Abstract
The 8p11 myeloproliferative syndrome (EMS) is associated with translocations that disrupt the FGFR1 gene. To date, 8 fusion partners of FGFR1 have been identified. However, no primary leukemia cell lines were identified that contain any of these fusions. Here, we screened more than 40 acute myeloid leukemia cell lines for constitutive phosphorylation of STAT5 and applied an immunoaffinity profiling strategy to identify tyrosine-phosphorylated proteins in the KG-1 cell line. Mass spectrometry analysis of KG-1 cells revealed aberrant tyrosine phosphorylation of FGFR1. Subsequent analysis led to the identification of a fusion of the FGFR1OP2 gene to the FGFR1 gene. Small interfering RNA (siRNA) against FGFR1 specifically inhibited the growth and induced apoptosis of KG-1 cells. Thus, the KG-1 cell line provides an in vitro model for the study of FGFR1 fusions associated with leukemia and for the analysis of small molecule inhibitors against FGFR1 fusions.
Introduction
Disruption of the fibroblast growth factor receptor-1 (FGFR1) gene is associated with a disease entity known as the 8p11 myeloproliferative syndrome (EMS)/stem cell leukemia-lymphoma syndrome. It is characterized by myeloid hyperplasia that rapidly transforms to acute myelogenous leukemia and/or lymphoma within a year of original diagnosis.1,2 To date, 8 different translocations have been described, which result in fusion of distinct aminoterminal partner proteins with the cytoplasmic tyrosine kinase domain of FGFR1. These include ZNF198,3 CEP110,4 FOP,5 BCR,6 HERV-K,7 FGFR1OP2,8 TIF1,9 and MYO18A,10 respectively. The resulting fusion proteins have constitutive tyrosine kinase activity and play an essential role in the pathogenesis of EMS.11-13 However, no cell lines have been available for the study of EMS. We screened more than 40 acute myeloid leukemia (AML) cell lines for constitutive phosphorylation of STAT5 and subjected phospho-STAT5–positive cells to phosphotyrosine profiling. This phosphoproteomic analysis identified the KG-1 cell line as an in vitro model for the FGFR1OP2-FGFR1 fusion. Furthermore, this study describes a tyrosinephosphorylation signaling network for FGFR1 fusions, which will benefit targeted signal transduction therapy for EMS.
Materials and methods
Cell culture
The KG-1 cell line was obtained from DSMZ (Braunschweig, Germany). The KG-1a cell line was obtained from American Type Culture Collection (Manassas, VA). All cell lines were grown in RPMI-1640 medium with 10% fetal bovine serum (FBS).
Phosphopeptide immunoprecipitation
Phosphopeptides were prepared using the PhosphoScan Kit (Cell Signaling Technology, Danvers, MA).
Analysis by liquid chromatography–tandem mass spectrometry
Analysis was performed as previously described.14
siRNA and siRNA transfection
FGFR1 SMARTpool siRNA duplexes (proprietary target sequences) and siControl nontargeting SiRNA pool were purchased from Dharmacon Research (Lafayette, CO). Cells were transfected with the siRNAvia electroporation.14 The number of viable cells was determined 48 hours later with the CellTiter 96 AQueous One solution cell proliferation assay (Promega, Madison, WI). The percentage of apoptotic cells at 48 hours was determined by flow cytometric analysis of cleaved caspase-3 (Cell Signaling Technology).
RACE
The RNeasy Mini Kit (Qiagen, Valencia, CA) was used to extract RNA from human leukemia cell lines. Rapid amplification of cDNA ends (RACE) was performed with the use of the 5′ RACE system (Invitrogen, Carlsbad, CA) with primers FGFR1-P1 (5′-ATGGACAGGTCCAGGTACTCC) for cDNA synthesis and FGFR1-P2 (5′-ACTCTGGTGGGTGTAGATCCG) and FGFR1-P3 (5′-CTTGGAGGCATACTCCACGAT) for a nested polymerase chain reaction (PCR).
RT-PCR assay
For reverse transcriptase (RT)–PCR, first-strand cDNA was synthesized from 2.5 μg total RNA with the use of SuperScript III first-strand synthesis system (Invitrogen) with oligo (dT)20. Then, the FGFR1OP2-FGFR1 fusion gene was amplified with the use of primer pairs OP2-F3 (5′-ATCAGTCGGCCTTGGAACTTA) and FGFR1-R9 (5′-AGAAGAACCCCAGAGTTCATG). The reciprocal fusion was detected with the use of primer pairs FGFR1-F7 (5′-AACTCTATCGGACTCTCCCAT) and OP2-R7 (5′-GTACTTTCCGACGCATCATCT). Wild-type FGFR1OP2 and FGFR1 were amplified with the use of primer pairs OP2-F3 and OP2-R580 (5′-TTGTTCCTTGCAACCCTGTTGCTC), and FGFR1-F7 and FGFR1-R9, respectively.
Western blotting
Cells were lysed in 1 × cell-lysis buffer (Cell Signaling Technology) supplemented with Protease Arrest (G Biosciences, St Louis, MO) and separated by electrophoresis. All antibodies and reagents for immunoblotting were from Cell Signaling Technology.
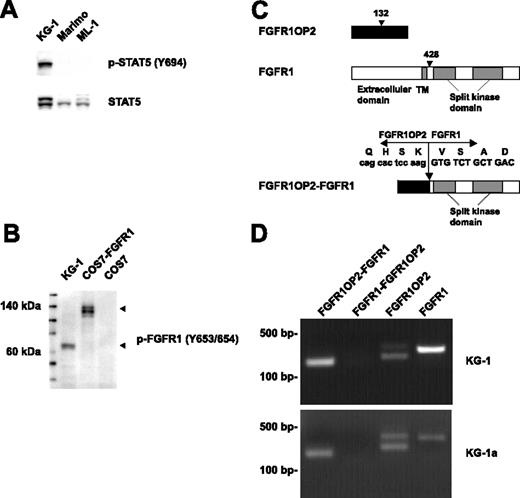
Fusion of FGFR1OP2 to FGFR1 in the KG-1 cell line. (A) Western blot analysis of whole-cell lysates revealed constitutive tyrosine phosphorylation of STAT5 in the KG-1 cell line. Detection of phospho-STAT5 is shown in the top panel, and total STAT5 is shown below. (B) Detection of a truncated form of activated FGFR1 in KG-1 cells by Western blot. COS7 cells transfected with full-length FGFR1 were used as a control. (C) Schematic representation of FGFR1OP2, FGFR1, and FGFR1OP2-FGFR1 proteins. Arrowhead indicates the position of the breakpoint. (D) Detection of FGFR1OP2-FGFR1, FGFR1OP2, and FGFR1 transcripts in the KG-1 and KG-1a cell lines.
Fusion of FGFR1OP2 to FGFR1 in the KG-1 cell line. (A) Western blot analysis of whole-cell lysates revealed constitutive tyrosine phosphorylation of STAT5 in the KG-1 cell line. Detection of phospho-STAT5 is shown in the top panel, and total STAT5 is shown below. (B) Detection of a truncated form of activated FGFR1 in KG-1 cells by Western blot. COS7 cells transfected with full-length FGFR1 were used as a control. (C) Schematic representation of FGFR1OP2, FGFR1, and FGFR1OP2-FGFR1 proteins. Arrowhead indicates the position of the breakpoint. (D) Detection of FGFR1OP2-FGFR1, FGFR1OP2, and FGFR1 transcripts in the KG-1 and KG-1a cell lines.
Results and discussion
Activated tyrosine kinases are frequently involved in various malignancies, including AML. They phosphorylate and activate STAT5, which plays an important role in leukemogenesis.15 We screened more than 40 AML cell lines for constitutive phosphorylation of STAT5 by Western blot. The KG-1 cell line, derived from a patient with AML,16 has constitutive tyrosine phosphorylation of STAT5 (Figure 1A). To identify protein tyrosine kinases responsible for the constitutive phosphorylation of STAT5 in KG-1, cell lysates were trypsin digested, and phosphopeptides were immunoprecipitated with phosphotyrosine antibody (pY-100), and analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS).17 LC-MS/MS identified 450 phosphotyrosine sites in 304 proteins (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). Among these proteins, more than 15 tyrosine kinases were tyrosine phosphorylated (Table 1), including FGFR1, which was phosphorylated at multiple sites, including Y653/654 within the activation loop. Based on this data, FGFR1 could be aberrantly activated in KG-1 cells. Western blot analysis confirmed the robust phosphorylation of FGFR1 at tyrosines 653/654 (Figure 1B). Two forms of FGFR1 have been described (a 120-kDa immature form, and a 145-kDa glycosylated form). Interestingly, we observed a 60-kDa truncated form of FGFR1. To determine whether a chimeric FGFR1 transcript was present, we performed 5′ RACE on the sequence encoding the kinase domain of FGFR1. Sequence analysis of the resultant product revealed that the kinase domain of FGFR1 was fused to FGFR1OP2 gene (Figure 1C). In agreement with a previous observation in a patient diagnosed with FGFR1OP2-FGFR1 EMS,8 the KG-1 cell line expressed an in-frame FGFR1OP2-FGFR1 fusion transcript (Figure 1D), with the fusion of exon 4 of FGFR1OP2 to exon 9 of FGFR1. No reciprocal FGFR1-FGFR1OP2 fusion gene was detected. The KG-1 cell line did express both wild-type FGFR1OP2 and wild-type FGFR1. Similar results were observed in KG-1a, a less differentiated variant of KG-1 (Figure 1D).18 FGFR1OP2 is a protein of unknown function. It is expressed in several different tissues, including thymus, spleen, and bone marrow. Its 2 potential coil-coiled domains were retained in FGFR1OP2-FGFR1.8 It is noteworthy that FGFR1OP2 was phosphorylated at a novel site, tyrosine 46 (Table S1). Whether this tyrosine residue is important for the oncogenic properties of the FGFR1OP2-FGFR1 fusion remains to be determined.
Phosphorylated tyrosine kinases identified by mass spectrometry analysis
Protein name . | NCBI accession no. . | NCBI site . |
|---|---|---|
| Protein kinase, tyrosine (nonreceptor) | ||
| Abl iso1 | NP_005148 | 393* |
| Arg | NP_005149 | 403 |
| Ack | NP_005772 | 859* |
| Ack | NP_005772 | 518* |
| Btk | NP_038510 | 344 |
| Btk | NP_038510 | 223* |
| FAK | NP_005598 | 598* |
| Fyn | NP_002028 | 420* |
| Src | NP_005408 | 419* |
| Yes | NP_005424 | 426* |
| Hck | NP_002101 | 411* |
| Lyn | NP_002341 | 397* |
| Jak2 | NP_004963 | 570* |
| Jak3 | NP_000206 | 785* |
| Lck | NP_005347 | 192* |
| Lck | NP_005347 | 394* |
| Lck | NP_005347 | 505* |
| Lyn | NP_002341 | 193* |
| Lyn | NP_002341 | 306 |
| Lyn | NP_002341 | 473* |
| Lyn | NP_002341 | 265 |
| Lyn | NP_002341 | 316 |
| Lyn | NP_002341 | 508* |
| Pyk2 | NP_004094 | 579* |
| Syk | NP_003168 | 323* |
| Syk | NP_003168 | 352* |
| Tyk2 | NP_003322 | 292* |
| Protein kinase, tyrosine (receptor) | ||
| FGFR1 | NP_000595 | 585* |
| FGFR1 | NP_000595 | 463* |
| FGFR1 | NP_000595 | 583* |
| FGFR1 | NP_000595 | 653* |
| FGFR1 | NP_000595 | 654* |
| Mer | NP_006334 | 749* |
| Mer | NP_006334 | 753* |
| Tyro3 | NP_006284 | 681 |
| Tyro3 | NP_006284 | 685 |
Protein name . | NCBI accession no. . | NCBI site . |
|---|---|---|
| Protein kinase, tyrosine (nonreceptor) | ||
| Abl iso1 | NP_005148 | 393* |
| Arg | NP_005149 | 403 |
| Ack | NP_005772 | 859* |
| Ack | NP_005772 | 518* |
| Btk | NP_038510 | 344 |
| Btk | NP_038510 | 223* |
| FAK | NP_005598 | 598* |
| Fyn | NP_002028 | 420* |
| Src | NP_005408 | 419* |
| Yes | NP_005424 | 426* |
| Hck | NP_002101 | 411* |
| Lyn | NP_002341 | 397* |
| Jak2 | NP_004963 | 570* |
| Jak3 | NP_000206 | 785* |
| Lck | NP_005347 | 192* |
| Lck | NP_005347 | 394* |
| Lck | NP_005347 | 505* |
| Lyn | NP_002341 | 193* |
| Lyn | NP_002341 | 306 |
| Lyn | NP_002341 | 473* |
| Lyn | NP_002341 | 265 |
| Lyn | NP_002341 | 316 |
| Lyn | NP_002341 | 508* |
| Pyk2 | NP_004094 | 579* |
| Syk | NP_003168 | 323* |
| Syk | NP_003168 | 352* |
| Tyk2 | NP_003322 | 292* |
| Protein kinase, tyrosine (receptor) | ||
| FGFR1 | NP_000595 | 585* |
| FGFR1 | NP_000595 | 463* |
| FGFR1 | NP_000595 | 583* |
| FGFR1 | NP_000595 | 653* |
| FGFR1 | NP_000595 | 654* |
| Mer | NP_006334 | 749* |
| Mer | NP_006334 | 753* |
| Tyro3 | NP_006284 | 681 |
| Tyro3 | NP_006284 | 685 |
NCBI indicates National Center for Biotechnology Information.
Published tyrosine phosphorylation sites.
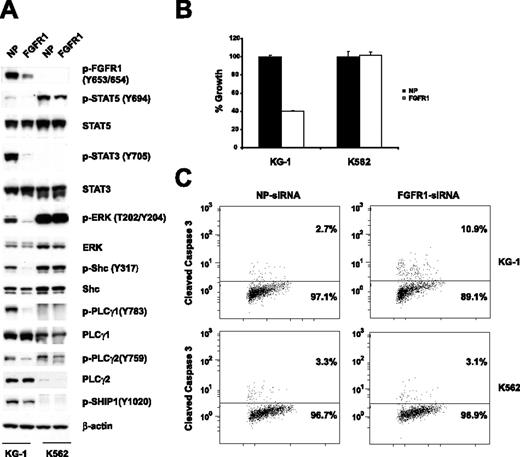
To find out whether the FGFR1OP2-FGFR1 fusion contributes to the growth and viability of KG-1 cells, expression of the FGFR1 fusion was down-regulated with FGFR1 siRNA. Western blot analysis revealed that expression of the FGFR1 fusion was specifically and significantly reduced 48 hours following transfection of FGFR1 siRNA. This is accompanied by a decrease in the phosphorylation of STAT5, STAT3, ERK, Shc, PLCγ1, and PLCγ2 (Figure 2A). Down-regulation of FGFR1 resulted in inhibition of cell growth (Figure 2B). Moreover, treatment with FGFR1 siRNA resulted in increased apoptosis as measured by a cleaved caspase-3 flow cytometric assay (Figure 2C). Thus, our results suggest that the FGFR1OP2-FGFR1 fusion is essential for the growth and proliferation of KG-1 cells. In conclusion, we identified KG-1 as the first cell line expressing the FGFR1 fusion gene. This makes KG-1 a valuable in vitro model for screening FGFR1 inhibitors in a human cell line expressing the fusion protein from its endogenous promoter. The KG-1 cell line provides a unique model for studying FGFR1 fusion–mediated signaling pathways. Indeed, this phosphoproteomic analysis provides the most comprehensive tyrosine-phosphorylation signaling profile reported for FGFR1 fusions to date. In our study, we observed tyrosine phosphorylation of known downstream effectors of FGFR1 (Figure 2A).19,20 Notably, PLCγ1 plays an important role in FGFR1 fusion–mediated EMS in vivo.12 Here, we confirmed the tyrosine phosphorylation of PLCγ1 at multiple sites, including several novel sites (Y379, Y481, Y506, and Y977). Moreover, we identified many novel tyrosine-phosphorylated proteins that could be regulated by FGFR1 fusions. SHIP1 is a hematopoietic-specific phosphatase that regulates cell survival, growth, cell-cycle arrest, and apoptosis.21 We show that phosphorylation of SHIP1 at tyrosine 1020 is inhibited by siRNA against FGFR1 (Figure 2A), suggesting that SHIP1 could be a novel downstream target of the FGFR1 fusion. This information may be useful for developing novel therapeutic approaches for the effective treatment of AML containing FGFR1 fusions, as well as other human disorders associated with deregulated FGFR1 activity.22,23
Effects of FGFR1 siRNA on the KG-1 cell line. (A) siRNA-induced silencing of FGFR1 was assessed by immunoblots of whole-cell lysates 48 hours after siRNA transfection. Down-regulation of the FGFR1OP2-FGFR1 fusion resulted in decreased phosphorylation of STAT5, STAT3, ERK, Shc, PLC-γ1, PLCγ2, and SHIP1. NP and FGFR1 indicate siControl nontargeting siRNA pool and FGFR1 SMARTpool siRNA, respectively. (B) Electroporation with FGFR1 siRNA into KG-1 cells resulted in a decrease of proliferation. Mean value plus SD of experiments performed in duplicate is represented. (C) Induction of apoptosis. Percentage of total population is indicated.
Effects of FGFR1 siRNA on the KG-1 cell line. (A) siRNA-induced silencing of FGFR1 was assessed by immunoblots of whole-cell lysates 48 hours after siRNA transfection. Down-regulation of the FGFR1OP2-FGFR1 fusion resulted in decreased phosphorylation of STAT5, STAT3, ERK, Shc, PLC-γ1, PLCγ2, and SHIP1. NP and FGFR1 indicate siControl nontargeting siRNA pool and FGFR1 SMARTpool siRNA, respectively. (B) Electroporation with FGFR1 siRNA into KG-1 cells resulted in a decrease of proliferation. Mean value plus SD of experiments performed in duplicate is represented. (C) Induction of apoptosis. Percentage of total population is indicated.
Authorship
Contributions: T.-L.G. designed research, performed research, and wrote the paper; V.L.G., C.R., L.P., J.N., J.M., and Y.W. performed research; D.K.W. and B.J.D. provided vital reagents; J.R. and M.J.C. designed research; and R.D.P. designed research and revised the paper.
Conflict-of-interest disclosure: Several of the authors (T.G., V.L.G., C.R., L.P., J.N., J.M., Y.W., J.R., M.J.C., and R.D.P.) are employed by a company (Cell Signaling Technology, Inc.) whose products were studied in the present work.
Prepublished online as Blood First Edition Paper, August 31, 2006; DOI 10.1182/blood-2006-06-026666.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Wendy Colpoys and Albrecht Moritz for providing research reagents. We thank Jessica Cherry for proofreading the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal