs control key developmental processes by inhibiting just one or a few mRNA targets. As we shall see, however, the typical miRNA can potentially regulate dozens or scores of genes,20,21 and most targets contain isolated miRNA recognition sites that may be inadequate for complete gene silencing. These considerations prompted Bartel and Chen to propose the presence of selective pressure on many genes to avoid undesirable interference from miRNAs that are both widespread and have limited complementarity requirements.22 Stark and colleagues recently reported that genes that seem to avoid miRNA regulation (“antitargets”) tend to function in processes common to all cells, whereas putative targets are more commonly implicated in developmental processes, especially organogenesis.23 In several tested examples in Drosophila embryos, miRNAs and some predicted targets are indeed expressed in nonoverlapping adjacent distributions, whereas miRNAs and antitargets are coexpressed. Farh et al24 found similar results for targets and antitargets of mammalian miRNAs, although in their analyses the predicted targets were present at lower levels in tissues with the cognate miRNAs and not excluded altogether. Furthermore, in a temporal analysis of mouse myotube differentiation, they showed that predicted targets of tissue-specific miR-1 and miR-133 were highly expressed before these miRNAs accumulated and subsequently, these targets were down-regulated. Together, these observations raise the possibility that some transcripts are predisposed to appear or persist in domains where they are unnecessary or potentially harmful; miRNAs may have evolved to fine-tune gene expression programs and to eliminate gene products more rapidly than might occur by intrinsic decay. This particular role may be especially pertinent in hematopoiesis. In a process known as multilineage gene priming, immature blood progenitors initially activate a leaky and promiscuous transcriptional program; as cells commit to a restricted fate, unilineage gene expression is reinforced and transcripts affiliated with alternative blood cell types disappear.25 It is tempting to speculate that miRNAs drive the latter phase, an idea that is as yet untested.
Animal genomes not only encode a large number of tissue-restricted miRNAs, but these miRNAs can be expressed in high, albeit variable, abundance, with estimates in C elegans ranging from 800 to 50 000 molecules per cell.26 Coupled with the limited evolutionary constraints on small non–protein-coding genes, miRNAs may be positioned ideally to regulate tissue-specific gene expression. As the latter function lies at the heart of cell differentiation, it is fair to speculate that miRNAs as a class play a vital role in generating and maintaining distinctive cell lineages in all multicellular species. However, forward genetic screens in C elegans and Drosophila have yielded very few miRNAs, possibly reflecting the small size of the coding unit, subtlety in mutant phenotypes, and a substantial degree of functional redundancy. Instead, current catalogs of miRNAs derive largely from the results of cDNA cloning. In May 2006, a unified database (miRBase)27 recognized 462 and 340 individual miRNAs in the human and mouse, respectively. Nomenclature and annotation for all species are assigned and managed centrally, with the “miR” designation followed by a unique identifying number.11,28 Identical miRNAs are given the same number, regardless of organism, as are nearly identical sequences that are presumed to represent orthologous miRNAs. Highly similar miRNAs within a species also are assigned the same number, with their genes distinguished by letter suffixes (eg, miR-181a).
miRNA biogenesis and mechanisms of posttranscriptional gene repression
Mature miRNAs are 18 to 26 nucleotides in length, with 5′-phosphate and 3′-hydroxyl groups. Their immediate precursors, called pre-miRNAs, are characteristic stem-loop structures 60 to 80 bases in length with a 2-nucleotide 3′ overhang. In turn, pre-miRNAs derive from long primary miRNA transcripts (pri-miRNAs), which can exceed 1 kb and typically are products of RNA polymerase II activity.29,30 Most pri-miRNAs are independent transcriptional units, although many mammalian miRNAs are encoded within mRNA introns and appear to share the same primary transcripts as their protein-coding host genes. MicroRNAs show highest evolutionary conservation in the stem, with variable divergence within the loop.26,31 Although conservation drops sharply in sequences flanking the miRNA hairpin,31 these regions are nevertheless required for nuclear processing of the primary transcript.15
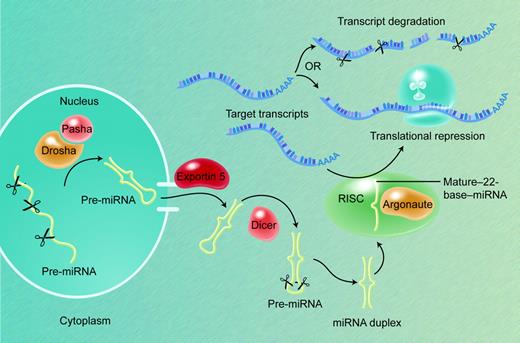
Pri-miRNAs are first processed in the nucleus into 50- to 80-base pre-miRNA stem-loops by the Microprocessor complex of nucleases and associated factors, including the RNase III Drosha and its partner DGCR8/Pasha (Figure 2).32,33 Pre-miRNA molecules, in association with the Ran-GTP–dependent factor exportin-5, are actively transported into the cytoplasm, where a second RNase III–like nuclease, Dicer, processes them further into duplexes that contain the 20- to 24-nucleotide mature miRNA and an oligonucleotide of similar size from the other arm of the hairpin.34-37 The nascent miRNA may be present on either arm of the stem-loop precursor, but each pre-miRNA yields a single mature miRNA product. Gaps, bulges, and mismatches in the secondary structure of pre-miRNAs are predicted to reduce their stability and to influence the efficiency by which they are processed to produce mature miRNAs.38 The functional element in the precursor duplex is determined when, after unwinding, one of the 2 strands is loaded asymmetrically into the RNA-induced silencing complex (RISC); this tends to be the strand in which the 5′ end is less tightly paired to its complement.39,40
Similar thermodynamic considerations apply to the interactions between mature miRNAs and their protein-coding RNA targets. Thus, the ability of a miRNA to repress cognate mRNAs is determined in large part by the free energy of binding of nucleotides 2 to 8, counted from the 5′ end, with substantially lesser contribution from the 3′ region of the miRNA.41 Nucleotides 2 to 8 are hence regarded as the “seed” for miRNA target recognition, and evolutionary conservation of complementarity with this relatively small region has served as a key basis for computational prediction of target genes. Conserved adenosine residues tend to flank the complementary region, which implies that primary sequence determinants and base-pairing combine to specify target recognition.21,42
In their size and chemistry, miRNAs resemble siRNAs, which in invertebrates recognize targets by precise complementarity and elicit their decay. As depicted in Figure 1, these 2 classes of small, noncoding RNAs derive from distinct classes of immediate precursors: long (hundreds to thousands of bases) double-stranded molecules in the case of siRNA and, in the case of miRNA, shorter (60- to 80-nucleotide) pre-miRNA stem-loop products. However, both the production and functions of these diverse small RNAs require many common factors: endonucleases such as Dicer, which specifically cleave double-stranded RNA, and small RNA-binding proteins of the Argonaute family, which direct target molecules into the RISC ribonuclear complex.34 Moreover, the mRNA degradation apparatus, including RISC complexes, localizes in discrete cytoplasmic “processing” or “P” bodies (also known as GW bodies), where transcripts targeted by miRNAs were shown recently to concentrate in a miRNA-dependent manner.43 Thus, like siRNA-induced mRNA degradation, apparent repression of translation by miRNAs also involves delivery of transcripts to P-bodies, which lack ribosomal components; this relocation could be a consequence or, alternatively, serve as the basis for inhibiting protein translation.
miRNA biogenesis and action. Most pri-mRNAs are the products of independent genes, transcribed by RNA polymerase II. The nuclear Microprocessor protein complex, which contains the RNase III Drosha and its partner DGCR8/Pasha, cleaves pri-miRNAs into 50- to 80-base pre-miRNA stem-loop moieties. The Ran-GTP–dependent factor exportin-5 actively transports pre-miRNAs into the cytoplasm, where the nuclease Dicer processes them further into duplexes that contain the 20- to 24-nucleotide mature miRNA; each pre-miRNA usually yields a single mature miRNA product. The functional strand is determined when one of the 2 strands of the duplex is loaded into the RISC, which contains Argonaute and related proteins and localizes in cytoplasmic P-bodies. Recognition of target mRNAs by partial sequence complementarity to the miRNA results in posttranscriptional gene repression by some combination of transcript degradation and translational inhibition.
miRNA biogenesis and action. Most pri-mRNAs are the products of independent genes, transcribed by RNA polymerase II. The nuclear Microprocessor protein complex, which contains the RNase III Drosha and its partner DGCR8/Pasha, cleaves pri-miRNAs into 50- to 80-base pre-miRNA stem-loop moieties. The Ran-GTP–dependent factor exportin-5 actively transports pre-miRNAs into the cytoplasm, where the nuclease Dicer processes them further into duplexes that contain the 20- to 24-nucleotide mature miRNA; each pre-miRNA usually yields a single mature miRNA product. The functional strand is determined when one of the 2 strands of the duplex is loaded into the RISC, which contains Argonaute and related proteins and localizes in cytoplasmic P-bodies. Recognition of target mRNAs by partial sequence complementarity to the miRNA results in posttranscriptional gene repression by some combination of transcript degradation and translational inhibition.
Early work suggested that siRNAs operate to eliminate their mRNA targets, whereas miRNAs commonly preserve target transcript levels and were thereby inferred to block protein translation. The distinctions between siRNAs and miRNAs continue to blur, and miRNAs are now recognized to facilitate both translational inhibition and mRNA degradation.44,45 Localization of the RISC complex in P-bodies may help in understanding the apparent differences, and miRNA-mediated transcript degradation may represent a secondary consequence of sequestration within P-bodies.3,43 Steady-state levels of transcripts with a high intrinsic decay rate may overtly reveal a substantial decline, whereas inherently stable mRNAs might persist in the cell; however, both transcript classes are probably equally subject to translational repression.
An insightful study recently shed light on several questions that are pertinent to this review. After delivering miR-124 into HeLa cells, Lim and colleagues observed a shift in the expression profile toward that of brain, the site where endogenous miR-124 is most abundant; similarly, transfection of miR-1 shifted the profile toward that of muscle, where miR-1 is preferentially expressed.45 Taken at face value, even in the absence of a clear molecular mechanism, these results implicate selected miRNAs in aspects of gene regulation that are central to cellular identity. Notably, in each case, the levels of dozens of transcripts declined within 12 hours, and the 3′-UTRs of affected mRNAs were enriched for sequences complementary to the 5′ ends of the cognate miRNAs. Thus, some of the down-regulated transcripts may be targets for direct miRNA-mediated degradation, an effect distinct from the purportedly main role of miRNAs in inhibiting protein translation. Moreover, at least miR-1 and miR-124, but probably other tissue-specific miRNAs, seem to regulate a large number of transcripts directly. It is thus conceivable that establishing or maintaining cell identity depends in part on fine regulation of lineage-specific gene expression by a few key miRNAs.
Cardinal differentiation functions have traditionally been attributed to lineage-restricted transcription factors (TFs); either the TFs themselves or their tissue-specific outputs, or both, are hence the best candidates for direct miRNA regulation. In this light, it is interesting that although current predictions of putative miRNA targets cover a broad range of molecular functions,20,21 the early sets are enriched for TF-coding genes,42,46 and validated examples of miRNA function also emphasize their roles in regulating tissue-restricted TFs such as NF-I, Hand2, and HoxB8.18,19,44,47 However, these examples reflect a certain investigation bias and miRNA functions clearly extend beyond targeting of TF transcripts, as would be expected if miRNAs as a class can affect hundreds or thousands of genes.23,24 A well-studied counterexample is offered by the evolutionarily conserved, pancreatic islet cell–specific miR-375, which seems to inhibit glucose-induced insulin secretion by targeting the myotrophin message and a resultant effect on insulin exocytosis.48
Independent of the possibly widespread role of miRNAs in suppressing TF genes, functional parallels between TFs and miRNAs as molecular regulators are noteworthy, as articulated in a recent review.49 Both classes of molecules exert their activity through composite cis-regulatory elements and function in combinations; single TFs or miRNAs may regulate many different genes, as we encountered for miR-1 and miR-124. TFs and miRNAs also may act cooperatively on their target genes, which typically harbor multiple cis-regulatory DNA elements for transcriptional activation and more than 1 putative miRNA recognition site for mRNA down-regulation.20,21,50 Thus, both TFs and miRNAs may generate combinatorial codes that dictate cellular identities and phenotypes. Understanding the putative networks of these interactions is a central challenge in studying miRNA functions in cell differentiation.
How many miRNAs? How many targets?
To appreciate the breadth of miRNA functions, it would be useful to have reliable estimates of the total number of miRNA genes in a species. After the initial cloning of more than 100 miRNAs,7-10 cloning efforts soon achieved a degree of saturation, with recurring identification of previously cloned species. However, miRNAs that are expressed at low abundance, restricted to a few cells in an organ, developmentally regulated, or technically resistant to cloning might elude detection in the common strategies. Likewise, even the most sensitive algorithms to predict miRNA genes make assumptions in order to enforce stringency in computational searches. Nevertheless, a combination of comprehensive cloning and computer prediction has expanded the number of animal miRNAs greatly. Most cloned miRNAs show sequence conservation among closely related species and also across large evolutionary distances: among the initially identified mouse and human miRNAs, more than half have identifiable homologs in fish, and about half of these have homologs in flies or worms.51 It is this evolutionary conservation, coupled with consideration of stem-loop structures, that provides an important basis for computational prediction of hundreds of miRNAs.52,53 In an especially thorough approach that integrated bioinformatic prediction, microarray analysis, and sequence-directed cloning, Bentwich and coworkers identified nearly 100 new miRNAs and predicted the presence of at least 800 miRNAs in aggregate.54 However, the scope of potential miRNA regulation remains uncertain, and predicted miRNAs should be regarded cautiously until they are confirmed by experiments.
To add further complexity, miRNAs share limited sequence complementarity with their mRNA targets, so that each miRNA could possibly interact with scores of genes; conversely, a single gene can harbor multiple miRNA recognition sites within 1 or more potential 3′-UTRs. Published studies in vertebrate species center on the results of bioinformatic predictions, and no mammalian miRNA target has yet been identified prospectively without the benefit of computationally derived clues. The algorithms used to predict miRNA targets typically develop scoring schemes based on sequence complementarity, free energy calculations of RNA duplex formation, and phylogenetic conservation. In a 4-genome analysis of 3′-UTRs, Lewis et al used the TargetScanS algorithm to identify 13 000 putative regulatory relationships conserved among the 4 species, thereby implicating more than 5 000 human genes (30% of the tested gene set) as candidate miRNA targets.42 Using the independent PicTar algorithm, Krek et al reached similar conclusions on the scope of conserved miRNA targeting in mammals, with a set of predictions that largely overlaps that of the TargetScanS analysis.20 By contrast, John et al have predicted about 2 000 human gene targets, which currently have little overlap with those of TargetScanS and PicTar.46 Lewis et al detected additional targeting in open reading frames and concluded that more than one-third of human genes are under selective pressure to maintain pairing to miRNAs.42 Furthermore, there is increasing experimental and computational evidence for a large number of nonconserved targets.23,24,45 However, individual predictions of miRNA targets should be regarded as provisional, and attempts at biological investigation should be preceded by careful experimental validation of putative miRNA-target combinations.
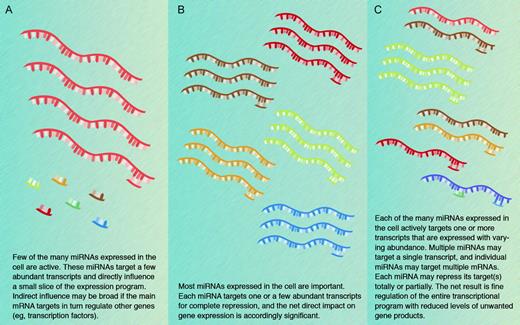
The possibility that a single miRNA may target multiple transcripts and that individual transcripts may be subject to regulation by multiple miRNAs amplifies the scope of putative miRNA regulation in potentially unfathomable terms. The conceptual and bioinformatic problems presented by these considerations are formidable and may be compared with those that follow from genome-scale identification of protein-protein interactions. In the latter example, however, mapping the domains of gene expression reduces the magnitude of the challenge as proteins can legitimately interact only when they are coexpressed. In contrast, miRNAs may be coexpressed with their targets to dampen gene expression, or the 2 may appear in complementary distributions when the miRNA serves to eliminate target gene expression. Resolution of the underlying questions will require rigorous experimentation and confident assessment of low levels of miRNA and mRNA products. In addition, it is likely that not all miRNAs function similarly: some may indeed regulate a single gene tightly, whereas others may have a wider influence over gene expression programs associated with selected developmental or physiologic states (Figure 3).
Before reviewing what little is known about miRNA influences in hematopoiesis, it is worthwhile to explore the general lessons we might learn from the few reported examples of mammalian miRNA functions. Mouse miR-196, which is encoded at 3 paralogous locations in the mammalian A, B, and C Hox gene clusters, represses HoxB8 mRNA and downstream Sonic hedgehog signaling, factors that distinguish developing forelimbs from hind-limbs.18,44 In a different scenario, the miR-1 genes are direct transcriptional targets of muscle differentiation TFs such as serum response factor, MyoD, and Mef2, and their miRNA products appear to regulate levels of Hand2, a TF that promotes cardiomyocyte expansion in the cardiac ventricles; overexpression of miR-1 in the developing heart reduces the pool size of proliferating ventricular cardiomyocytes.19 In addition to a third instance that is described in the next section, “miRNA expression and functions in hematopoiesis,”47 these examples share certain common themes that may be instructive. First, in each case, the miRNA under detailed investigation shows significant differential expression in a manner that permitted generation of cogent and testable hypotheses. miR-1 is highly enriched in cardiac and skeletal muscle precursors, whereas miR-196 expression is at least 20-fold higher in hindlimb buds than in the forelimbs. A second, related point is that in each case there was already considerable information about the tissue under study so that the investigators could test for consistency and rely on experimental systems that are well validated. Third, identification of miRNA targets remains heavily reliant on computational prediction, again coupled with prior knowledge that helped focus on plausible candidates. These themes will likely continue to guide further work in biological miRNA functions into the foreseeable future. Of course, these examples illustrate the seminal role of selected factors that may have a limited repertoire of TF targets. This scenario is quite possibly the exception rather than the rule for general miRNA functions. It will require new experimental paradigms to gain better insights into cases where a few or many miRNAs operate in combination with innumerable targets to fine-regulate biological processes.
Models for the biological significance of miRNA functions. Three nonmutually exclusive models are presented, and the truth is likely to incorporate elements from each. Different modes may also operate in different cells or at different stages in development. Computational predictions form the basis for much of the current thinking and experimental evidence favoring each of the models remains limited.
Models for the biological significance of miRNA functions. Three nonmutually exclusive models are presented, and the truth is likely to incorporate elements from each. Different modes may also operate in different cells or at different stages in development. Computational predictions form the basis for much of the current thinking and experimental evidence favoring each of the models remains limited.
miRNA expression and functions in hematopoiesis
Hematopoietic cells were not included in the earliest reported efforts to identify tissue-restricted miRNAs.10 Subsequently, Chen and colleagues15 cloned about 100 unique miRNAs from mouse bone marrow. Whereas many had been identified previously from nonhematopoietic tissues, a select few, including miR-142, miR-181a, and miR-223, are expressed preferentially in blood cells and a limited number of other sites. miR-223 and miR-142 showed the most restricted distribution, virtually confined to bone marrow and spleen; some miR-142 expression is also seen in thymus, where miR-181a is especially abundant.15 Other results from miRNA microarray and Northern analysis of assorted blood cell (mainly lymphocyte) populations suggest that few miRNAs are truly restricted in distribution.55 Indeed, tissue specificities reported for miRNA expression in general probably reflect enrichment, which can even be considerable, in certain cells and not necessarily exclusion from all other sites. Nevertheless, some miRNAs are remarkably responsive to cellular manipulations: miR-150 is rapidly down-regulated upon T-cell stimulation under T helper 1 (Th1) or Th2 conditions, whereas miR-146 is selectively increased in Th1 lymphocytes.55 In a recent expression analysis of megakaryocyte cultures derived from CD34+ bone marrow cells, Garzon and colleagues identified 19 miRNAs that were reduced by 2- to 50-fold and none that increased with megakaryocytic differentiation.56 Although 2 of these miRNAs, miR-10a and miR-130a, are thought to target the transcription factor genes HOXA1 and MAFB, respectively,56 the true significance of most miRNA differential expression or of changes induced in hematopoiesis is not known.
To interrogate miRNA functions in hematopoiesis, Chen et al15 introduced the blood cell–specific miRNAs they identified into murine Lin– hematopoietic progenitor cells, followed by in vitro differentiation or transplantation into lethally irradiated hosts. Overexpression of miR-181a induced a 2-fold increase in B lymphocyte numbers without affecting the T-cell lineage in vitro, although the mouse transplantation studies yielded a nearly 2-fold reduction in the number of circulating T lymphocytes and about a 90% decrease in the CD8+ subpopulation. In contrast, miR-142s and miR-223, both evaluated only in vitro, modestly increased T-cell and not B-cell numbers; miR-30, which is not preferentially expressed in blood cells, did not affect hematopoiesis as judged by flow cytometry for lineage markers. Whereas miR-181a hence seems capable of modulating lymphoid cell lineages, it is important to note that these conclusions rely exclusively on studies of overexpressed miRNAs. Indeed, experimental constraints limit the ability to assess miRNA functions rapidly through loss of function, and in any event, it is likely that regulation by miRNAs encompasses considerable redundancy.
In contrast to the specific requirement of selected miRNAs, the general requirement for miRNAs in a hematopoietic organ has been evaluated in mice with selective deletion of the dcr-1 (Dicer) gene in the thymus.57 These mice displayed a severe block in development of peripheral CD8+ T cells and reduced numbers of CD4+ T lymphocytes, which also proliferated poorly or died by apoptosis quickly after agonist stimulation. Dicer-deficient CD4+ T cells preferentially expressed interferon-γ, a cytokine affiliated with the Th1 lineage. These observations represent the net effect of miRNA deficiency and perhaps other unknown Dicer functions, possibly including processing of siRNA. Accordingly, it remains unclear whether they reflect tissue-specific absence of a single critical miRNA, of a few regulators that control a limited number of mRNA targets, or the functional output of a complex pathway consisting of many miRNAs, each with a defined individual role. Indeed, the fundamental limitation of both the miR-181 overexpression and Dicer gene deletion studies is that the identity and activities of the primary mRNA targets are unknown. Target prediction algorithms identify no fewer than 340 conserved candidate targets for miR-181a, with equivalent degrees of confidence for dozens of them. Thus, dissecting the regulatory circuit poses an enormous challenge. By contrast, the miRanda algorithm,46 whose predicted targets overlap minimally with others', hints at the human HOXA5 gene as a candidate miR-181 target. This putative target gene is expressed in hematopoietic cells, where it has previously been implicated in regulating the balance between myeloid and erythroid cell precursors.58,59 The latter studies did not, however, report lymphocyte ratios; miR-181a overexpression does not influence erythroid cell differentiation;15 and interaction between miR-181a and HOXA5 remains speculative at best.
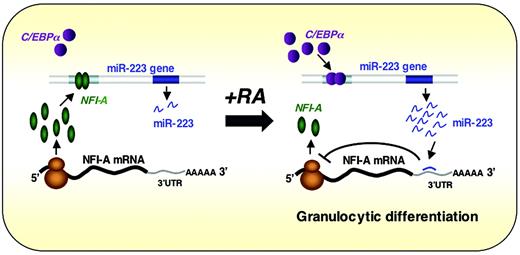
miR-223 is another blood cell–restricted miRNA, which achieves highest levels in CD34– bone marrow cells, is not present in T or B lymphocytes, and is induced by retinoic acid treatment of myeloid precursors through the transcriptional activator C/EBPα.47 C/EBPα is known to be important in myelopoiesis as it regulates many myeloid cell–specific genes and promotes granulocyte-specific genes at the expense of the monocytic differentiation program in bipotential precursors60 ; C/EBPα knockout mice show selective failure of neutrophil and eosinophil differentiation.61 Overexpression of miR-223 in the undifferentiated NB4 cell line induced morphologic changes consistent with maturation and expression of CD11b and the granulocyte colony-stimulating factor receptor. Conversely, in one of the few examples where a vertebrate miRNA function has been evaluated through experimental reduction in its levels, miR-223–complementary locked nucleic acid oligonucleotides induced modest reductions in the amounts of both miR-223 and CD11b.47 These miR-223 effects are mediated in part through translational inhibition of mRNA for the TF NFI-A. In a particularly provocative twist, NFI-A and C/EBPα compete for binding to partially overlapping DNA sequences in the miR-223 gene promoter, and as long as NFI-A is present, this miR gene is inactive. With the appearance of C/EBPα, which occurs upon retinoic acid–induced commitment to granulocytic differentiation, miR-223 expression is activated, resulting in repression of its NFI-A mRNA target. Thus, miR-223 appears to be a key player in a simple regulatory circuit of TFs that control granulopoiesis and may help stabilize the neutrophil phenotype induced by C/EBPα (Figure 4). As NFI-A mRNA is just one among literally hundreds of possible miR-223 targets, the particular circuit described here may represent only a facet of its full molecular effects. In principle, miR-223 could serve to down-regulate many genes that would ordinarily be expressed in multipotential progenitors or nongranulocytic cells and thus help refine the neutrophil-specific gene expression program. Although this hypothesis remains to be tested, it is consistent both with emerging concepts in the field and with the demonstration that particular miRNAs can influence discrete aspects of hematopoiesis.
Working model for the regulatory circuit involving miR-223, C/EBPα, and NFI-A in neutrophil differentiation. Experimental results47 are consistent with the idea that NFI-A and C/EBPα compete for binding to partially overlapping DNA sequences in the miR-223 gene promoter, and as long as NFI-A is present, this miR gene is inactive. Cells committed to granulocytic differentiation express C/EBPα, which activates miR-223 expression, resulting in repression of its NFI-A mRNA target. Thus, miR-223 appears to participate in a simple regulatory circuit of TFs that control granulopoiesis and may help stabilize the neutrophil phenotype induced by C/EBPα. Reprinted from Fazi et al47 with permission.
Working model for the regulatory circuit involving miR-223, C/EBPα, and NFI-A in neutrophil differentiation. Experimental results47 are consistent with the idea that NFI-A and C/EBPα compete for binding to partially overlapping DNA sequences in the miR-223 gene promoter, and as long as NFI-A is present, this miR gene is inactive. Cells committed to granulocytic differentiation express C/EBPα, which activates miR-223 expression, resulting in repression of its NFI-A mRNA target. Thus, miR-223 appears to participate in a simple regulatory circuit of TFs that control granulopoiesis and may help stabilize the neutrophil phenotype induced by C/EBPα. Reprinted from Fazi et al47 with permission.
In a similar vein, Felli and colleagues62 found high expression of miR-221 and miR-222 in human cord blood–derived CD34+ hematopoietic progenitor cells; unilineage erythroid differentiation of these cells in culture caused reduction in the levels of these 2 miRNAs, which have virtually identical sequence in the seed region of target pairing. Bioinformatic analysis suggested c-kit as a possible target transcript, and c-kit mRNA levels correlate inversely with those of miR-221 and miR-222 in cultured cells. Forced expression of these miRNAs in TF-1 erythroleukemia cells or in erythroid culture of CD34+ cord blood cells slowed cell growth concomitant with reduced c-kit protein levels. Although these results do not address the true functions of the X-linked miR-221/miR-222 cluster in vivo, the authors' approach and the internal consistency of the findings highlight current experimental paradigms to link specific, tissue-restricted miRNAs with candidate mRNA targets. In the future, such studies will need to be extended in 2 complementary ways: targeted disruption of individual miRNA genes and targeted mutation of putative miRNA-binding sites in the 3′-UTRs of candidate targets. While redundancy among miRNA regulators may limit the force of such experiments, a thorough understanding of miRNA functions will only emerge from integrated consideration of complementary results. miRNAs implicated to date in specific hematopoietic functions are summarized in Table 1.
miRNAs implicated to date in hematopoietic functions
miRNA . | Candidate target genes . | Function . | Source . |
|---|---|---|---|
| miR-181a | None identified | Lymphopoiesis | Chen et al15 |
| miR-223 | NFI-A | Granulopoiesis | Fazi et al47 |
| miR-221, miR-222 | c-kit | Erythropoiesis | Felli et al62 |
| miR-130a | MafB | Megakaryopoiesis (assumed) | Garzon et al56 |
| miR-10a | HoxA1 | Megakaryopoiesis (assumed) | Garzon et al56 |
miRNA . | Candidate target genes . | Function . | Source . |
|---|---|---|---|
| miR-181a | None identified | Lymphopoiesis | Chen et al15 |
| miR-223 | NFI-A | Granulopoiesis | Fazi et al47 |
| miR-221, miR-222 | c-kit | Erythropoiesis | Felli et al62 |
| miR-130a | MafB | Megakaryopoiesis (assumed) | Garzon et al56 |
| miR-10a | HoxA1 | Megakaryopoiesis (assumed) | Garzon et al56 |
miRNA expression and functions in leukemia
Naturally, there has been considerable interest in any connection between miRNAs and cancer, including leukemias. The miR-15a/miR-16 locus lies within a region of human chromosome 13 that is also the site of the most common structural abnormality in B-cell chronic lymphocytic leukemia (CLL) and mantle cell lymphoma; both miRNAs are deleted or down-regulated in nearly two-thirds of CLL samples.63 Other miRNA genes also are frequently located at fragile sites, minimal regions of loss of heterozygosity or amplification, and common breakpoint regions previously identified in a range of human malignancies.64 Calin and colleagues65 identified germline or somatic mutations in 5 of 42 sequenced miRNAs in 11 of 75 patients with CLL and no such mutations in 160 subjects without cancer. Whereas these observations provoke speculation about direct miRNA functions in leukemogenesis, other advances center on more general features of miRNA expression in the disease context. The mixed-lineage leukemia (MLL) 1 transcriptional regulator binds to transcription-start sites of innumerable genes, including 30% of all tested miRNA loci,66 which suggests that broad dysregulation of both mRNA and miRNA expression may underlie leukemic phenotypes. Lu and coworkers67 report widespread down-regulation of miRNAs in tumors compared with their normal tissue counterparts, much as others found reduced levels of specific miRNAs in colon cancer specimens.68 In one study, miRNA expression profiles were more successful than mRNA expression in classifying poorly differentiated tumors.67 Furthermore, a unique expression profile consisting of 13 miRNA genes could distinguish cases of CLL with distinct molecular markers and clinical outcomes.65 The broader role of miRNAs in classifying tumors or contributing to disease progression remains to be defined and is likely to be an area of intense investigation.
Concluding remarks
Progress in the area of miRNAs seems to have occurred faster than in any major biological discipline in recent memory. The field has moved from virtual ignorance about an abundant class of regulatory molecules to a reasonably advanced understanding of the mechanisms of miRNA biogenesis and an emerging consensus about the numbers of miRNAs and their targets in several species, including humans. The next series of challenges is already becoming evident: how is transcription of miRNA genes controlled? Do miRNAs operate more commonly through single, critical target transcripts, as the examples reported to date would imply, or do they regulate broad programs of unique gene expression through minor effects on multiple targets, as suggested by evolutionary considerations and some experimental results? What is the larger role of miRNAs in human disease and do they provide novel therapeutic opportunities? As these questions begin to capture investigators' attention, it is worth recalling the seminal contributions of hematologists and their particular models in solving problems in genetics, cell differentiation, and biochemistry. Inas-much as miRNAs may control molecular aspects of cell specialization, hematopoiesis offers an ideal model system in which ideas about their mechanisms and functions can be tested incisively. Meanwhile, the apparent scope of miRNA regulation has disturbed simple and conventional views about the classes of candidate regulators in hematopoiesis and hematologic disorders. No doubt the field will rapidly embrace the new biology and endeavor to understand how and to what degree miRNAs influence different aspects of blood cell renewal and specialization. By the time these problems are solved, we can look forward to other discoveries that might inform us about the functions of the vast majority of the genome that encodes neither proteins nor other known regulatory molecules like miRNAs.
Authorship
The author declares no competing financial interests.
Prepublished online as Blood First Edition Paper, August 1, 2006; DOI 10.1182/blood-2006-01-030015.
I am grateful to Jochen Hartner, David Nathan, and Stuart Orkin for critical review of the manuscript and to Alice Chen for help in preparing figures. As Blood guidelines limit the number of cited references, I regret the highly selective (and sometimes arbitrary) citation process I needed to adopt in writing a succinct review.
This work was supported in part by grants from the National Institutes of Health.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal