Abstract
Both CD4+ T cells and macrophages are major reservoirs of HIV-1. Previous study showed that HIV-1–specific cytolytic T lymphocytes (CTLs) hardly recognize HIV-1–infected CD4+ T cells because of Nef-mediated HLA class I down-regulation, suggesting that HIV-1 escapes from HIV-1–specific CTLs and continues to replicate in HIV-1–infected donors. On the other hand, the CTL recognition of HIV-1–infected macrophages and the effect of Nef-mediated HLA class I down-regulation on this recognition still remain unclear. We show a strong HIV-1 antigen presentation by HIV-1–infected macrophages. HIV-1–specific CTLs had strong abilities to suppress HIV-1R5 virus replication in HIV-1–infected macrophages and to kill HIV-1R5–infected macrophages. Nef-mediated HLA class I down-regulation minimally influenced the recognition of HIV-1–infected macrophages by HIV-1–specific CTLs. In addition, HIV-1–infected macrophages had a stronger ability to stimulate the proliferation of HIV-1–specific CTLs than HIV-1–infected CD4+ T cells. Thus, the effect of Nef-mediated HLA class I down-regulation was less critical with respect to the recognition by HIV-1–specific CTLs of HIV-infected macrophages than that of HIV-1–infected CD4+ T cells. These findings support the idea that the strong HIV-1 antigen presentation by HIV-1–infected macrophages is one of the mechanisms mediating effective induction of HIV-1–specific CTLs in the acute and early chronic phases of HIV-1 infection.
Introduction
HIV-1–specific CD8+ T cells play a critical role in the control of HIV-1 infections.1,2 However, HIV-1–infected individuals develop AIDS if they are not treated with antiretroviral therapy. HIV-1 escape occurs during acute and chronic phases of an HIV-1 infection.3 Several hypotheses concerning HIV-1 mechanisms affording escape from the host immune system have been proposed.4–6 One of these is impaired activity of HIV-1–specific cytotoxic T lymphocytes (CTLs) to kill HIV-1–infected CD4+ T cells and to suppress HIV-1 replication by Nef-mediated down-regulation of HLA class I molecules. Previous studies reported that HIV-1–specific CTL clones failed to kill CD4+ T cells infected with Nef+ HIV-1.7,8 Our previous studies using NL-432 X4 clone and NL-M20A lacking Nef function for HLA class I molecules showed that most HIV-1–infected CTLs failed to kill NL-432–infected CD4+ T cells and partially suppressed NL-432 replication but that they could effectively kill NL-M20A–infected CD4+ T cells and completely suppress NL-M20A replication,8,9 indicating that Nef-mediated HLA class I down-regulation critically affects recognition of HIV-1–infected CD4+ T cells by HIV-1–specific CTLs. The effects of Nef-mediated HLA class I down-regulation on these antiviral activities of HIV-1–specific CTLs varied among CTLs specific for various HIV-1 epitopes.9
CD4+ T cells and macrophages are major targets of HIV-1.10,11 Macrophages are persistently infected with HIV-1 and serve as a reservoir of the M-tropic/R5 strain of HIV-1.11,12 HIV-1–infected macrophages are detected in the various tissues of individuals infected with HIV-1, disseminating HIV-1 throughout the body.13 Therefore, the ability of CTLs to suppress HIV-1 replication in macrophages may be an important factor in the control of HIV-1 infections. Previous studies showed that HIV-1–specific CTLs can kill HIV-1–infected alveolar macrophages derived from HIV-1–infected individuals.14,15 However, it still remains unclear whether such CTLs effectively suppress HIV-1 replication in macrophages and whether Nef-mediated HLA class I down-regulation critically affects HIV-1–specific CTL recognition of HIV-1–infected macrophages as it does that of HIV-1–infected CD4+ T cells.
The X4 virus infects CD4+ T cells and weakly infects macrophages, whereas the R5 virus infects both macrophages and CD4+ T cells. The X4 virus dominantly appears in late phase of HIV-1 infection, whereas the R5 virus involves in the transmission and replicates in the early phase. Analysis of CTL responses to X4 virus-infected CD4+ T cells, R5 virus-infected CD4+ T cells, and R5 virus-infected macrophages is important to understand CTL-mediated immune responses in both early and late phases of HIV-1 infection.
In this study, we tested the ability of HIV-1–specific CD8+ T cells to kill HIV-1R5 virus-infected macrophages and to suppress the replication of HIV-1R5 virus in macrophages, and we also investigated the effect of Nef-mediated HLA class I down-regulation on the recognition by HIV-1–specific CD8+ T cells of HIV-1R5 virus-infected macrophages. In addition, we compared the antiviral activities of these cells against HIV-1R5 virus-infected macrophages with those against HIV-1-R5 virus-infected or X4 virus-infected CD4+ T cells. Finally, we investigated the mechanisms underlying the effective recognition of HIV-1–infected macrophages by HIV-1–specific CTLs.
Materials and methods
This study was approved by Kumamoto University Ethics Committee. Informed consent was obtained from all subjects, in accordance the Declaration of Helsinki.
Isolation and culture of macrophages and CD4+ T cells
Monocytes were isolated from peripheral blood mononuclear cells (PBMCs) of HLA-B*5101+ or HLA-B*3501+ healthy donors by an adherence method as previously described.16 CD4+ T cells were purified from nonadherent cells by means of anti–human CD4 monoclonal antibody (mAb)–coated magnetic beads (magnetic-activated cell sorting [MACS] beads; Miltenyi Biotec, Bergisch Galdbach, Germany). These cultured macrophages and CD4+ T cells were infected with HIV-1 clones as previously described.8
HIV-1–specific CTL clones
HIV-1–specific CTL clones (HLA-B*5101-restricted CTL clones: Pol743-8-40, Pol283-8-237, -240, -320, and -340; Gag327-9-131, -142, -148, and -287; Rev71-11-8, -17, and -55; and HLA-B*3501-restricted CTL clones: Env77-9-110, Pol273-9-2; and an HLA-A*1101-restricted CTL clone: Gag349-11-18 and -22 as mismatched CTL clone) were generated as previously described.17–19 These CTL clones predominantly showed CD27−CD28−CD45RA− phenotype (data not shown).
HIV-1 clones
An infectious proviral clone of HIV-1, pNL-432, and its Nef mutant, pNL-M20A (containing a substitution of Ala for Met at residue 20 of Nef), were reported previously.20 pJRFL and its Nef-defective mutant, pJR-Xh, which has a frame shift at a XhoI site (44th amino acid of Nef protein), were kindly donated by Dr Koyanagi (Kyoto University, Kyoto, Japan). pJRFLNL-432 Nef and JRFLNL-M20A Nef were constructed by exchanging the Nef region of JRFL for that of NL-432 or NL-M20A.
CTL assay
The cytotoxicity of CTL clones against HIV-1–infected macrophages or CD4+ T cells (40-50% p24 antigen-positive cells) was determined by a standard 51Cr-release assay, as previously described.8
Flow cytometric analysis
Cells infected with HIV-1 clone were stained to assess the expression of HLA class I in HIV-1–infected macrophages or CD4+ T cells, as previously described.9 For detection of intracellular cytokines, HIV-1–specific CTL clones were cocultured with HIV-1–infected cells for 6 hours at an effector-stimulator (E/S) ratio of 1:4. Then, brefeldin A was added (10 μg/mL). After a 6-hour incubation, the cells were stained with FITC-labeled anti–human IFN-γ, PE-labeled anti–human MIP-1β, PerCp-labeled anti–human CD8, or APC-labeled anti–human TNF-α mAbs (BD Biosciences, San Jose, CA), as previously described.21
Suppression of HIV-1 replication by HIV-1–specific CTLs
The ability of HIV-1–specific CTLs to suppress HIV-1 replication was examined as previously described.8 Briefly, CD4+ T cells or macrophages were incubated with a given HIV-1 clone for 12 hours at 37°C. After several washes with R10 medium, the cells were cocultured with HIV-1–specific CTL clones. From day 3 to 12 after infection, 10 μL culture supernatant was collected, and the concentration of p24 antigen was measured by use of an enzyme immunoassay (HIV-1 p24 antigen enzyme-linked immunosorbent assay [ELISA] kit; ZeptMetrix, Buffalo, NY). The percentage of suppression of HIV-1 replication was calculated as follows: % suppression = (1 − concentration of p24 antigen in the supernatant of HIV-1–infected CD4+ T cells cultured with HIV-1–specific CTLs/concentration of p24 antigen in the supernatant of HIV-1–infected CD4+ T cells culture without the CTLs) × 100.
Western blot analysis
Cells were lysed in lysis buffer (1% Triton X-100, 50 mM Tris HCl, pH 7.6, 1 mM MgCl2, 150 mM NaCl, and 0.1 mM EDTA) containing a mixture of protease inhibitors (Boehringer Mannheim, Mannheim, Germany). Samples were boiled in sodium dodecyl sulfate (SDS) sample buffer, separated by SDS-10% polyacrylamide gel electrophoresis (PAGE), and transferred to an Immun-Blot PVDF membrane (Bio-Rad, Hercules, CA). Protein detection was performed after incubation with appropriate first and secondary antibodies by using a Chromogenic Western Blot Immunodetection Kit (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. The first antibodies for p24 and Nef were purchased from ZeptoMetrix and Advanced Biotechnologies, respectively. Quantification was performed by using National Institutes of Health Image.
Proliferation assay
HIV-1–infected CD4+ T cells or macrophages were irradiated and cocultured with thawed HIV-1–specific CTL clones (5 × 103 cells/well) for 3 days in triplicate in 96-well plates at an E/S ratio of 1:4. Then 0.5 μCi/well (0.0185 MBq) of 3H [thymidine] was added, and the cells were subsequently incubated for an additional 16 hours. The incorporation was measured by a scintillation counter.
Results
Strong abilities of HIV-1–specific CTL clones to suppress HIV-1 replication in macrophages and to kill HIV-1–infected macrophages
To investigate CTL recognition of HIV-1–infected macrophages, we measured the ability of HIV-1–specific CTLs to suppress the replication of HIV-1 R5 strain JRFL and X4 strain NL-432 in HIV-1–infected macrophages and CD4+ T cells, respectively. We used CTL clones specific for 4 HLA-B*5101-restricted epitopes (Pol743-9, Pol283-8, Gag327-9, and Rev71-11) and 2 HLA-B*3501-restricted epitopes (Env77-9 and Pol273-9). Previous studies using these epitope-specific CTL clones demonstrated that the 2 B*5101-restricted Pol-specific CTL clones completely suppressed the replication of HIV-1 X4 strain NL-432 but that other CTL clones only partially suppressed it.8,9 We measured the ability of these 6 CTL clones to suppress the replication of JRFL and its Nef-defective mutant JR-Xh in HIV-1–infected macrophages. The surface expression of HLA-B*5101 molecules was down-regulated in JRFL-infected macrophages and NL-432–infected CD4+ T cells but not in JR-Xh–infected macrophages and NL-M20A–infected CD4+ T cells (Figure 1). The down-regulation of HLA-B*3501 molecules was also found in only JRFL-infected macrophages and NL-432–infected CD4+ T cells (data not shown). SF2-Rev71-11-8 partially suppressed the replication of JRFL, whereas the other 5 clones completely suppressed the replication of both JRFL and JR-Xh (Figure 2A). On the other hand, only Pol743-9– and Pol283-8–specific CTLs completely suppressed the replication of both NL-432 and NL-M20A in HIV-1–infected CD4+ T cells (Figure 2B). These CTL clones showed similar effects in terms of their cytolytic activity toward HIV-1–infected macrophages and CD4+ T cells (Figure 2C-D). These results of the CTL clones for JRFL-infected macrophages contrast with those for NL-432–infected CD4+ T cells.8,9
Expression of HLA class I molecules on macrophages or CD4+ T cells infected with Nef+ or Nef− HIV-1. Macrophages established from monocytes and CD4+ T cells of an HLA-B*5101+ donor were infected with HIV-1 JRFL or JR-Xh and NL-432 or NL-M20A, respectively, and then cultured for 6 days. The cultured macrophages and CD4+ T cells were stained with anti-p24 and 4D12 anti–HLA-B5 mAbs. The surface expression of HLA-B*5101 on p24+ or p24− cells is shown as the mean fluorescence intensity (MFI) in each figure.
Expression of HLA class I molecules on macrophages or CD4+ T cells infected with Nef+ or Nef− HIV-1. Macrophages established from monocytes and CD4+ T cells of an HLA-B*5101+ donor were infected with HIV-1 JRFL or JR-Xh and NL-432 or NL-M20A, respectively, and then cultured for 6 days. The cultured macrophages and CD4+ T cells were stained with anti-p24 and 4D12 anti–HLA-B5 mAbs. The surface expression of HLA-B*5101 on p24+ or p24− cells is shown as the mean fluorescence intensity (MFI) in each figure.
Strong abilities of HIV-1–specific CTLs to suppress HIV-1 replication in HIV-1–infected macrophages and to kill them. (A-B) Ability of HIV-1–specific CTL clones to suppress HIV-1 replication in HIV-1–infected macrophages and in HIV-1–infected CD4+ T cells. Macrophages and CD4+ T cells from an HLA-B*5101+ donor and an HLA-B*3501+ donor were infected with JRFL or JR-Xh and NL-432 or NL-M20A, respectively, and then cocultured with each HIV-1–specific CTL clone at an E/T ratio of 1:1. HIV-1 p24 antigens in the supernatant were measured on day 9 after infection by use of an enzyme immunoassay. Data shown in the figure are averages of triplicate assays for each HIV-1–specific CTL clone. (C) Cytotoxic activity against HIV-1–infected macrophages. Macrophages from an HLA-B*5101+ donor and an HLA-B*3501+ donor were infected with JRFL or JR-Xh. JRFL-infected (56% of total cells were p24 antigen-positive), JR-Xh–infected (48% of total cells were p24 antigen-positive) macrophages were used as target cells at an E/T ratio of 2:1. Data shown in the figure are averages ± SD of triplicate assays for each HIV-1–specific CTL clones. (D) Cytotoxic activity against HIV-1–infected CD4+ T cells. CD4+ T cells from an HLA-B*5101+ donor were infected with NL-432 or NL-M20A. NL-432–infected (81.6% of total cells were p24 antigen-positive) and NL-M20A–infected (79.1% of total cells were p24 antigen-positive) were used as target cells at an E/T ratio of 2:1.
Strong abilities of HIV-1–specific CTLs to suppress HIV-1 replication in HIV-1–infected macrophages and to kill them. (A-B) Ability of HIV-1–specific CTL clones to suppress HIV-1 replication in HIV-1–infected macrophages and in HIV-1–infected CD4+ T cells. Macrophages and CD4+ T cells from an HLA-B*5101+ donor and an HLA-B*3501+ donor were infected with JRFL or JR-Xh and NL-432 or NL-M20A, respectively, and then cocultured with each HIV-1–specific CTL clone at an E/T ratio of 1:1. HIV-1 p24 antigens in the supernatant were measured on day 9 after infection by use of an enzyme immunoassay. Data shown in the figure are averages of triplicate assays for each HIV-1–specific CTL clone. (C) Cytotoxic activity against HIV-1–infected macrophages. Macrophages from an HLA-B*5101+ donor and an HLA-B*3501+ donor were infected with JRFL or JR-Xh. JRFL-infected (56% of total cells were p24 antigen-positive), JR-Xh–infected (48% of total cells were p24 antigen-positive) macrophages were used as target cells at an E/T ratio of 2:1. Data shown in the figure are averages ± SD of triplicate assays for each HIV-1–specific CTL clones. (D) Cytotoxic activity against HIV-1–infected CD4+ T cells. CD4+ T cells from an HLA-B*5101+ donor were infected with NL-432 or NL-M20A. NL-432–infected (81.6% of total cells were p24 antigen-positive) and NL-M20A–infected (79.1% of total cells were p24 antigen-positive) were used as target cells at an E/T ratio of 2:1.
Comparison between abilities of HIV-1–specific CTLs to suppress R5 virus replication in CD4+ T cells and macrophages
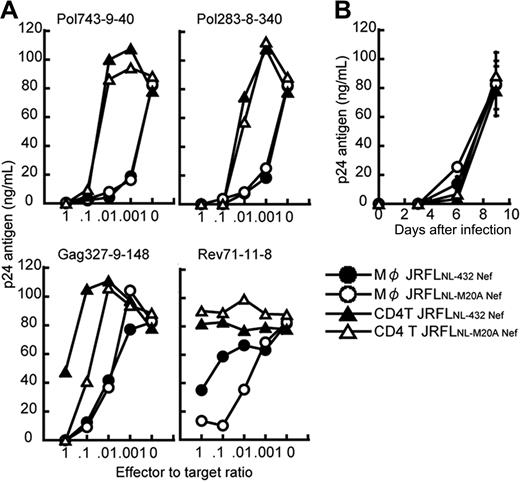
It remains possible that the strong ability to suppress the replication of JRFL and the effect of Nef-mediated HLA class I down-regulation were a strain-dependent effect. To exclude this possibility, we generated 2 R5 chimera viruses, specifically, JRFLNL-432Nef (JRFL carrying NL432-derived Nef) and JRFLNL-M20ANef (JRFL carrying NL-M20A–derived Nef), and then investigated the ability of 4 HLA-B*5101-restricted CTL clones to suppress the replication of these chimera viruses in HIV-1–infected CD4+ T cells and macrophages. The down-regulation of HLA class I molecules was found to occur in JRFLNL-432 Nef-infected cells, but not in JRFLNL-M20A Nef-infected cells (data not shown). The Rev71-11-8 CTL clone suppressed the replication of JRFLNL-M20A Nef in JRFLNL-M20A Nef-infected macrophages more strongly than that of JRFLNL-432 Nef in JRFLNL-432 Nef-infected macrophages, whereas other CTL clones showed the same ability to suppress the replication of the 2 JRFL chimera viruses (Figure 3A).
Comparison between abilities of HIV-1–specific CTLs to suppress HIV-1 replication in CD4+ T cells and macrophages infected with HIV-1 R5 strain. (A) The ability of HIV-1–specific CTL clones to suppress JRFLNL-432 Nef and JRFLNL-M20A Nef replication in CD4+ T cells and macrophages infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. CD4+ T cells and macrophages from HLA-B*5101+ donor were infected with JRFLNL-432 Nef or JRFLNL-M20A Nef and then cocultured with HLA-B*5101-restricted HIV-1–specific CTL clones at various E/T ratios. The amount of HIV-1 p24 antigen in the supernatant on day 9 after infection was measured by using an enzyme immunoassay. (B) Kinetics of JRFLNL-432 Nef and JRFLNL-M20A Nef replication in CD4+ T cells and macrophages infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. The amount of HIV-1 p24 antigen in the supernatant on days 3 to 9 after infection was measured by the enzyme immunoassay. Data shown in the figure are averages of triplicate assays for each time point. The experiments shown in panels A and B were performed simultaneously.
Comparison between abilities of HIV-1–specific CTLs to suppress HIV-1 replication in CD4+ T cells and macrophages infected with HIV-1 R5 strain. (A) The ability of HIV-1–specific CTL clones to suppress JRFLNL-432 Nef and JRFLNL-M20A Nef replication in CD4+ T cells and macrophages infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. CD4+ T cells and macrophages from HLA-B*5101+ donor were infected with JRFLNL-432 Nef or JRFLNL-M20A Nef and then cocultured with HLA-B*5101-restricted HIV-1–specific CTL clones at various E/T ratios. The amount of HIV-1 p24 antigen in the supernatant on day 9 after infection was measured by using an enzyme immunoassay. (B) Kinetics of JRFLNL-432 Nef and JRFLNL-M20A Nef replication in CD4+ T cells and macrophages infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. The amount of HIV-1 p24 antigen in the supernatant on days 3 to 9 after infection was measured by the enzyme immunoassay. Data shown in the figure are averages of triplicate assays for each time point. The experiments shown in panels A and B were performed simultaneously.
These CTL clones revealed more than 10- to 100-fold stronger ability to suppress the replication of the chimera viruses in HIV-1–infected macrophages than in HIV-1–infected CD4+ T cells (Figure 3A). The replication kinetics of JRFLNL-432 Nef and JRFLNL-M20A Nef between these 2 cell types were similar, as shown in Figure 3B, thus indicating that the difference in the ability of the specific CTL clones to suppress the chimera virus replication was unrelated to replication kinetics. Thus, these results indicate that HIV-1–specific CTLs could recognize HIV-1–infected macrophages more effectively than HIV-1–infected CD4+ T cells.
Ability of HIV-1–infected macrophages to stimulate HIV-1–specific CTLs
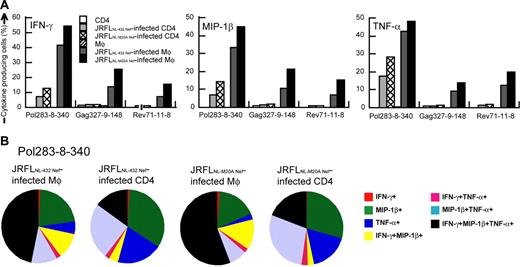
We further analyzed the cytokine production from the HLA-B*5101-restricted CTL clones after having stimulated them with either HIV-1–infected CD4+ T cells or HIV-1–infected macrophages. IFN-γ–, MIP-1β–, or TNF-α–producing CTL clones were much more detectable after the clones had been stimulated with HIV-1 chimera virus-infected macrophages than after stimulation with HIV-1 chimera virus-infected CD4+ T cells (Figure 4A-B). We considered p24− cells to be HIV-1–uninfected cells because only p24+ cells showed down-regulation of CD4 (data not shown). HIV-1–infected cells might exist in p24− cells, but they should express very low level of HIV-1 proteins and can hardly stimulate HIV-1–specific CTLs. Therefore we counted p24+ cells as HIV-1–infected cells. Frequencies of Pol283-8-340 CTL clones producing at least one cytokine were 69.6% and 72.5%, after the clones had been stimulated with JRFLNL-432 Nef-infected and JRFLNL-M20A Nef-infected macrophages, respectively, whereas they were 29.7% and 43.8% after stimulation with JRFLNL-432 Nef-infected and JRFLNL-M20A Nef-infected CD4+ T cells, respectively. HIV-1–specific CTL clones stimulated with HIV-1–infected CD4+ T cells predominantly produced a single or 2 cytokines, whereas cells producing 3 cytokines were more frequently found among those stimulated with HIV-1–infected macrophages (Figure 4B). Similar results were found for other CTL clones tested (Figure 4A). These results suggest that HIV-1–infected macrophages can stimulate HIV-1–specific CD8+ T cells more strongly than HIV-1–infected CD4+ T cells in vivo.
Ability of HIV-1–infected CD4+ T cells and HIV-1–infected macrophages to stimulate cytokine production by HIV-1–specific CTLs. Cultured CD4+ T cells and macrophages were infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. JRFLNL-432 Nef-infected macrophages (22.7% p24 antigen-positive) and CD4+ T cells (19.9% p24 antigen-positive), as well as JRFLNL-M20A Nef-infected macrophages (21.5% p24 antigen-positive) and CD4+ T cells (22.4% p24 antigen-positive) were used to stimulate 3 HLA-B*5101-restricted CTL clones at an effector-stimulator (E/S) ratio of 1:4. (A) The frequency of cells expressing each cytokine is shown as a percentage of the total number of CD8+ cells. (B) The frequency of cells expressing these cytokines among total cytokine-producing cells is also shown in this pie chart.
Ability of HIV-1–infected CD4+ T cells and HIV-1–infected macrophages to stimulate cytokine production by HIV-1–specific CTLs. Cultured CD4+ T cells and macrophages were infected with JRFLNL-432 Nef or JRFLNL-M20A Nef. JRFLNL-432 Nef-infected macrophages (22.7% p24 antigen-positive) and CD4+ T cells (19.9% p24 antigen-positive), as well as JRFLNL-M20A Nef-infected macrophages (21.5% p24 antigen-positive) and CD4+ T cells (22.4% p24 antigen-positive) were used to stimulate 3 HLA-B*5101-restricted CTL clones at an effector-stimulator (E/S) ratio of 1:4. (A) The frequency of cells expressing each cytokine is shown as a percentage of the total number of CD8+ cells. (B) The frequency of cells expressing these cytokines among total cytokine-producing cells is also shown in this pie chart.
HIV-1–specific CTLs are frequently found in HIV-1–infected individuals, although their number decreases in the late chronic phase of an HIV-1 infection.22 Since we found the ability of HIV-1 antigen presentation by HIV-1–infected CD4+ T cells to be much weaker than that by HIV-1–infected macrophages and macrophages are well known to be professional antigen-presenting cells, we speculated that HIV-1–infected macrophages would induce the proliferation of HIV-1–specific CD8+ T cells more effectively than HIV-1–infected CD4+ T cells. To test this possibility, we analyzed the capacities of HIV-1–infected macrophages and HIV-1–infected CD4+ T cells to induce the proliferation of HLA-B*5101-restricted HIV-1–specific CTL clones (Pol283-8-340, Gag327-9-148, Rev71-11-17, and HLA-mismatched CTL clones; Figure 5A). All 3 HLA-B*5101-restricted CTL clones cocultured with JRFLNL-M20A Nef-infected or JRFLNL-432 Nef-infected macrophages or JRFLNL-M20A Nef-infected CD4+ T cells showed significantly higher proliferation than those cocultured with uninfected macrophages or uninfected CD4+ T cells, respectively, whereas when the clones were cocultured with JRFLNL-432–infected CD4+ T cells, only Pol283-8-40 showed significantly higher proliferation compared with that obtained with uninfected CD4+ T cells. The proliferation abilities of these CTL clones stimulated with JRFLNL-M20A Nef-infected macrophages were significantly higher than those of the clones stimulated with JRFLNL-M20A Nef-infected CD4+ T cells. Similar results were found for the proliferation of all 3 HLA-B*5101-restricted HIV-1–specific CTL clones stimulated with JRFLNL-432 Nef-infected macrophages or JRFLNL-432 Nef-infected CD4+ T cells. To confirm these results, we measured the proliferation of other CTL clones with the same specificity (Figure 5B). A higher proliferation of the CTL clones stimulated with HIV-1–infected macrophages than with HIV-1–infected CD4+ T cells was confirmed for Pol283-8– and Gag327-9–specific CTL clones but not for the Rev71-11–specific CTL clones. Furthermore, they showed a higher proliferation when they were stimulated with JRFLNL-M20A Nef-infected cells than with JRFLNL-432 Nef-infected cells, but the influence of Nef-mediated down-regulation of HLA class I molecules was less crucial for the stimulation with HIV-1–infected macrophages than for that with HIV-1–infected CD4+ T cells. These results strongly suggest that HIV-1–infected macrophages can much more effectively induce proliferation of HIV-1–specific CTLs than can HIV-1–infected CD4+ T cells in vivo and support our idea that HIV-1–specific CTLs are strongly induced by HIV-1–infected macrophages in the acute and early chronic phases but that they are weakly induced in the late chronic phase, since the X4 virus predominantly replicates in this phase.
Ability of HIV-1–infected CD4+ T cells and HIV-1–infected macrophages to induce proliferation of HIV-1–specific CTLs. Eleven HLA-B*5101-restricted CTL clones (Pol283-8-340, -320, -237, and -240; Gag327-9-148, -142, -287, and -131; Rev71-11-8, -55, and -17) were cocultured for 96 hours with uninfected macrophages, irradiated JRFLNL-432 Nef-infected macrophages (17.8% p24 antigen-positive), JRFLNL-M20A Nef-infected macrophages (23.2% p24 antigen-positive), uninfected CD4+ T cells, JRFLNL-432 Nef-infected CD4+ T cells (20.8% p24 antigen-positive), or JRFLNL-M20A Nef-infected CD4+ T cells (24.8% p24 antigen-positive) at an E/S ratio of 1:4. The incorporation was measured after an additional 16-hour incubation. (A) Typical example of 3H-incorporation in HLA-B*5101-restricted CTL clones (Pol283-8-340, Gag327-9-148, and Rev71-11-17), and HLA-mismatched CTL clone. Data shown in this figure are averages ± SD of triplicate assays. (B) Average ± SD of proliferation in triplicate assays for 4 Pol283-8–, 4 Gag327-9–, or 3 Rev71-11–specific CTL clones.
Ability of HIV-1–infected CD4+ T cells and HIV-1–infected macrophages to induce proliferation of HIV-1–specific CTLs. Eleven HLA-B*5101-restricted CTL clones (Pol283-8-340, -320, -237, and -240; Gag327-9-148, -142, -287, and -131; Rev71-11-8, -55, and -17) were cocultured for 96 hours with uninfected macrophages, irradiated JRFLNL-432 Nef-infected macrophages (17.8% p24 antigen-positive), JRFLNL-M20A Nef-infected macrophages (23.2% p24 antigen-positive), uninfected CD4+ T cells, JRFLNL-432 Nef-infected CD4+ T cells (20.8% p24 antigen-positive), or JRFLNL-M20A Nef-infected CD4+ T cells (24.8% p24 antigen-positive) at an E/S ratio of 1:4. The incorporation was measured after an additional 16-hour incubation. (A) Typical example of 3H-incorporation in HLA-B*5101-restricted CTL clones (Pol283-8-340, Gag327-9-148, and Rev71-11-17), and HLA-mismatched CTL clone. Data shown in this figure are averages ± SD of triplicate assays. (B) Average ± SD of proliferation in triplicate assays for 4 Pol283-8–, 4 Gag327-9–, or 3 Rev71-11–specific CTL clones.
High expression of HIV-1 proteins in HIV-1–infected macrophages
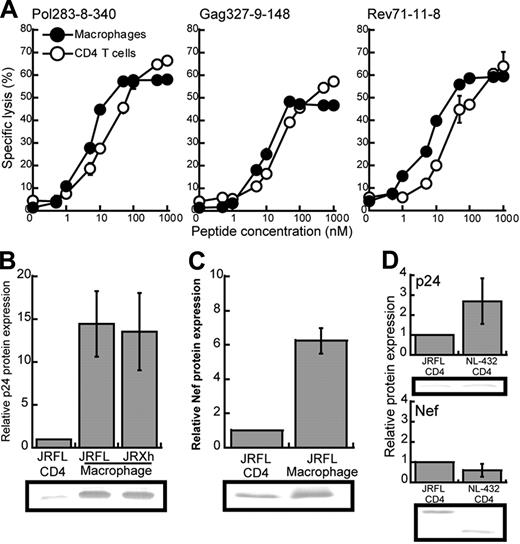
We speculated that the difference in the suppressive effect of HIV-1–specific CTLs on JRFL replication between macrophages and CD4+ T cells may have resulted from a difference in the amount of surface expression of HLA class I molecules between these HIV-1–infected cells. Flow cytometric analysis using mAb specific for HLA-B*5101 revealed that the surface expression of HLA-B*5101 on CD4+ T cells was approximately 2-fold lower than that on macrophages (data not shown). To investigate the effect of this difference in surface expression of HLA-B*5101 on the recognition by HIV-1–specific CTLs, we measured the killing activity of 3 HLA-B*5101–restricted CTL clones toward HLA-B*5101+CD4+ T cells or macrophages prepulsed with the appropriate epitope peptides (Figure 6A). The ability of CTLs to kill the peptide-pulsed macrophages (LL50, peptide concentration providing a half of maximum percent specific lysis) was approximately 3-fold lower (2.67 ± 0.53) than that to kill the peptide-pulsed CD4+ T cells. These results suggest that the difference in surface expression of HLA-B*5101 molecules between macrophages and CD4+ T cells may partially influence the recognition of these cells by HIV-1–specific CD8+ T cells. Another possibility is that HIV-1 antigens are much more expressed in HIV-1–infected macrophages than in HIV-1–infected CD4+ T cells. To examine this possibility, we measured the amount of p24 and Nef proteins in HIV-1–infected macrophages and HIV-1–infected CD4+ T cells. The amount of p24 was approximately 13-fold larger in either JRFL- or JR-Xh–infected macrophages than in JRFL-infected CD4+ T cell (Figure 6B), and that of Nef protein was more than 7-fold larger in JRFL-infected macrophages than in JRFL-infected CD4+ T cells (Figure 6C). There was no difference in the amount of p24 or Nef protein between NL-432–infected CD4+ T cell and JRFL-infected CD4+ T cells (Figure 6D). Such results indicate that HIV-1–infected macrophages can synthesize much more HIV-1 protein than HIV-1–infected CD4+ T cells. Thus, it is likely that the difference in HIV-1 antigen presentation between the 2 cells resulted from the difference in the production of HIV-1 epitope peptide, because the difference in HIV-1 protein expression was much larger than that in HLA class I expression. These results suggest that HIV-1–infected macrophages can present a sufficient amount of peptide-MHC class I complexes for CTL recognition in spite of Nef-mediated down-regulation of HLA class I molecules.
Different expression of HLA class I molecules and HIV-1 proteins between CD4+ T cells and macrophages infected with HIV-1. (A) Comparison of the susceptibility between CD4+ T cells and macrophages for cytotoxic activity of HIV-1–specific CTL clones. Cytotoxic activity of HLA-B*5101-restricted CTL clones was examined for CD4+ T cells and macrophages prepulsed with each epitope peptide at an E/T ratio of 2:1. Data shown in the figure are averages of triplicate assays for each CTL clone. (B-C) The expression of p24 and Nef proteins in JRFL-infected CD4+ T cells and macrophages. After p24+ cells had become 20% to 30% of the total cell population, these cells were lysed. The cell lysates (6 μg) were analyzed by Western blotting with anti-p24 or anti-Nef mAb. Relative protein expression indicates the ratio of the amount of the p24 and Nef proteins in JRFL or JR-Xh–infected macrophages to that in JRFL-infected CD4+ T cells per equal cell number. Data are shown as the average for 3 independent experiments. (D) The expression of p24 and Nef proteins in CD4+ T cells infected with either NL-432 or JRFL. Data are shown as the average ± SD for 3 independent experiments.
Different expression of HLA class I molecules and HIV-1 proteins between CD4+ T cells and macrophages infected with HIV-1. (A) Comparison of the susceptibility between CD4+ T cells and macrophages for cytotoxic activity of HIV-1–specific CTL clones. Cytotoxic activity of HLA-B*5101-restricted CTL clones was examined for CD4+ T cells and macrophages prepulsed with each epitope peptide at an E/T ratio of 2:1. Data shown in the figure are averages of triplicate assays for each CTL clone. (B-C) The expression of p24 and Nef proteins in JRFL-infected CD4+ T cells and macrophages. After p24+ cells had become 20% to 30% of the total cell population, these cells were lysed. The cell lysates (6 μg) were analyzed by Western blotting with anti-p24 or anti-Nef mAb. Relative protein expression indicates the ratio of the amount of the p24 and Nef proteins in JRFL or JR-Xh–infected macrophages to that in JRFL-infected CD4+ T cells per equal cell number. Data are shown as the average for 3 independent experiments. (D) The expression of p24 and Nef proteins in CD4+ T cells infected with either NL-432 or JRFL. Data are shown as the average ± SD for 3 independent experiments.
Discussion
Previous studies showed that HIV-1–specific CTLs can kill HIV-1–infected alveolar macrophages derived from HIV-1–infected individuals but that they failed to kill HIV-1–infected CD4+ T cells.7,14,15 These results imply that HIV-1–infected macrophages can present HIV-1 antigens more effectively than HIV-1–infected CD4+ T cells. Our previous studies using NL-432 X4 clone and NL-M20A lacking Nef function for HLA class I molecules showed that most HIV-1–infected CTLs failed to kill NL-432–infected CD4+ T cells and partially suppressed NL-432 replication but that they could effectively kill NL-M20A–infected CD4+ T cells and completely suppress NL-M20A replication,8,9 indicating that Nef-mediated HLA class I down-regulation critically affects recognition of HIV-1–infected CD4+ T cells by HIV-1–specific CTLs. These studies together suggest that the assay measuring the ability of HIV-1–specific CTLs to suppress HIV-1 replication is more sensitive than the cytotoxic assay and imply that the effect of Nef-mediated HLA class I down-regulation is much stronger on the recognition by HIV-1–specific CTLs of HIV-1–infected CD4+ T cells than that of HIV-1–infected macrophages. In fact, we here demonstrated that HIV-1–specific CTLs much more strongly suppressed JRFL replication in the culture of HIV-1–specific CTLs with JRFL-infected macrophages than that in those of HIV-1–specific CTLs with JRFL-infected CD4+ T cells. Thus, the present study indicates that Nef-mediated HLA class I down-regulation only partially affected recognition of HIV-1–infected macrophages by HIV-1–specific CTLs.
The difference in the suppressive effect of HIV-1–specific CTLs on HIV-1 replication between macrophages and CD4+ T cells may be explained by several mechanisms such as differences of HLA class I surface expression and HIV-1 protein expression between macrophages and CD4+ T cells. The present study demonstrated that the surface expression of HLA class I molecules on macrophages was approximately 2-fold higher than that on CD4+ T cells and that this difference weakly influenced ability of HIV-1–specific CTL clones to kill these cells prepulsed with the epitope peptides. These results suggest that the difference in HLA class I surface expression between these 2 cells only partially influenced that in the suppressive effect of HIV-1–specific CTLs on HIV-1 replication. On the other hand, we demonstrated that HIV-1 antigens were much more expressed in HIV-1–infected macrophages than in HIV-1–infected CD4+ T cells. Thus, it is likely that the difference in HIV-1 protein expression between the 2 cells resulted in the difference in HIV-1 antigen presentation since the difference in HIV-1 protein expression was much larger than that in HLA class I expression. Because macrophages are also known to carry costimulatory molecules and function as professional antigen-presenting cells, HIV-1–specific CTLs can effectively proliferate when stimulated by HIV-1–infected macrophages.
A previous study showed that most HIV-1–specific CTLs partially suppress NL-432 replication in NL-432–infected CD4+ T cells,8,9 whereas the present study exhibited that they also had similar ability to suppress JRFL replication in JRFL-infected CD4+ T cells, indicating that HIV-1–specific CTLs fail to suppress HIV-1 replication in CD4+ T cells in early and late phases of HIV-1 infection. In contrast, HIV-1–specific CTLs strongly suppressed HIV-1 replication in macrophages. These observations imply that HIV-1 replication is more controlled by the CTLs in the early phase than in the late stage.
It is well known that dendritic cells (DCs) play an important role in the transmission of HIV-1 to CD4+ T cells and in antigen presentation.23 DCs can present antigens to naïve T cells, whereas macrophages present antigens only to memory and effector T cells.24–27 These findings suggest that DCs present HIV-1 antigens to naïve T cells, so that HIV-1–specific effector and memory T cells are induced in the early stage of an HIV-1 infection. On the other hand, HIV-1–infected macrophages may play a role in maintenance of HIV-1–specific memory and effector T cells, because macrophages can stimulate memory and effector T cells but not naïve T cells. In fact, the present study demonstrated that HIV-1–infected macrophages stimulated HIV-1–specific CTL clones much more strongly than did HIV-1–infected CD4+ T cells, indicating that HIV-1–specific CD8+ T cells are maintained in HIV-1–infected donors due to stimulation by HIV-1–infected macrophages but not due to that by HIV-1–infected CD4+ T cells. In HIV-1–infected individuals, the number of DCs is decreased and their functional impairment is observed,28,29 suggesting that HIV-1–specific memory and effector T cells may be maintained by antigen presentation by HIV-1–infected macrophages rather than by DCs. Since the X4 virus, which infects only CD4+ T cells, dominantly appears in the late phase, the antigen presentation by HIV-1–infected macrophages would not be expected in this phase. In addition to the loss of HIV-1–specific helper T cells and DCs,28–32 this may be one of the mechanisms that mediates the reduction in the number of HIV-1–specific T cells and failure of suppression of HIV-1 replication in the late phase.
In the present study, we demonstrated a strong HIV-1 antigen presentation by HIV-1–infected macrophages and less effect of Nef-mediated HLA class I down-regulation on the recognition of HIV-1–infected macrophages by HIV-1–specific CD8+ T cells. HIV-1 R5 virus-infected macrophages could induce higher proliferation of HIV-1–specific CTLs. Antigen presentation by HIV-1–infected macrophages and DCs are major pathways for the induction of HIV-1–specific T cells in HIV-1–infected donors. Because HIV-1–infected macrophages are frequently detected in various tissues,33–36 they may be considered to be involved in the maintenance of HIV-1–specific acquired immunity in acute and early chronic phases of an HIV-1 infection. However, because HIV-1 expression depends on the activation statuses of the cells, it still remains unclear that HIV-1–infected macrophages can strongly express HIV-1 proteins and can strongly stimulate HIV-1–specific CTLs in vivo. Further studies of HIV-1–infected macrophages in vivo are necessary to clarify whether HIV-1–infected macrophages are strong professional antigen-presenting cells in vivo.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases launched as a project commissioned by the Ministry of Education, Science, Sports and Culture, Japan; by a grant-in aid for scientific research from the Ministry of Health, Japan; by a grant-in aid (no. 18390141) for scientific research from the Ministry of Education, Science, Sports and Culture, Japan; and a grant from the Japan Health Science Foundation. M.F. is a Japanese Society for the Promotion of Science (JSPS) Research Fellow.
The authors thank Dr Tomiyama, Hirokazu Koizumi, and Tomohiro Akahoshi for technical assistance, Dr Suzu for gift of M-CSF, and Sachiko Sakai for secretarial assistance.
Authorship
Contribution: M.F. performed experiments, analyzed data, and helped to write the manuscript; M.T. designed experiments and helped to write the manuscript.
Conflict-of-interest disclosure: The authors declare they have no competing financial interests.
Correspondence: Masafumi Takiguchi, Division of Viral Immunology, Center for AIDS Research, Kumamoto University, 2-2-1 Honjo, Kumamoto 860-0811, Japan; e-mail: masafumi@kumamoto-u.ac.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal