Patients with acute myeloid leukemia (AML) and normal karyotype are classified in an intermediate-risk group, albeit this subset is heterogeneous for clinical outcome. A recent complementary DNA microarray study identified a gene-expression signature that—when used to cluster normal karyotype patients—separated them into 2 prognostically relevant subgroups. We sought the first independent validation of the prognostic value of this signature. Using oligonucleotide microarrays to measure gene expression in samples from uniformly treated adults with karyotypically normal AML, we performed cluster analysis based on the previously identified signature. We also developed a well-defined classification rule using the signature to predict outcome for individual patients. Cluster analysis confirmed the prognostic utility of the signature: patient clusters differed in overall (P = .001) and disease-free (P = .001) survival. The signature-based classifier identified groups with differences in overall (P = .02) and disease-free (P = .05) survival. A strong association of the outcome classifier with the prognostically adverse FLT3 internal tandem duplication (FLT3 ITD) potentially explained the prognostic significance of the signature. However, in the subgroup of patients without FLT3 ITD there was a moderate difference in survival for the classifier-derived groups. Our analysis confirms the applicability of the gene-expression profiling strategy for outcome prediction in cytogenetically normal AML.
Introduction
Cytogenetic abnormalities detected at diagnosis are among the most important factors predicting clinical outcome in acute myeloid leukemia (AML).1 However, approximately 45% of adult de novo AML patients present with a normal karyotype.1-4 Importantly, patients in this cytogenetic subgroup are not homogeneous with respect to clinical outcome due to a high degree of molecular heterogeneity.5 Several groups have reported subsets of cytogenetically normal AML with different outcomes based on the presence of mutations (eg, MLL,6,7 FLT3,8-14 CEBPA,13,15 and NPM116-18 ) and/or overexpression (eg, BAALC13,14,19 and ERG20 ) of distinct genes involved in tissue homeostatic regulatory pathways, and it is likely that other aberrantly expressed genes (eg, EVI121 ) will be added to this list in the future. Because these biomarkers are often not mutually exclusive, the prognostic weight of each is affected by the concurrent presence of the others.13,14,16,20 Thus, rather than performing multiple assays testing for a restricted number of currently known prognostic biomarkers, gene-expression profiling was explored as a strategy to capture a global view of the molecular heterogeneity of cytogenetically normal AML.22,23 The goal of these studies has been to recognize characteristic patterns of gene activation and silencing, the so-called “expression signatures,” that can identify subsets of patients with different outcomes. Although intriguing, the hitherto reported results remain preliminary and require independent validation.24-26
Recently, Bullinger et al22 reported for the first time a complementary DNA (cDNA) microarray-based expression signature that separated AML patients with a normal karyotype into 2 cluster-derived prognostically relevant subgroups. In the current study, we applied this signature to the gene-expression profiles of a different group of AML patients with a normal karyotype. The gene-expression profiles investigated in this study were generated with oligonucleotide arrays. We were able to confirm the prognostic significance of the signature in AML patients with a normal karyotype, even with the use of a different microarray platform.
Materials and methods
Samples
Sixty-four adult patients younger than 60 years of age, with primary AML and normal cytogenetics at diagnosis, treated on Cancer and Leukemia Group B (CALGB) 962127 were included in our analysis. Pretreatment cytogenetic analyses of marrow (BM) were performed by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion,28 and were centrally reviewed, as previously reported.2 All patients signed IRB approved consent for the treatment study CALGB 9621, the cytogenetic study CALGB 8461 and the clinical sample collection CALGB 9665. To be considered cytogenetically normal, at least 20 metaphase cells had to be analyzed and the karyotype found to be normal in each case. Patient samples were also centrally reviewed for confirmation of the diagnosis of AML and French-American-British (FAB) morphologic classification. The presence or absence of the MLL partial tandem duplication, FLT3 internal tandem duplication (ITD), mutations in the CEBPA and NPM1 genes, and BAALC levels were also determined centrally in pretreatment samples as described previously.6,12,15,16,19 Patients were considered to be FLT3 ITD positive if the mutation was detectable by genescan analysis, regardless of the FLT3 ITD/wild-type allele ratio.12
Treatment on CALGB 9621 consisted of induction chemotherapy with cytarabine, daunorubicin, and etoposide with (ADEP) or without (ADE) the multidrug resistance protein modulator, PSC-833, intensification with autologous peripheral stem cell transplantation or with an alternative regimen, and maintenance with interleukin-2 as previously detailed.27 The clinical endpoints considered for this study (ie, complete remission, overall survival, and disease-free survival) have been previously defined.20
Gene-expression profiling
Pretreatment blood samples were analyzed using Affymetrix U133 plus 2.0 GeneChips (Affymetrix, Santa Clara, CA). Total RNA extraction, double-stranded cDNA preparation, and biotinylated RNA in vitro transcription, labeling, and hybridization to the U133 plus 2.0 GeneChip were previously described.20 Scanned images were converted to cell intensity (CEL) files using GeneChip Operating Software (Affymetrix). Invariant set normalization and model-based expression index (MBEI) computation were performed using dChip version 1.3 (Harvard University, Cambridge, MA). The log2 MBEIs were exported to BRB-ArrayTools v3.2.3 (National Cancer Institute, Bethesda, MD) for further analysis.
The expression signature identified by Bullinger et al22 using cDNA arrays manufactured by the Stanford Functional Genomics Facility consisted of 133 genes represented by 149 cDNAs (see their supplementary table 6 at www.ncbi.nlm.nih.gov/geo/; accession no. GSE425). We found 101 named genes from this signature on the Affymetrix U133 plus 2.0 GeneChip, represented by 256 probe sets. Of the 256 probe sets, 157 were expressed (ie, received an Affymetrix “Present” call) in 25% or more of our patient specimens, resulting in 81 of the 101 genes being represented among these 157 probe sets (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The log2 MBEI values of these 157 probe sets comprised the prognostic molecular signature we used for validation purposes (hereafter referred to as the “Bullinger validation signature”).
Statistical analysis
Hierarchical cluster analysis29 was performed on patient specimens with respect to the Bullinger validation signature. Expression values for each probe set were median-centered across patients prior to clustering. Average linkage was used and the dissimilarity measure was one minus correlation. We cut the resulting dendrogram at a height that created 2 clusters of specimens and tested for a difference in survival between the clusters using a permutation method based on the log-rank test (Document S1, available at the Blood website; see the Supplemental Materials link at the top of the online article).
We investigated the ability of the Bullinger validation signature to predict poor or good outcome for individual patients using a class prediction algorithm, compound covariate prediction (CCP).30 We used the Bullinger validation signature to compute a compound covariate (a linear combination of the log-expression values for the 157 probe sets) for each specimen in our data set. The weight of each probe set in the linear combination was taken to be the 2-sample t statistic of the corresponding gene from Bullinger et al's gene-expression data22 comparing the cluster-based poor and good outcome groups within the subset of normal karyotype patients (data not shown).
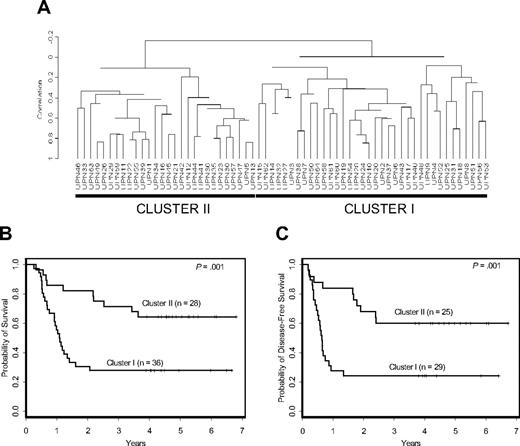
Prognostic significance of signature-based clusters in cytogenetically normal AML. (A) The Bullinger validation signature dichotomized uniformly treated CALGB patients with cytogenetically normal AML into 2 distinct clusters. Patients included in cluster I had a worse overall (B) and disease-free (C) survival compared with those in cluster II.
Prognostic significance of signature-based clusters in cytogenetically normal AML. (A) The Bullinger validation signature dichotomized uniformly treated CALGB patients with cytogenetically normal AML into 2 distinct clusters. Patients included in cluster I had a worse overall (B) and disease-free (C) survival compared with those in cluster II.
For overall survival, patients were dichotomized into poor and good outcome classes based on whether they died or were alive at last follow-up, respectively. This dichotomization is fitting since all currently living patients have been followed for at least 4.6 years and all patients who died did so within the first 3.6 years. A similar dichotomization was used for disease-free survival as all patients who relapsed (the poor outcome class) did so within 2.4 years and all patients currently in continuous complete remission (the good outcome class) have a follow-up time of at least 4.5 years. CCP was then performed using a leave-one-out cross-validation. Of note, since genes and weights were completely specified by the previous gene-expression study,22 the only step that was cross-validated in this analysis was the computation of the classification threshold. The patient to be predicted was removed from the analysis, the mean compound covariate value for each class (ie, poor and good outcome classes) among the remaining patients was computed and the classification threshold was defined as the midpoint of the means of these 2 classes. The outcome group for the left-out patient was predicted by comparing its compound covariate value to the classification threshold. We measured overall prediction accuracy and compared survival curves for the predicted outcome classes using a permutation method based on the log-rank test (Document S1).
Fisher 2-sided exact or Wilcoxon rank-sum tests were used to measure the association between predicted outcome groups and categorical or continuous pretreatment clinical features, respectively. Proportional hazards models were fit for overall and disease-free survival using the predicted outcome group and FLT3 ITD status variables, both with and without their interaction term. The proportional hazards assumption was checked. All analyses were performed by the CALGB Statistical Center.
Results
Prognostic significance of the Bullinger validation signature in AML with a normal karyotype
Patients were initially clustered with respect to expression of the Bullinger validation signature. The resulting dendrogram contained 2 distinct clusters of specimens: cluster I, corresponding to the poor-outcome group, and cluster II, corresponding to the good-outcome group previously identified22 (Figure 1A). Cluster I patients had inferior overall survival (P = .001; Figure 1B) and disease-free survival (P = .001; Figure 1C), with estimated 5-year overall and disease-free survival rates of 28% and 24% compared with 64% and 60%, respectively, for cluster II patients.
Outcome prediction by a classifier based on the Bullinger validation signature
Validation of a signature must move beyond assessing whether the same genes are prognostic in a subsequent study; a well-defined classifier needs to be developed that can be applied to individual patients in future studies for assessment of the signature's predictive value and clinical relevance.31 Indeed, studies of potentially important gene-expression signatures for breast cancer32,33 and lymphoma34 followed this approach. Bullinger et al22 also attempted this but did not have success when focusing on patients with a normal karyotype. Thus, we explored here the ability of CCP30 to create such a classifier based on the Bullinger validation signature using our larger group of patients.
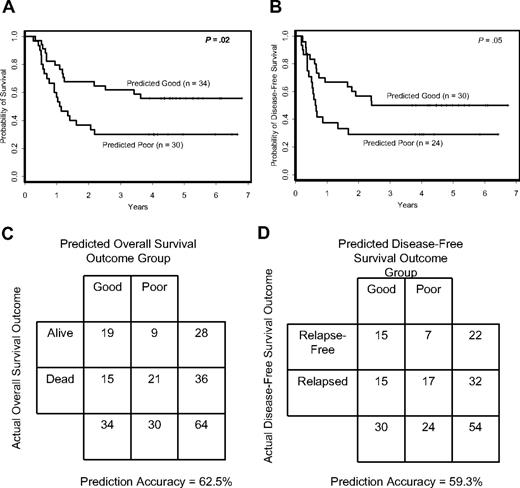
For overall survival, CCP predicted 30 patients to have poor outcome and 34 patients to have good outcome. With regard to pretreatment features (Table 1), patients in the predicted poor-outcome class for overall survival had higher incidence of the FLT3 ITD (P < .001), a different distribution of FAB subtypes (P = .02) and less extramedullary involvement (P = .02) than the predicted good-outcome class. The difference in overall survival between the 2 classes was significant (P = .02; Figure 2A). Patients in the predicted good-outcome class had an estimated 5-year survival rate of 56% compared with only 30% for the patients in the predicted poor-outcome class. The overall prediction accuracy for outcome class was 62.5% (Figure 2C).
Baseline characteristics of predicted good- and poor-outcome groups for overall survival
Characteristic . | Predicted good outcome, n = 34 . | Predicted poor outcome, n = 30 . | P . |
|---|---|---|---|
| Age, y, median (range) | 46 (21-59) | 44 (21-59) | .99 |
| Male sex, no. (%) | 19 (56) | 16 (53) | 1.00 |
| Race, no. (%) | .77 | ||
| White | 29 (85) | 27 (90) | |
| Hispanic | 3 (9) | 1 (3) | |
| African American | 1 (3) | 1 (3) | |
| Oriental | 0 (0) | 1 (3) | |
| Other | 1 (3) | 0 (0) | |
| Hemoglobin, g/L, median (range)* | 95 (68-121) | 86 (60-129) | .28 |
| Platelet count, × 109/L, median (range) | 48 (16-378) | 54 (12-250) | .89 |
| WBC count, × 109/L, median (range) | 37.7 (2.1-146.0) | 36.2 (3.7-295.0) | .37 |
| Percentage of PB blasts, median (range) | 62 (2-97) | 71 (24-95) | .09 |
| Percentage of BM blasts, median (range) | 61.5 (30-88) | 71.5 (37-90) | .16 |
| FAB, no. (%)† | .02 | ||
| M0 | 0 (0) | 1 (4) | |
| M1 | 8 (24) | 8 (29) | |
| M2 | 6 (18) | 7 (25) | |
| M4 | 10 (30) | 12 (43) | |
| M5 | 9 (27) | 0 (0) | |
| Extramedullary involvement, no. (%) | 19 (56) | 8 (27) | .02 |
| CNS | 0 (0) | 0 (0) | NA |
| Hepatomegaly | 3 (9) | 1 (3) | .61 |
| Splenomegaly | 3 (9) | 1 (3) | .61 |
| Lymphadenopathy | 6 (18) | 3 (10) | .48 |
| Skin infiltrates | 7 (21) | 2 (7) | .16 |
| Gingival hypertrophy | 10 (29) | 3 (10) | .07 |
| FLT3 status, no. (%)‡ | < .001 | ||
| Wild-type | 27 (82) | 10 (33) | |
| Internal tandem duplication | 6 (18) | 20 (67) | |
| MLL PTD positive, no. (%) | 1 (3) | 3 (10) | .33 |
| BAALC level, no. (%)§ | .58 | ||
| Low | 14 (58) | 13 (48) | |
| High | 10 (42) | 14 (52) | |
| CEBPA status, no. (%)¶ | .15 | ||
| Not mutated | 26 (79) | 28 (93) | |
| Mutated | 7 (21) | 2 (7) | |
| NPM1 status, no. (%)|| | .17 | ||
| Not mutated | 12 (36) | 6 (20) | |
| Mutated | 21 (64) | 24 (80) |
Characteristic . | Predicted good outcome, n = 34 . | Predicted poor outcome, n = 30 . | P . |
|---|---|---|---|
| Age, y, median (range) | 46 (21-59) | 44 (21-59) | .99 |
| Male sex, no. (%) | 19 (56) | 16 (53) | 1.00 |
| Race, no. (%) | .77 | ||
| White | 29 (85) | 27 (90) | |
| Hispanic | 3 (9) | 1 (3) | |
| African American | 1 (3) | 1 (3) | |
| Oriental | 0 (0) | 1 (3) | |
| Other | 1 (3) | 0 (0) | |
| Hemoglobin, g/L, median (range)* | 95 (68-121) | 86 (60-129) | .28 |
| Platelet count, × 109/L, median (range) | 48 (16-378) | 54 (12-250) | .89 |
| WBC count, × 109/L, median (range) | 37.7 (2.1-146.0) | 36.2 (3.7-295.0) | .37 |
| Percentage of PB blasts, median (range) | 62 (2-97) | 71 (24-95) | .09 |
| Percentage of BM blasts, median (range) | 61.5 (30-88) | 71.5 (37-90) | .16 |
| FAB, no. (%)† | .02 | ||
| M0 | 0 (0) | 1 (4) | |
| M1 | 8 (24) | 8 (29) | |
| M2 | 6 (18) | 7 (25) | |
| M4 | 10 (30) | 12 (43) | |
| M5 | 9 (27) | 0 (0) | |
| Extramedullary involvement, no. (%) | 19 (56) | 8 (27) | .02 |
| CNS | 0 (0) | 0 (0) | NA |
| Hepatomegaly | 3 (9) | 1 (3) | .61 |
| Splenomegaly | 3 (9) | 1 (3) | .61 |
| Lymphadenopathy | 6 (18) | 3 (10) | .48 |
| Skin infiltrates | 7 (21) | 2 (7) | .16 |
| Gingival hypertrophy | 10 (29) | 3 (10) | .07 |
| FLT3 status, no. (%)‡ | < .001 | ||
| Wild-type | 27 (82) | 10 (33) | |
| Internal tandem duplication | 6 (18) | 20 (67) | |
| MLL PTD positive, no. (%) | 1 (3) | 3 (10) | .33 |
| BAALC level, no. (%)§ | .58 | ||
| Low | 14 (58) | 13 (48) | |
| High | 10 (42) | 14 (52) | |
| CEBPA status, no. (%)¶ | .15 | ||
| Not mutated | 26 (79) | 28 (93) | |
| Mutated | 7 (21) | 2 (7) | |
| NPM1 status, no. (%)|| | .17 | ||
| Not mutated | 12 (36) | 6 (20) | |
| Mutated | 21 (64) | 24 (80) |
WBC indicates white blood cell; PB, peripheral blood; BM, bone marrow; FAB, French-American-British classification; CNS, central nervous system; MLL PTD, partial tandem duplication of the MLL gene; NA, not applicable
For 1 patient with predicted poor outcome, hemoglobin level was unknown
For 1 patient with predicted good outcome, and 2 patients with predicted poor outcome, FAB classification was unknown
For 1 patient with predicted good outcome, FLT3 status was unknown
For 10 patients with predicted good outcome, and 3 patients with predicted poor outcome, BAALC level was unknown
For 1 patient with predicted good outcome, CEBPA status was unknown
For 1 patient with predicted good outcome, NPM1 status was unknown
For disease-free survival, 24 patients were predicted to have poor outcome and 30 patients good outcome. Differences in pretreatment features between the 2 disease-free survival predicted outcome classes were similar to those identified for overall survival (Table 2). There was a significant difference in disease-free survival between CCP predicted outcome classes (P = .05; Figure 2B), with estimated 5-year disease-free survival rates of 50% and 29% for the predicted good- and poor-outcome classes, respectively. The overall prediction accuracy for outcome class was 59.3% (Figure 2D).
Baseline characteristics of predicted good- and poor-outcome groups for disease-free survival
Characteristic . | Predicted good outcome, n = 30 . | Predicted poor outcome, n = 24 . | P . |
|---|---|---|---|
| Age, y, median (range) | 45 (24-59) | 41 (21-59) | .55 |
| Male sex, no. (%) | 16 (53) | 12 (50) | 1.00 |
| Race, no. (%) | .80 | ||
| White | 25 (83) | 21 (88) | |
| Hispanic | 3 (10) | 1 (4) | |
| African American | 1 (3) | 1 (4) | |
| Oriental | 0 (0) | 1 (4) | |
| Other | 1 (3) | 0 (0) | |
| Hemoglobin, g/L, median (range)* | 97 (68-121) | 84 (60-129) | .15 |
| Platelet count, × 109/L, median (range) | 46 (16-311) | 55 (12-250) | .24 |
| WBC count, × 109/L, median (range) | 37.7 (8.0-146.0) | 32.9 (9.4-295.0) | .53 |
| Percentage of PB blasts, median (range) | 69.5 (2-97) | 70 (24-95) | .30 |
| Percentage of BM blasts, median (range) | 63 (32-88) | 71.5 (37-90) | .27 |
| FAB, no. (%)† | .04 | ||
| M0 | 0 (0) | 1 (5) | |
| M1 | 8 (28) | 7 (32) | |
| M2 | 5 (17) | 4 (18) | |
| M4 | 8 (28) | 10 (45) | |
| M5 | 8 (28) | 0 (0) | |
| Extramedullary involvement, no. (%) | 17 (57) | 6 (25) | .03 |
| CNS | 0 (0) | 0 (0) | NA |
| Hepatomegaly | 2 (7) | 0 (0) | .49 |
| Splenomegaly | 2 (7) | 0 (0) | .49 |
| Lymphadenopathy | 5 (17) | 3 (13) | .72 |
| Skin infiltrates | 6 (20) | 1 (4) | .12 |
| Gingival hypertrophy | 9 (30) | 3 (13) | .19 |
| FLT3 status, no. (%) | < .001 | ||
| Wild-type | 25 (83) | 8 (33) | |
| Internal tandem duplication | 5 (17) | 16 (67) | |
| MLL PTD positive, no. (%) | 0 (0) | 3 (13) | .08 |
| BAALC level, no. (%)‡ | .76 | ||
| Low | 13 (62) | 11 (55) | |
| High | 8 (38) | 9 (45) | |
| CEBPA status, no. (%) | .27 | ||
| Not mutated | 23 (77) | 22 (92) | |
| Mutated | 7 (23) | 2 (8) | |
| NPM1 status, no. (%) | .13 | ||
| Not mutated | 11 (37) | 4 (17) | |
| Mutated | 19 (63) | 20 (83) |
Characteristic . | Predicted good outcome, n = 30 . | Predicted poor outcome, n = 24 . | P . |
|---|---|---|---|
| Age, y, median (range) | 45 (24-59) | 41 (21-59) | .55 |
| Male sex, no. (%) | 16 (53) | 12 (50) | 1.00 |
| Race, no. (%) | .80 | ||
| White | 25 (83) | 21 (88) | |
| Hispanic | 3 (10) | 1 (4) | |
| African American | 1 (3) | 1 (4) | |
| Oriental | 0 (0) | 1 (4) | |
| Other | 1 (3) | 0 (0) | |
| Hemoglobin, g/L, median (range)* | 97 (68-121) | 84 (60-129) | .15 |
| Platelet count, × 109/L, median (range) | 46 (16-311) | 55 (12-250) | .24 |
| WBC count, × 109/L, median (range) | 37.7 (8.0-146.0) | 32.9 (9.4-295.0) | .53 |
| Percentage of PB blasts, median (range) | 69.5 (2-97) | 70 (24-95) | .30 |
| Percentage of BM blasts, median (range) | 63 (32-88) | 71.5 (37-90) | .27 |
| FAB, no. (%)† | .04 | ||
| M0 | 0 (0) | 1 (5) | |
| M1 | 8 (28) | 7 (32) | |
| M2 | 5 (17) | 4 (18) | |
| M4 | 8 (28) | 10 (45) | |
| M5 | 8 (28) | 0 (0) | |
| Extramedullary involvement, no. (%) | 17 (57) | 6 (25) | .03 |
| CNS | 0 (0) | 0 (0) | NA |
| Hepatomegaly | 2 (7) | 0 (0) | .49 |
| Splenomegaly | 2 (7) | 0 (0) | .49 |
| Lymphadenopathy | 5 (17) | 3 (13) | .72 |
| Skin infiltrates | 6 (20) | 1 (4) | .12 |
| Gingival hypertrophy | 9 (30) | 3 (13) | .19 |
| FLT3 status, no. (%) | < .001 | ||
| Wild-type | 25 (83) | 8 (33) | |
| Internal tandem duplication | 5 (17) | 16 (67) | |
| MLL PTD positive, no. (%) | 0 (0) | 3 (13) | .08 |
| BAALC level, no. (%)‡ | .76 | ||
| Low | 13 (62) | 11 (55) | |
| High | 8 (38) | 9 (45) | |
| CEBPA status, no. (%) | .27 | ||
| Not mutated | 23 (77) | 22 (92) | |
| Mutated | 7 (23) | 2 (8) | |
| NPM1 status, no. (%) | .13 | ||
| Not mutated | 11 (37) | 4 (17) | |
| Mutated | 19 (63) | 20 (83) |
WBC, white blood cell; PB, peripheral blood; BM, bone marrow; FAB, French-American-British classification; CNS, central nervous system; NA, not applicable; MLL PTD, partial tandem duplication of the MLL gene; NA, not applicable
For 1 patient with predicted poor outcome, hemoglobin level was unknown
For 1 patient with predicted good outcome, and 2 patients with predicted poor outcome, FAB classification was unknown
For 9 patients with predicted good outcome, and 4 patients with predicted poor outcome, BAALC level was unknown
A strong association was observed between the signature-based outcome classifications and FLT3 ITD status. Of the patients constituting the predicted good-outcome class, fewer than 20% were FLT3 ITD positive, whereas 67% of the patients in the predicted poor-outcome class were FLT3 ITD positive (Tables 1 and 2). Because of this association and the well-established prognostic importance of FLT3 ITD, we examined the impact of the predicted outcome classes on outcome when adjusting for FLT3 ITD status. Multivariable models for overall and disease-free survival showed that the signature-based class membership did not predict outcome independently of FLT3 ITD status (P = .69 and P = .82 for overall and disease-free survival, respectively). However, there was a moderate interaction effect between the 2 variables with respect to overall survival (P = .10), and a significant interaction effect with respect to disease-free survival (P = .03). Whereas in patients with FLT3 ITD the signature-based classification failed to result in an appropriate separation of the outcome curves of the predicted poor- and good-outcome classes (Figure 3A), in patients with wild-type FLT3, the signature-based classification produced a moderate separation in curves (Figure 3B). This suggested that the signature-based classifier may be able to distinguish a subset of patients with poor outcome when analysis is restricted to those with wild-type FLT3 and normal cytogenetics.
Compound covariate prediction (CCP) algorithm predicts outcome for individual patients. Overall (A) and disease-free (B) survival according to CCP class membership. Accuracy rates of the signature-based CCP classifier for overall (C) and disease-free (D) survival.
Compound covariate prediction (CCP) algorithm predicts outcome for individual patients. Overall (A) and disease-free (B) survival according to CCP class membership. Accuracy rates of the signature-based CCP classifier for overall (C) and disease-free (D) survival.
Discussion
We successfully confirmed, using a different microarray platform, the prognostic significance of the expression signature identified by Bullinger et al22 in karyotypically normal AML patients. Cluster analysis of our patients with respect to expression of the Bullinger validation signature resulted in 2 distinct clusters with differences in survival. Furthermore, we developed a well-defined classifier based on the signature that predicts dichotomized outcome for individual patients. The prognostic significance of the Bullinger validation signature was maintained with the shift in analysis strategies as the predicted outcome classes exhibited a significant difference in both overall and disease-free survival.
Our use of a classifier to validate the prognostic significance of the expression signature complements the cluster-based results of the previous study. Bullinger et al22 acknowledged the limitations of cluster-based classification and successfully implemented a class prediction algorithm (the prediction analysis of microarrays method of nearest shrunken centroids) for their whole patient group that included both cytogenetically normal and abnormal patients, but were unable to maintain prognostic significance in the subset with a normal karyotype.22 Their lack of success in the subset of karyotypically normal patients was likely not due to the use of a different prediction algorithm but, rather, to inadequate sample size as testing of their initial results was performed on only 22 AML patients with a normal karyotype.22 In contrast, as the expression signature was already defined using data from the earlier study,22 we did not have to divide our 64 patients into separate training and test sets but could treat the whole patient cohort as a validation set. Furthermore, the median duration of follow-up for survivors in our patient group (4.7 years) was considerably longer than that of less than 2 years reported in the Bullinger et al study.22
Interaction between FLT3 ITD status and CCP class membership. (A) Overall (i) and disease-free (ii) survival for patients with FLT3 ITD according to poor and good CCP class membership. (B) Overall (i) and disease-free (ii) survival for patients with wild-type FLT3 according to poor and good CCP class membership.
Interaction between FLT3 ITD status and CCP class membership. (A) Overall (i) and disease-free (ii) survival for patients with FLT3 ITD according to poor and good CCP class membership. (B) Overall (i) and disease-free (ii) survival for patients with wild-type FLT3 according to poor and good CCP class membership.
Confirming the prognostic significance of the Bullinger validation signature in our study is noteworthy not only because the 2 studies included patients treated on different protocols at different institutions, but also because different microarray platforms were used. The previous study employed a common reference design using cDNA arrays manufactured by the Stanford Functional Genomics Facility,22 whereas we used the commercially available Affymetrix U133 plus 2.0 GeneChip. We were unable to create a perfect representation of the signature identified in the previous study using our microarray data because the original signature contained expressed sequence tags that we could not match to probe sets on the Affymetrix oligonucleotide array. Furthermore, some named genes in the original signature were represented by multiple probe sets on the oligonucleotide array and therefore appeared more than once in the validation signature. Even with this lack of precision in our representation of the original signature, the validation signature was robust enough so that its prognostic value was maintained in our patient set.
While we confirmed the expression signature reported by Bullinger et al,22 the prediction accuracy of the classifier for dichotomized outcome classes was modest for both overall and disease-free survival. The outcome of approximately 40% of the patients could not be correctly predicted. Despite this limitation and the association of the signature-based classifier with FLT3 ITD status, the classifier showed some ability to identify a subset of patients with wild-type FLT3 who fare poorly. Furthermore, while Bullinger et al22 also noted a strong association of FLT3 status with the 2 major clusters of normal karyotype patients identified by the expression signature, the prognostic significance of the signature was shown to be independent of FLT3 when considering the whole patient set (including patients with cytogenetic aberrations).
In conclusion, despite some limitations of the signature-based classifier, our analysis validates the use of the gene-expression profiling strategy for outcome prediction in cytogenetically normal AML. The goal of future studies is to refine this strategy and assess whether it is possible to identify different classifiers that predict outcome for individual patients with cytogenetically normal AML more accurately than the one based on the only hitherto reported expression signature with prognostic significance.
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study: North Shore Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R. K. Koduru (grant no. CA35279); The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield and Karl S. Theil (grant no. CA77658); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (grant no. CA02599); Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Wendy L. Flejter, and Mark J. Pettenati (grant no. CA03927); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Mazin B. Qumsiyeh (grant no. CA47577); Vermont Cancer Center, Burlington, VT: Hyman B. Muss and Elizabeth F. Allen (grant no. CA77406); University of Puerto Rico School of Medicine, San Juan, PR: Enrique Velez-Garcia, Paola Dal Cin, Cynthia C. Morton, and Leonard L. Atkins; Christiana Care Health Services Inc, Newark, DE: Stephen S. Grubbs, Jeanne M. Meck, and Digamber S. Borgaonkar (grant no. CA45418); University of California at San Diego: Stephen L. Seagren and Marie L. Dell'Aquila (grant no. CA11789); University of Chicago Medical Center, Chicago, IL: Gini Fleming, Michelle M. LeBeau, and Diane Roulston (grant no. CA41287); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim Huang (grant no. CA12046); Weill Medical College of Cornell University, New York, NY: Scott Wadler and Prasad R. K. Koduru (grant no. CA07968); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff and Thuluvancheri K. Mohandas (grant no. CA04326); Eastern Maine Medical Center CCOP, Bangor, ME: Philip L. Brooks and Laurent J. Beauregard (grant no. CA35406); Ft Wayne Medical Oncology/Hematology, Ft Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Georgetown University Medical Center, Washington, DC: Edward P. Gelmann and Jeanne M. Meck (grant no. CA77597); Medical University of South Carolina, Charleston, SC: Mark R. Green and G. Shashidhar Pai (grant no. CA03927); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (grant no. CA04457); Southern Nevada Cancer Research Foundation, Las Vegas, NV: John Ellerton and Renée Bernstein (grant no. CA35421); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano and Constance K. Stein (grant no. CA21060); University of Illinois at Chicago: Thomas E. Lad and Maureen M. McCorquodale (grant no. CA74811); University of Tennessee Memphis Cancer Center: Harvey B. Niell and Sugandhi A. Tharapel (grant no. CA47555); Walter Reed Army Medical Center, Washington, DC: Joseph J. Drabeck and Digamber S. Borgaonkar (grant no. CA26806); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett and Michael S. Watson (grant no. CA77440).
Prepublished online as Blood First Edition Paper, May 2, 2006; DOI 10.1182/blood-2006-02-005538.
A list of Cancer and Leukemia Group B members who participated in this study appears in “Appendix.”
Supported in part by National Cancer Institute, Bethesda, MD (grants CA101 140, CA77 658, CA102 031, CA31 946, CA09 512, CA16 058, and CA90 469), and The Coleman Leukemia Research Foundation, St. Paul, MN. M.A.C. is chair of the CALGB Leukemia Correlative Science Committee, Chicago, IL, and Director of the CALGB Leukemia Tissue Bank, Columbus, OH.
M.D.R. and G.M. cowrote this paper, with input from C.D. Bloomfield, K.M., and A.S.R. M.D.R. validated statistical methods used, performed data analysis, and participated in the results interpretation. G.M. reviewed the data, participated in the results interpretation and patient clinical care, and provided laboratory technical support. A.S.R. did the statistical analysis of clinical outcome. K.M. participated in cytogenetic central review and the results interpretation. S.P.W., P.P., T.V., and C. D. Baldus performed the molecular biology analysis of, respectively, the FLT3, NPM1, CEBPA, and BAALC genes. J.W.V. supervised the central pathology review. J.E.K. was the principal investigator of the CALGB 9621 treatment protocol, and participated in patient clinical care. R.A.L. participated in the development and conduct of CALGB treatment protocols. C. D. Bloomfield designed the study and supervised data analysis and interpretation, participated in the coordination of CALGB correlative laboratory studies, and provided funding for this project. All authors commented on drafts of the manuscript and approved its final version.
M.D.R. and G.M. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal