Abstract
Of 11 children with juvenile myelomonocytic leukemia (JMML) carrying RAS mutations (8 with NRAS mutations, 3 with KRAS2 mutations), 5 had a profound elevation in either or both the white blood cells and spleen size at diagnosis. Three patients had no or modest hepatosplenomegaly and mild leukocytosis at presentation but subsequently showed a marked increase in spleen size with or without hematologic exacerbation, for which nonintensive chemotherapy was initiated. The other three patients with NRAS or KRAS2 glycine to serine substitution received no chemotherapy, but hematologic improvement has been observed during a 2- to 4-year follow up. In the third group, all hematopoietic cell lineages analyzed had the RAS mutations at the time of hematologic improvement, whereas DNA obtained from the nails had the wild type. Additionally, numbers of circulating granulocyte-macrophage progenitors were significantly reduced during the clinical course. Thus, some patients with JMML with specific RAS mutations may have spontaneously improving disease.
Introduction
Somatic point mutations of the RAS genes at codons 12, 13, and 61 (NRAS and KRAS2) are found in approximately 20% of patients with juvenile myelomonocytic leukemia (JMML).1,2 Other patients show inactivation of NF1 or PTPN11 mutations.3–5
Although most patients with JMML die from progressive disease unless treated with hematopoietic stem cell transplantation, there are a few patients who have been reported to spontaneously recover without intervention.6,7 Some of these children have JMML associated with Noonan syndrome, but others do not. So far, the individual prognosis in JMML-carrying specific genetic aberrations remains unclear. We report the clinical course in 11 patients with RAS mutations.
Materials and methods
This study was approved by the Institutional Review Board of Shinshu University. Informed consent was obtained from the guardians of the patients following institutional guidelines.
Cell preparation
We used peripheral blood (PB) or bone marrow (BM) mononuclear cells (MNCs) that had been frozen with liquid nitrogen. CD3- and CD56-positive PB cells were separated immunomagnetically.8 Ninety-nine percent of the isolated cells were CD3- or CD56-positive according to a flow cytometric analysis.
Clonal cell culture
Twenty thousand PB or BM MNCs were plated in a dish containing methylcellulose medium supplemented with or without 0.01 to 10 ng/mL of granulocyte-macrophage colony-stimulating factor (GM-CSF).9 To examine the clonal derivation of myeloid and erythroid lineages, 2000 CD34+ PB cells harvested immunomagnetically were cultured in methylcellulose medium supplemented with GM-CSF, stem cell factor, interleukin 3, and erythropoietin. Twelve days after incubation in 5% CO2, GM colonies, erythroid colonies, and mixed colonies were individually lifted and prepared as single cell suspensions. Sequence analyses were then performed on individual colony-constituent cells.
Detection of NRAS and KRAS2 mutations
DNA was extracted from PB or BM MNCs and nails. Exon 1 (codons 12 and 13) and exon 2 (codon 61) of NRAS and KRAS2 genes were amplified by polymerase chain reaction (PCR) using primer pairs described previously.10,11 The PCR products were subjected to direct sequencing from both directions on an automatic DNA sequencer.
Results and discussion
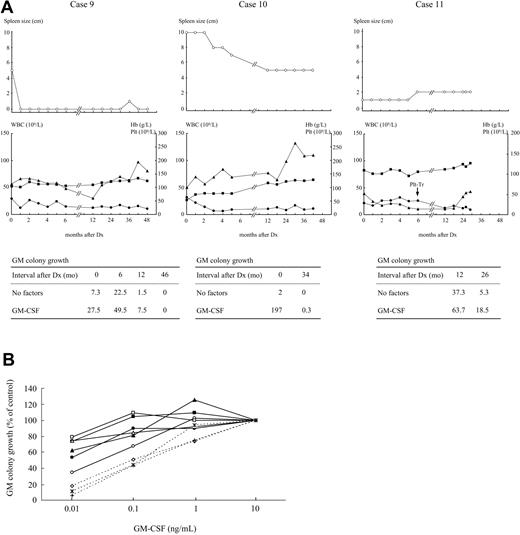
Among a total of 80 children with JMML, 13 children had RAS mutations (10 had NRAS mutations, three had KRAS2 mutations) with the wild type of the PTPN11 gene. All the patients with RAS mutations met the criteria proposed by the International JMML Working Group12 but did not have clinical features of neurofibromatosis type I. Clinical characteristics of 11 patients for whom all clinical information was available are presented in Table 1. Five patients (cases no. 1 to 5) were treated with allogenic bone marrow transplantation or nonintensive chemotherapy, including 6-mercaptopurine (6-MP), at or soon after admission, based on marked leukocytosis/splenomegaly (>6 cm) alone or in combination with the existence of a related donor. Hematologic abnormalities and hepatosplenomegaly in three patients (cases no. 6 to 8) were modest at first presentation. A marked enlargement of spleen size, however, was noted within 2 to 3 months after diagnosis in all 3 patients, followed by deterioration of leukocytosis in case no. 7 and of anemia/thrombocytopenia in case no. 8. Accordingly, oral 6-MP was administered to these three patients 3 to 9 months after diagnosis. In three other patients (cases no. 9 to 11), the hematologic findings were similar to those of cases no. 6 to 8 at first presentation. There was no significant change or a decrease of spleen size within 3 month of diagnosis in cases no. 9 to 11, as shown in Figure 1A. Surprisingly, in these patients, hematologic improvement was observed over a 2- to 4 year follow-up despite no chemotherapy. Thus, patients with JMML carrying RAS mutations appear to be heterogeneous with respect to the hematologic progression.
Clinical characteristics of 11 patients with JMML with RAS mutations
| Case (no) . | Mutations . | Age (mo) . | Sex . | Liver (cm) . | Spleen (cm) . | WBC (109/L) . | Mono (%) . | Hb (g/dL) . | Plt (109/L) . | HbF (%) . | Treatment (Interval after diagnosis) . | Outcome (Interval after diagnosis) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NRAS, codon 61 CAA (Gln) > CTA (Leu) | 1 | M | 3 | 2 | 151.0 | 31.6 | 8.4 | 34 | 65 | famil BMT (2 mo) | dead (2 mo) |
| 2 | KRAS, codon 13 GGC (Gly) > GAC (Asp) | 7 | F | 6 | 10 | 17.9 | 9 | 6.4 | 15 | 16.8 | sib BMT (5 mo) | alive (+43 mo) |
| 3 | NRAS, codon 12 GGT (Gly) > GAT (Asp) | 12 | F | 10 | 8 | 61.0 | 21 | 6.6 | 12 | 50.4 | 6-MP+Ara-C+VP-16 (0 mo), UCBT (2 mo) | alive (+22 mo) |
| 4 | NRAS, codon 13 GGT (Gly) > GAT (Asp) | 4 | M | 3 | 10 | 72.3 | 21 | 8.1 | 65 | 13.2 | 6-MP (0 mo), UBMT in preparation | alive (+44 mo) |
| 5 | NRAS, codon 12 GGT (Gly) > GAT (Asp) | 2 | M | 2 | 6.5 | 84.0 | 22 | 11.6 | 110 | 25.1 | 6-MP (1 mo) | alive (+4 mo) |
| 6 | KRAS, codon 12 GGT (Gly) > GAT (Asp) | 22 | F | 0 | 0 | 28.8 | 11 | 11.6 | 41 | 20.1 | 6-MP (3 mo), Pred (6 mo) | dead (6 mo) |
| 7 | NRAS, codon 12 GGT (Gly) > TGT (Cys) | 2 | M | 4 | 1.5 | 32.4 | 19 | 10.6 | 90 | 8.9 | 6-MP (6 mo), UBMT (36 mo) | dead (47 mo) |
| 8 | NRAS, codon 13 GGT (Gly) > GAT (Asp) | 12 | M | 0 | 0 | 14.6 | 10 | 8.8 | 52 | 4.5 | 6-MP (9 mo), UCBT (11 mo) | alive (+16 mo) |
| 9 | NRAS, codon 12 GGT (Gly) > AGT (Ser) | 10 | M | 4 | 5 | 29.4 | 16.5 | 10.5 | 113 | 0.5 | None | alive (+50 mo) |
| 10 | NRAS, codon 12 GGT (Gly) > AGT (Ser) | 10 | M | 5 | 10 | 31.8 | 20 | 5.4 | 100 | 1.7 | None | alive (+42 mo) |
| 11 | KRAS, codon 12 GGT (Gly) > AGT (Ser) | 4 | F | 4 | 1 | 21.2 | 8 | 11 | 52 | 8.8 | None | alive (+30 mo) |
| Case (no) . | Mutations . | Age (mo) . | Sex . | Liver (cm) . | Spleen (cm) . | WBC (109/L) . | Mono (%) . | Hb (g/dL) . | Plt (109/L) . | HbF (%) . | Treatment (Interval after diagnosis) . | Outcome (Interval after diagnosis) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NRAS, codon 61 CAA (Gln) > CTA (Leu) | 1 | M | 3 | 2 | 151.0 | 31.6 | 8.4 | 34 | 65 | famil BMT (2 mo) | dead (2 mo) |
| 2 | KRAS, codon 13 GGC (Gly) > GAC (Asp) | 7 | F | 6 | 10 | 17.9 | 9 | 6.4 | 15 | 16.8 | sib BMT (5 mo) | alive (+43 mo) |
| 3 | NRAS, codon 12 GGT (Gly) > GAT (Asp) | 12 | F | 10 | 8 | 61.0 | 21 | 6.6 | 12 | 50.4 | 6-MP+Ara-C+VP-16 (0 mo), UCBT (2 mo) | alive (+22 mo) |
| 4 | NRAS, codon 13 GGT (Gly) > GAT (Asp) | 4 | M | 3 | 10 | 72.3 | 21 | 8.1 | 65 | 13.2 | 6-MP (0 mo), UBMT in preparation | alive (+44 mo) |
| 5 | NRAS, codon 12 GGT (Gly) > GAT (Asp) | 2 | M | 2 | 6.5 | 84.0 | 22 | 11.6 | 110 | 25.1 | 6-MP (1 mo) | alive (+4 mo) |
| 6 | KRAS, codon 12 GGT (Gly) > GAT (Asp) | 22 | F | 0 | 0 | 28.8 | 11 | 11.6 | 41 | 20.1 | 6-MP (3 mo), Pred (6 mo) | dead (6 mo) |
| 7 | NRAS, codon 12 GGT (Gly) > TGT (Cys) | 2 | M | 4 | 1.5 | 32.4 | 19 | 10.6 | 90 | 8.9 | 6-MP (6 mo), UBMT (36 mo) | dead (47 mo) |
| 8 | NRAS, codon 13 GGT (Gly) > GAT (Asp) | 12 | M | 0 | 0 | 14.6 | 10 | 8.8 | 52 | 4.5 | 6-MP (9 mo), UCBT (11 mo) | alive (+16 mo) |
| 9 | NRAS, codon 12 GGT (Gly) > AGT (Ser) | 10 | M | 4 | 5 | 29.4 | 16.5 | 10.5 | 113 | 0.5 | None | alive (+50 mo) |
| 10 | NRAS, codon 12 GGT (Gly) > AGT (Ser) | 10 | M | 5 | 10 | 31.8 | 20 | 5.4 | 100 | 1.7 | None | alive (+42 mo) |
| 11 | KRAS, codon 12 GGT (Gly) > AGT (Ser) | 4 | F | 4 | 1 | 21.2 | 8 | 11 | 52 | 8.8 | None | alive (+30 mo) |
Liver and spleen sizes are given in cm below the costal margin. Case no. 1 had monosomy 7 at diagnosis. Case nos. 4 and 7 had a normal karyotype at presentation, but monosomy 7 approximately 3 years after diagnosis. The other 8 patients had normal karyotype. Ara-C, cytarabine; famil BMT, related nonsibling bone marrow transplantation; Hb F, hemoglobin F; mo, month(s); Mono, monocyte; 6-MP, 6-mercaptopurine; Plt, platelet count; Pred, prednisolone; sib BMT, sibling bone marrow transplantation; UBMT, unrelated bone marrow transplantation; UCBT, unrelated cord blood transplantation; VP-16, etoposide; WBC, white blood cell count.
Clinical course and dose response to GM-CSF of colony growth in patients with JMML withRASglycine to serine substitution. (A) Clinical course and time course study of circulating GM progenitors of cases no. 9 to 11. Mean numbers of GM colonies generated by 2 × 104 PB MNCs under stimulation with or without 10 ng/mL of GM-CSF are shown. Mean numbers of GM colonies in 4 normal controls were 0 ± 0 for no factors and 0.1 ± 0.1 for GM-CSF. Dx, diagnosis; Plt-Tr, platelet transfusion. •, WBC; ■, Hb values; ▴, platelet counts. (B) Comparison of proliferative response of GM progenitors to low doses of GM-CSF among patients with JMML with NRAS or KRAS2 glycine to serine substitution, patients with JMML with the other mutations, and normal controls. Twenty thousand PB or BM MNCs were cultured in a well-containing methylcellulose culture medium supplemented with GM-CSF at concentrations of 0.01 ng/mL to 10 ng/mL. The data are expressed as a percentage of the colony growth obtained with 10 ng/mL of GM-CSF in each case. The colony growth stimulated with 0.01 ng/mL and 0.1 ng/mL of GM-CSF was significantly higher in 3 patients with Gly12Ser mutation in the RAS gene than in normal controls but comparable to the value obtained from the 3 other mutants. ∘, case no. 5; □, case no. 6; ▵, case no. 8; •, case no. 9; ■, case no. 11; ▴, The other patient with NRAS glycine to serine substitution; *, normal control 1; ◇, normal control 2; †, normal control 3.
Clinical course and dose response to GM-CSF of colony growth in patients with JMML withRASglycine to serine substitution. (A) Clinical course and time course study of circulating GM progenitors of cases no. 9 to 11. Mean numbers of GM colonies generated by 2 × 104 PB MNCs under stimulation with or without 10 ng/mL of GM-CSF are shown. Mean numbers of GM colonies in 4 normal controls were 0 ± 0 for no factors and 0.1 ± 0.1 for GM-CSF. Dx, diagnosis; Plt-Tr, platelet transfusion. •, WBC; ■, Hb values; ▴, platelet counts. (B) Comparison of proliferative response of GM progenitors to low doses of GM-CSF among patients with JMML with NRAS or KRAS2 glycine to serine substitution, patients with JMML with the other mutations, and normal controls. Twenty thousand PB or BM MNCs were cultured in a well-containing methylcellulose culture medium supplemented with GM-CSF at concentrations of 0.01 ng/mL to 10 ng/mL. The data are expressed as a percentage of the colony growth obtained with 10 ng/mL of GM-CSF in each case. The colony growth stimulated with 0.01 ng/mL and 0.1 ng/mL of GM-CSF was significantly higher in 3 patients with Gly12Ser mutation in the RAS gene than in normal controls but comparable to the value obtained from the 3 other mutants. ∘, case no. 5; □, case no. 6; ▵, case no. 8; •, case no. 9; ■, case no. 11; ▴, The other patient with NRAS glycine to serine substitution; *, normal control 1; ◇, normal control 2; †, normal control 3.
As presented in Figure 1B, there was no significant difference in GM colony growth stimulated with low doses of GM-CSF between the patients with the NRAS or KRAS2 glycine to serine substitution and the other mutants of RAS, including glycine to aspartic acid substitution. Interestingly, numbers of GM colonies grown from 2×104 PB MNCs in the absence or presence of 10 ng/mL GM-CSF were significantly reduced approximately 2 to 4 years after presentation, compared with the values at diagnosis in cases no. 9 and 10 or with the values 1 year after diagnosis in case no. 11 (Figure 1A).
A possible explanation for the spontaneous improvement of hematologic findings in cases no. 9 to 11 is a decline in the number of cells harboring RAS mutation and an increase in the number of normal cells during the clinical course. Sequence analyses at the time of hematologic improvement revealed that the mutation was present in PB CD3+ cells and CD56+ cells as well as CD3−CD56− cells in the 3 patients. The RAS mutations were also detected in all of 15 to 25 GM colonies, 11 to 25 erythroid colonies, and 3 to 5 mixed colonies derived from PB CD34+ cells harvested simultaneously. Identical mutations were detectable at mRNA levels in PB MNCs and individual GM/erythroid/mixed colonies in cases no. 10 and 11. Therefore, it is unlikely that the normal cell population coexisted with the abnormal clone and expanded.
A second possibility is that hematologic abnormalities are a manifestation of germline RAS mutations, because patients with Noonan syndrome and germline PTPN11 or RAS mutations have been reported to have a JMML-like myeloproliferative disease.13,14 Additionally, germline HRAS mutations are observed in individuals with Costello syndrome who often develop benign tumors and cancers.15 In cases no. 9 to 11, DNA obtained from their nails had wild type NRAS or KRAS2 gene, whereas hematopoietic cells had a point mutation. Thus, the 3 patients had a somatic and not germline Gly12Ser mutation in the RAS gene.
Niemeyer et al16 proposed age, platelet count, and fetal hemoglobin (HbF) at diagnosis as factors for predicting the length of survival. Cases no. 9 to 11 were younger than 24 months of age and had platelet counts of >33× 109/L and HbF of <15% at onset. On the other hand, case nos. 6 to 8 also had no risk factors at diagnosis except for the HbF value in case no. 6. One possible parameter distinguishing between the 2 groups is change of spleen size within 3 month of diagnosis.
Among 32 patients with JMML with NRAS or KRAS2 mutations reported by us and others,2,5,17,18 codon 12 in the NRAS gene was the most commonly affected and variably substituted. Braun et al19 reported that somatic activation of a latent KRASG12D allele, the mutation identical to case no. 6, rapidly induces a fatal myeloproliferative disorder in mice. On the other hand, clinical and hematologic abnormalities improved despite no chemotherapy in our 3 patients with NRAS or KRAS2 glycine to serine substitution. Given the evidence that different substitutions produce HRAS alleles with different strength in the transforming potential,20,21 Gly12Ser mutation in the RAS gene may be insufficient to confer the disease progression in JMML. Close follow up is requisite, because these children still harbor a clonal oncogenic abnormality in their bone marrow.
In conclusion, no chemotherapy may be a recommended management for patients with JMML with NRAS or KRAS2 glycine to serine substitution, but further large-scale studies are needed to assess accurately the relationship between the mutational spectrum of the RAS gene and the disease phenotype.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Grants-in-Aid No. 11,390,300 from the Ministry of Education and by Grant-in-Aid for Cancer Research 16-3 from the Ministry of Health, Labour, and Welfare of Japan.
We thank Ms Yumiko Oguchi for her technical support.
Authorship
Contribution: K.K. designed and performed research, analyzed data, and wrote the paper; K.M. performed research, analyzed data, and wrote the paper; A.S., A.O., A.W., S.Y., S.I., K.K., F.Y., K.K., M.Y., and A.K. collected data; Y.O., E.H., and K.Y. analyzed data; and N.Y., M.T., R.Y., Y.N., M.S., A.M., and S.K. collected and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenichi Koike, Department of Pediatrics, Shinshu University School of Medicine, 3-1-1, Asahi, Matsumoto, 390-8621, Japan; e-mail: koikeken@hsp.md.shinshu-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal