Abstract
Perforin is a cytolytic protein expressed mainly in activated cytotoxic lymphocytes and natural killer cells. Inherited perforin mutations account for 20% to 40% of familial hemophagocytic lymphohistiocytosis, a fatal disease of early childhood characterized by the absence of functional perforin. Aplastic anemia, the paradigm of immune-mediated bone marrow failure syndromes, is characterized by hematopoietic stem cell destruction by activated T cells and Th1 cytokines. We examined whether mutations in the perforin gene occurred in acquired aplastic anemia. Three nonsynonymous PRF1 mutations among 5 unrelated patients were observed. Four of 5 patients with the mutations showed some hemophagocytosis in the bone marrow at diagnosis. Perforin protein levels in these patients were very low or absent, and perforin granules were completely absent. Natural killer (NK) cell cytotoxicity from these patients was significantly decreased. Our data suggest that PRF1 genetic alterations help explain the aberrant proliferation and activation of cytotoxic T cells and may represent genetic risk factors for bone marrow failure.
Introduction
Aplastic anemia is characterized by peripheral blood pancytopenia and a hypocellular bone marrow.1 In most cases, aplastic anemia is an immune-mediated disease with active destruction of hematopoietic cells by activated cytotoxic T lymphocytes (CTLs) and increased IFN-γ levels2 ; natural killer (NK) cell numbers and NK cytolytic activity in vitro are decreased.3
NK cells and CTL can lyse tumor cells and virus-infected cells, but they also have activity against some normal cells that present self-antigens (such as immature dendritic cells), primarily to prevent autoimmunity. One of the pathways that NK cells and CTLs use to destroy target cells is release of cytolytic proteins. Perforin, a key component of this cytolytic process, is expressed mainly in CTLs and NK cells4,5 ; perforin is stored in cytoplasmic granules and is essential for killing by non–Fas-mediated mechanisms. Functional perforin is essential for normal CTL and NK cell function.6–9
We describe 4 unrelated patients with a mutation in exon 2 of PRF1 gene and 1 patient with a mutation in exon 3; all mutations were in the coding region of PRF1 and all but one gene carried also a polymorphism in exon 3. We hypothesize that genetic alterations in PRF1 gene may contribute to the development of acquired aplastic anemia.
Patients, materials, and methods
Patients and controls
Informed consent was acquired according to protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute for all patient samples.
DNA from 75 unrelated patients with acquired aplastic anemia were sequenced for PRF1. For controls, resequence analysis was performed in both the SNP500 Cancer set and the Human Genomic Diversity Panel (HGDP)10,11 (Document S1, available on the Blood website; see the Supplemental Document link at the top of the online article).
Nucleotide sequencing
Sequencing in DNA samples extracted from peripheral blood and buccal smear cells from patients and healthy controls was performed as previously described12 (Document S1).
Western blot analysis, confocal microscopy, and cytotoxicity
Statistical analysis
Differences in the frequencies of coding-sequence variations between samples from patients and those from healthy controls were evaluated by chi-square test using the Prism software (Prism Software, Irvine, CA).
Results and discussion
Patients with acquired aplastic anemia and perforin mutations
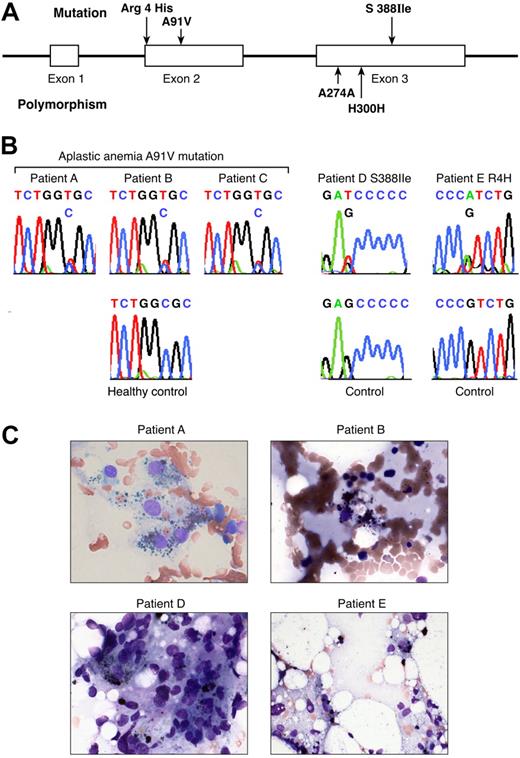
Of the 75 patients with acquired aplastic anemia examined, 3 novel, nonsynonymous mutations were identified in PRF1 in 5 patients (Document S1; Figure 1; Tables 1–2). In 3 patients, PRF1 sequencing revealed a heterozygous mutation in exon 2 in codon 91, with alanine replaced by valine (A91V). A previously known, common single nucleotide polymorphism in exon 3 (H300H) was also present. A novel mutation in exon 3 was identified in patient D: in codon 388, serine was replaced by isoleucine (S388I); this patient also carried a single nucleotide polymorphism in exon 3 (A274A). In patient E, a heterozygous mutation in exon 2 was identified: in codon 4, an arginine residue was replaced by histidine (R4H). The germ line origin of PRF1 mutations was established by their detection in DNA extracted from buccal mucosa specimens obtained from 4 of 4 patients (A, B, C, and D).
Perforin mutations in patients with aplastic anemia. (A) Linear structure of the PRF1 gene, which encodes the human perforin protein and aplastic anemia-associated mutations. The segments represent the exons. PRF1 has 3 exons, 2 of which (exons 2 and 3) contain coding sequences. In aplastic anemia, nonsynonymous mutations were found in exon 2 (codon 4 Arg/His and codon 91 Ala/Val) and exon 3 (codon 388 Ser/Ile). The polymorphisms in exon 3 are also shown (codons 274 and 300). R indicate arginine; H, histidine; I, isoleucine; V, valine; A, alanine. (B) Sequences of the patients carrying the mutations compared with sequences obtained from the controls. Green identifies adenine (A); red identifies thymidine (T); black identifies guanine (G); and blue identifies cytosine (C). (C) Bone marrow smear examination from the patients carrying PRF1 mutations. Four of 5 patients carrying mutations revealed hemophagocytosis in bone marrow at diagnosis.
Perforin mutations in patients with aplastic anemia. (A) Linear structure of the PRF1 gene, which encodes the human perforin protein and aplastic anemia-associated mutations. The segments represent the exons. PRF1 has 3 exons, 2 of which (exons 2 and 3) contain coding sequences. In aplastic anemia, nonsynonymous mutations were found in exon 2 (codon 4 Arg/His and codon 91 Ala/Val) and exon 3 (codon 388 Ser/Ile). The polymorphisms in exon 3 are also shown (codons 274 and 300). R indicate arginine; H, histidine; I, isoleucine; V, valine; A, alanine. (B) Sequences of the patients carrying the mutations compared with sequences obtained from the controls. Green identifies adenine (A); red identifies thymidine (T); black identifies guanine (G); and blue identifies cytosine (C). (C) Bone marrow smear examination from the patients carrying PRF1 mutations. Four of 5 patients carrying mutations revealed hemophagocytosis in bone marrow at diagnosis.
Screening for PRF1 gene mutations in patients with aplastic anemia and healthy controls
| Location of variation . | Patients with aplastic anemia n = 75 . | Healthy controls n = 1156 . | P . |
|---|---|---|---|
| Exon 2, codon 4 CGT/CAT (Arg/His), | |||
| No. of patients (% of allele frequency) | 1 (0.66) | 21 (0.96) | .7 |
| Exon 2, codon 91 GCG/GTG (Ala/Val) | |||
| No. of patients (% of allele frequency) | 3 (2) | 24 (1.1) | .27 |
| Exon 3, codon 388 AGC/ATC (Ser/Ile) | |||
| No. of patients (% of allele frequency) | 1 (0.66) | 0 (0) | .001 |
| Location of variation . | Patients with aplastic anemia n = 75 . | Healthy controls n = 1156 . | P . |
|---|---|---|---|
| Exon 2, codon 4 CGT/CAT (Arg/His), | |||
| No. of patients (% of allele frequency) | 1 (0.66) | 21 (0.96) | .7 |
| Exon 2, codon 91 GCG/GTG (Ala/Val) | |||
| No. of patients (% of allele frequency) | 3 (2) | 24 (1.1) | .27 |
| Exon 3, codon 388 AGC/ATC (Ser/Ile) | |||
| No. of patients (% of allele frequency) | 1 (0.66) | 0 (0) | .001 |
The controls included 1036 from the HGDP, 102 from the SNP500 Cancer set, and 18 anonymous healthy persons.
Clinical characteristics and laboratory profiles of patients carrying the PRF1 mutations
| Patient . | Mutation . | Age, y . | Race or ethnic group* . | Hemoglobin level, g/dL . | Absolute neutrophil count, × 10−3/mm3 . | Platelet count, 10−3/mm3 . | Bone marrow cellularity, % . | Hemophagocytosis . | Follow-up . |
|---|---|---|---|---|---|---|---|---|---|
| A | A91V/H300H | 31 | White | 8.0 | 25 | 5 000 | 5 | Yes | No response to immunosuppression |
| B | A91V/H300H | 77 | White | 8.8 | 95 | 11 000 | 15 | Yes | Relapse; CsA dependent |
| C | A91V/H300H | 78 | White | 10.0 | 940 | 7 000 | 10 | No | Transient response; relapse; CsA dependent |
| D | S388I/A274A | 33 | Hispanic | 5.8 | 950 | 9 000 | 10 | Yes | No response to immunosuppression |
| E | R4H | 21 | African American | 4.1 | 780 | 6 000 | 5 | Yes | No response to immunosuppression |
| Patient . | Mutation . | Age, y . | Race or ethnic group* . | Hemoglobin level, g/dL . | Absolute neutrophil count, × 10−3/mm3 . | Platelet count, 10−3/mm3 . | Bone marrow cellularity, % . | Hemophagocytosis . | Follow-up . |
|---|---|---|---|---|---|---|---|---|---|
| A | A91V/H300H | 31 | White | 8.0 | 25 | 5 000 | 5 | Yes | No response to immunosuppression |
| B | A91V/H300H | 77 | White | 8.8 | 95 | 11 000 | 15 | Yes | Relapse; CsA dependent |
| C | A91V/H300H | 78 | White | 10.0 | 940 | 7 000 | 10 | No | Transient response; relapse; CsA dependent |
| D | S388I/A274A | 33 | Hispanic | 5.8 | 950 | 9 000 | 10 | Yes | No response to immunosuppression |
| E | R4H | 21 | African American | 4.1 | 780 | 6 000 | 5 | Yes | No response to immunosuppression |
The peripheral blood and bone marrow counts at diagnosis (Document SI) are shown. All patients were men with a diagnosis of severe aplastic anemic. CsA indicates cyclosporine A.
Race or ethnic group was self-reported by the patients.
In the normal immune response, antigen-presenting cells (APCs) can be eliminated through perforin-dependent cytotoxicity. The pathophysiology of familial hemophagocytic lymphohistiocytosis (FHLH), a fatal disease of the early childhood, is explained by the absence of perforin.7,8 T cells receive activation signals through the APCs, but the APCs and the activated T cells are not eliminated, resulting in uncontrolled expansion of CTLs and CD34 destruction.13 Perforin gene mutations account for 20% to 40% of FHLH cases linked to 10q21-22 (FHLH2).14 Perforin consists of a leader and a lytic peptide, 2 regions of low homology, a conserved amphipathic α-helix, an EGF-like domain, a C2 domain, and a cleavable C terminus. Mutations have been identified in all domains but the cleavable C terminus9 ; approximately 48 different mutations in PRF1 have been identified in FHLH2.15 In idiopathic aplastic anemia, T cells are activated,1,2,16 and here we examined whether perforin was involved in this activated T-cell phenotype.
The A91V mutation in exon 2 has been argued to be functionally important despite that it has also been reported to be a polymorphism in the general population, with an allele frequency ranging between 3% and 17%.17–20 Our analysis of the A91V site in 52 worldwide populations with more than 1000 unrelated persons of diverse backgrounds revealed an overall prevalence of approximately 1% (we did not see an enrichment or increased prevalence in a particular geographic region or ethnic group). To our knowledge, we describe for the first time PRF1 gene mutations in adults older than 30 years of age who had developed a significant hematologic disease. A communication describes atypical presentation of FHLH in 2 siblings in their mid-20s carrying the A91V mutation.21 The A91V mutation may have a milder phenotype and accounting for later onset of clinical manifestations but, nevertheless, participate in the development of catastrophic immune-mediated syndromes.20,22 Of note, alanine at position 91 is conserved among human, mouse, and rat perforin protein.9
Although the number of patients in our study is too small to draw a definitive conclusion, 4 of 5 patients with aplastic anemia with PRF1 mutations showed evidence for hemophagocytosis in the bone marrow when first diagnosed, but there were no other typical clinical features of hemophagocytic syndrome9,15,23,24 (Figure 1C). Historically, some elements of hemophagocytosis in bone marrow aspirates in severe aplastic anemia has long been recognized but may be underdescribed because of the dramatic empty marrow appearance that is pathognomonic of the disease.25 Possibly PRF1 gene mutations are related to a more severe phenotype of marrow failure, for which hemophagocytosis may be a morphologic marker of prognosis, because none of our patients with PRF1 mutations experienced hematologic recovery with immunosuppressive treatment.
Perforin protein levels and cytolytic activity
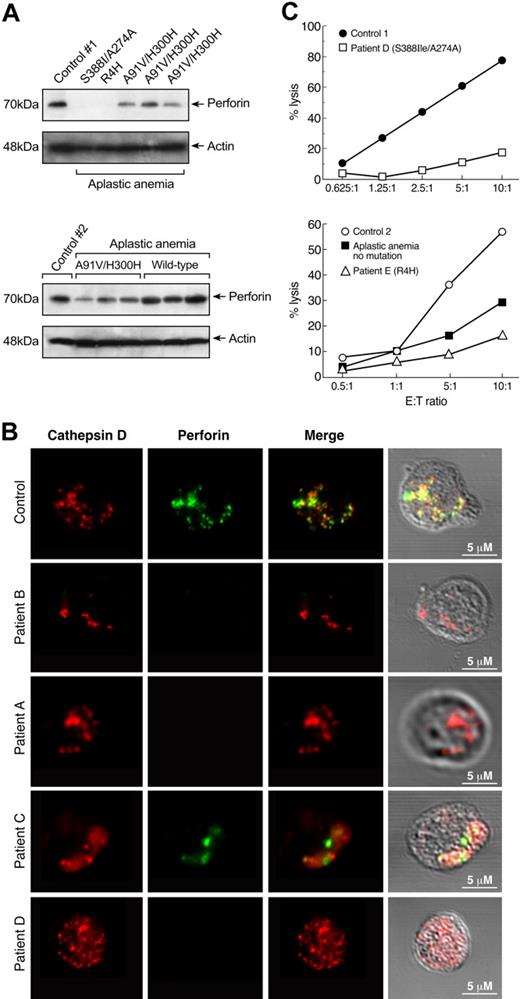
PRF1 mutations in patients with aplastic anemia appear to be functionally important because they associate with low perforin protein levels and impaired cytolytic activity. Perforin protein levels were markedly reduced or absent in all patients carrying the PRF1 mutations (n = 5) compared with the T cells of healthy controls (n = 8; Figure 2A). All but one of the patients with aplastic anemia without PRF1 mutations (n = 7) had similar amounts of perforin protein compared with healthy controls. Three patients with myelodysplastic syndrome and 2 patients with pure red cell aplasia, who served as disease controls, showed perforin protein levels comparable to those from healthy controls (data not shown).
Decreased perforin protein levels and cytolytic activity in patients carrying PRF1 mutations. (A) Cytoplasmic extracts from T cells of patients with aplastic anemia with PRF1 mutations (n = 5) and healthy controls (n = 8) were analyzed for perforin protein expression by immunoblot. All 5 patients carrying PRF1 mutations revealed decreased or absent perforin protein levels. All samples were run side by side. The upper panel (from left to right) shows the perforin protein levels from a healthy person and patients D, E, A, B, and C. The lower panel shows the immunoblot results from chronologically different samples obtained from the same patients (from left to right a healthy control, patient C, patient A, and patient B, and 3 patients with no mutations). Patients without PRF1 mutations revealed perforin protein levels comparable to that from healthy volunteers. (B) Decreased perforin granules in cytotoxic cells from patients with aplastic anemia with PRF1 mutations. CD8+ T cells from patients carrying PRF1 mutations and healthy controls were double stained in parallel with anti-cathepsin D (red, first column) and anti-perforin (green, second column) antibodies in at least 3 different experiments. Perforin and cathepsin D showed the expected pattern of colocalization in cytotoxic granules of control cells. Staining of the patients' cytotoxic cells revealed complete absence of perforin granules in patients A, B, and D (Figure 4); patient C showed slightly decreased staining of perforin granules, but they were abnormal in size. (Differences between perforin protein levels and perforin granule staining in patients A, B, and C, who have the same PRF1 mutation may be explained by different genetic background or additional unshared genetic alterations). The frequency of cathepsin-D in CD8+ T cells was similar in patients' and controls' samples. (C) Decreased cytolytic activity in patients with aplastic anemia. Natural killer cells were isolated from patients D and E, and their killing efficiency was determined in a standard Cr51-release assay against K562 target cells in comparison to cells from healthy donors. Patients carrying PRF1 mutations showed markedly decreased cytolytic activity compared with controls. A patient (not carrying any mutation or polymorphism) also showed decreased cytolytic activity but not as low as that observed in the patients with PRF1 mutations.
Decreased perforin protein levels and cytolytic activity in patients carrying PRF1 mutations. (A) Cytoplasmic extracts from T cells of patients with aplastic anemia with PRF1 mutations (n = 5) and healthy controls (n = 8) were analyzed for perforin protein expression by immunoblot. All 5 patients carrying PRF1 mutations revealed decreased or absent perforin protein levels. All samples were run side by side. The upper panel (from left to right) shows the perforin protein levels from a healthy person and patients D, E, A, B, and C. The lower panel shows the immunoblot results from chronologically different samples obtained from the same patients (from left to right a healthy control, patient C, patient A, and patient B, and 3 patients with no mutations). Patients without PRF1 mutations revealed perforin protein levels comparable to that from healthy volunteers. (B) Decreased perforin granules in cytotoxic cells from patients with aplastic anemia with PRF1 mutations. CD8+ T cells from patients carrying PRF1 mutations and healthy controls were double stained in parallel with anti-cathepsin D (red, first column) and anti-perforin (green, second column) antibodies in at least 3 different experiments. Perforin and cathepsin D showed the expected pattern of colocalization in cytotoxic granules of control cells. Staining of the patients' cytotoxic cells revealed complete absence of perforin granules in patients A, B, and D (Figure 4); patient C showed slightly decreased staining of perforin granules, but they were abnormal in size. (Differences between perforin protein levels and perforin granule staining in patients A, B, and C, who have the same PRF1 mutation may be explained by different genetic background or additional unshared genetic alterations). The frequency of cathepsin-D in CD8+ T cells was similar in patients' and controls' samples. (C) Decreased cytolytic activity in patients with aplastic anemia. Natural killer cells were isolated from patients D and E, and their killing efficiency was determined in a standard Cr51-release assay against K562 target cells in comparison to cells from healthy donors. Patients carrying PRF1 mutations showed markedly decreased cytolytic activity compared with controls. A patient (not carrying any mutation or polymorphism) also showed decreased cytolytic activity but not as low as that observed in the patients with PRF1 mutations.
To confirm the absence of perforin protein in patients carrying PRF1 mutations, we examined cultured CD8+ T cells from patients A, B, C, and D in comparison to healthy controls (n = 5) (sequencing for PRF1 in these controls did not reveal mutations or polymorphisms) by confocal microscopy. Staining of the patients' cytotoxic cells revealed complete absence of perforin granules in patients A, B, and D (Figure 2B); patient C showed slightly decreased staining of perforin granules (Figure 2B). Patients with PRF1 mutations also showed markedly decreased cytolytic activity (Figure 2C; aplastic anemia patients not carrying mutations (n = 2) also showed decreased cytolytic activity,3 although not as low as observed in the patients with PRF1 mutations).
Mechanistically, PRF1 gene mutations may help explain the aberrant proliferation and activation of CTLs. Additional genetic variations in genes implicated in the homeostasis of the immune system and the down-regulation of an immune response cannot be excluded.1 Here, we show that mutations in an immune regulatory mechanism previously identified in young children can manifest in adults without typically associated clinical findings or a suggestive family history, providing a further link between constitutional and acquired bone marrow failure syndromes.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all patients for donating blood samples. We thank our research nurses Olga Nunez and Barbara Weinstein for collection of the samples, and Faith Williams for help in preparing the figures. The help from all our laboratory members is appreciated.
This work was supported by the Intramural Research Program of the National Institutes of Health.
Authorship
Contribution: E.E.S. designed research, performed research, analyzed data, and wrote the paper; F.G. performed research and analyzed data; B.S. performed research and analyzed data; D.M. collected and analyzed data; M.B. performed research and analyzed data; V.V. performed research; S.G. provided technical support; R.C. analyzed data and revised the paper; S.J.C. designed research, analyzed data, and revised the paper; N.S.Y. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elena E Solomou, NIH, Hematology Branch, Building 10 CRC, Rm 3E-5216, 10 Center Dr, Bethesda, MD 20892; e-mail: solomoue@nhlbi.nih.gov or elenasolomou@hotmail.com.