Abstract
Three Gata transcription factors (Gata1, -2, and -3) are essential for hematopoiesis. These factors are thought to play distinct roles because they do not functionally replace each other. For instance, Gata2 messenger RNA (mRNA) expression is highly elevated in Gata1-null erythroid cells, yet this does not rescue the defect. Here, we test whether Gata2 and -3 transgenes rescue the erythroid defect of Gata1-null mice, if expressed in the appropriate spatiotemporal pattern. Gata1, -2, and -3 transgenes driven by β-globin regulatory elements, directing expression to late stages of differentiation, fail to rescue erythropoiesis in Gata1-null mutants. In contrast, when controlled by Gata1 regulatory elements, directing expression to the early stages of differentiation, Gata1, -2, and -3 do rescue the Gata1-null phenotype. The dramatic increase of endogenous Gata2 mRNA in Gata1-null progenitors is not reflected in Gata2 protein levels, invoking translational regulation. Our data show that the dynamic spatiotemporal regulation of Gata factor levels is more important than their identity and provide a paradigm for developmental control mechanisms that are hard-wired in cis-regulatory elements.
Introduction
The Gata transcription factor family in mammals is composed of 6 members (Gata1-6). These transcription factors are characterized by the presence of 2 zinc finger domains that mediate DNA binding to (T/A)GATA(A/G) consensus sequences.1 They have highly homologous zinc finger motifs, but little amino acid sequence homology is found outside these domains. Gata1, -2, and -3 constitute a subfamily because all 3 are expressed in hematopoietic cells.2 Gata1 is expressed in erythrocytes,3 megakaryocytes,4 eosinophils,5 and mast cells.4 Gene targeting studies show that Gata1 is required for normal erythroid differentiation. Loss of Gata1 activity results in a developmental arrest at the proerythroblast stage, causing these cells to undergo apoptosis.6 Consequently, Gata1-null mouse embryos die from severe anemia between gestational days 10.5 and 11.5 (E10.5—E11.5).7,8 A role for Gata1 in early erythroid-megakaryocyte progenitors has recently been demonstrated.9 Paradoxically, overexpression of Gata1 in the erythroid lineage also causes a lethal anemia. Gata1-overexpressing fetuses die around E12.5 to E13.5 because erythroid precursors fail to undergo terminal differentiation,10 suggesting a need for dynamic control of Gata1 activity during erythropoiesis.
The hematopoietic expression pattern of Gata2 overlaps extensively but not completely with that of Gata1. Gata2 is expressed in mast cells, megakaryocytes, and multilineage progenitors.3,11 Within the hematopoietic system, Gata3 expression is mainly in the T-cell lineage.12 Expression of Gata3 in multilineage progenitors is inferred from the hematopoietic deficiency in Gata3 knockout embryos.13 The overlapping expression patterns of Gata1, -2, and -3 suggest that in some hematopoietic cells, these transcription factors may have redundant functions. However, several studies show that a given Gata protein does not compensate for the absence of another. For example, in vitro hematopoietic differentiation of Gata1-null embryonic stem (ES) cells results in a 50-fold increase of Gata2 messenger RNA (mRNA) in erythroid cells, yet these cells are arrested in differentiation and die by apoptosis.6 Paradoxically, other Gata proteins can rescue the Gata1-null phenotype at least partially. In vitro erythroid differentiation of Gata1-null ES cells was restored when Gata3 and Gata4 were expressed under the control of the Gata1 promoter.14 The knockin of Gata3 complementary DNA (cDNA) into the Gata1 locus also partially rescues the Gata1-null phenotype,15 with increased survival of erythroid precursor cells and extension of embryo viability to E13.5. In addition, Gata2 and Gata3 transgenes under the control of Gata1 regulatory sequences can rescue the embryonic lethality of a Gata1 knockdown (G1.05) mutation.16 G1.05 mice die at E12.5. The compound transgene::G1.05 mice survive into adulthood. However, they show signs of anemia and present abnormal erythroid cells in peripheral blood. Because G1.05 mice express approximately 5% of wild-type Gata1 levels and, unlike Gata1-null proerythroblasts, G1.05 proerythroblasts do not undergo apoptosis,6,17 one interpretation of these results is that Gata2 and Gata3 contribute to the rescue of the G1.05 lethal phenotype but have no direct effect on erythroid differentiation. An alternative interpretation is that the correct spatiotemporal control of Gata activity is the most important parameter during erythroid differentiation, rather than the identity of the Gata factor expressed. This is supported by studies suggesting that the quantitative levels of Gata1 are important for erythroid lineage differentiation.10,17,18 Hence, to test during erythroid differentiation whether other factors can functionally substitute for Gata1 if expressed in a correct spatiotemporal and quantitative manner, we performed rescue experiments in mice carrying a complete Gata1-null mutation.19 We compared the potential of Gata1, Gata2, and Gata3 transgenes to rescue the Gata1-null phenotype when driven by 2 different transcriptional regulatory sequences: the β-globin promoter and Locus Control Region (βLCR), which is most highly active in late erythroid cells,20 or the Gata1 hematopoietic regulatory domain (HRD), which directs expression to the early stages of erythroid differentiation.16 We show here that the other Gata factors can rescue the Gata1-null phenotype when directed by the Gata1 HRD regulatory elements. Thus, we conclude that the correct spatiotemporal control of Gata activity is more important for erythroid development than the identity of the Gata factor expressed.
Materials and methods
Generation and genotyping of mice
Gata1-null mice were generated by breeding mice harboring a floxed Gata1 allele 19 with Zp3-Cre mice.21 Genomic DNA was analyzed by polymerase chain reaction (PCR) using primers for the Gata1 gene 5′G1: 5′-CGCCGAGCTGTGTGTAGTAA-3′ and 3′G1: 5′-TTCCTGTTTCTCCTCCTCCG-3′) located 5′ and 3′ of the first loxP site, and a primer located in the GFP gene (GFP: 5′-GGTGCTCAGGTAGTGGTTG-3′). Gata1 primers generate a 1.4-kilobase pair (kb) product (floxed Gata1 locus), whereas 5′G1 and GFP produce a 2.8-kb product (recombined Gata1 locus).
Myc-tagged Gata1, hemagglutinin (HA)-tagged Gata2, and HA-tagged Gata3 cDNAs were cloned in the pEV3 vector containing the human β-globin promoter and Locus Control Region 22 and used to generate transgenic mice. Genotyping was by Southern blot using specific probes for the cDNAs or by PCR using primers specific for human β-globin sequences (βIVS2-s: CAGTGTGGAAGTCTCAGGATCC; βIVS2-as GAATGGTGCAAAGAGGCATGA). Gata1.05 knockdown mice and HRD transgenic mice were described previously 16 ; here we used lines 801 (HRD-G1), 620 (HRD-G2) and 390 (HRD-G3).
Histologic staining
Cells were spun onto glass slides and air-dried. Slides were stained with 1% O-dianisidine (Sigma, St. Louis, MO) in methanol and Differential Quick Red and Blue staining.23 Images were captured with an Olympus BX40 microscope, 100×, 1.25 numerical aperture, oil immersion lens, fitted with a DP50 camera, using Viewfinder Lite software. Images were processed with Adobe Photoshop (Adobe Systems, Mountain View, CA).
Erythroid hanging drop cultures
Fetal livers were disrupted and cells were seeded at a density of 2.5 × 106 cells/mL in Dulbecco modified Eagle medium containing 20% fetal calf serum, 200 μmol/L hemin chloride (Sigma), 2 units/mL erythropoietin (Epo; a kind gift of Ortho-Biotech, Tilburg, The Netherlands), 5 μg/mL insulin and 100 μmol/L β-mercaptoethanol (Sigma). Cells were grown for 2 days in 20 μl drops containing 5 × 105 cells hanging from the lid of a culture dish.24
Primary mouse erythroblast cultures
Single cell suspensions were prepared from fetal livers and enriched for primary erythroblasts by repetitive centrifugation at 700 rpm. Primary erythroblasts were grown in serum-free medium (StemPro-34; Invitrogen, Carlsbad, CA) supplemented with 0.5 units/mL Epo, 100 ng/mL murine stem cell factor (SCF; R&D Systems, Minneapolis, MN), and 10−6 mol/L dexamethasone (Dex; Sigma).25,26 Terminal differentiation was triggered by washing the cells in phosphate-buffered saline and reseeding them at 1 to 2 × 106 cells/mL in serum-free medium, supplemented with 5 units/mL Epo and 1 mg/mL iron-saturated human transferrin (Sigma). Cell numbers and size distributions were determined using an electronic cell counter (CASY-1; Schärfe-Systems, Reutlingen, Germany), and differentiation was assessed by the size reduction of the cells. To test the stability of the Gata1 and Gata2 proteins, 20 μg/mL cycloheximide was added to the culture media, cells were harvested at various times after cycloheximide addition, and the levels of Gata1 and Gata2 were determined by Western blotting, using Npm1 (which has a half life of > 8 hours [data not shown]) as a loading control.
Fluorescence-activated cell sorting analysis
Cells were collected and incubated with PE-conjugated anti-mouse TER119, FITC-conjugated anti-mouse CD71 (BD Pharmingen, San Diego, CA) and 7-aminoactinomycin-D (7AAD; Invitrogen). Fifty-thousand cells were analyzed by fluorescence-activated cell sorting and dead cells (7AAD+) were removed from the analysis. Differentiation of erythroid cells was evaluated by the expression of TER119 (TER119+) and reduction of cell size caused by enucleation (FSClow).24
Western blotting
Nuclear protein extracts were prepared as described previously.27 For Western blot analysis, 20 to 50 μg of nuclear protein was loaded per lane, separated on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA). Blots were probed with rat anti-Gata1 mAb (N6), polyclonal rabbit anti-Gata2 (H-116), and anti-Gata3 (HG3-31) (Santa Cruz Biotechnology, Santa Cruz, CA), or mouse monoclonal anti-Npm1 (a kind gift of Dr. P. K. Chan, Baylor College of Medicine, Houston, TX). Second-step reagents were horseradish peroxidase-conjugated goat anti-rat Ig, goat anti-rabbit Ig, and goat anti-mouse Ig (Dako Denmark A/S, Glostrup, Denmark). Peroxidase activity was visualized by enhanced chemiluminescence using standard procedures.
Gene expression analysis
Results
Gata1 mutants and transgenic mice used in this study
The Gata1 germline mutants and transgenes used in this study, and the abbreviations used to indicate them, are presented in Figure 1. Gata1 knockout (G1KO), Gata1 knockdown (G1.05), and Gata cDNA transgenes under the control of the hematopoietic regulatory domain of the Gata1 gene (HRD-G) have been described previously.16,19,30
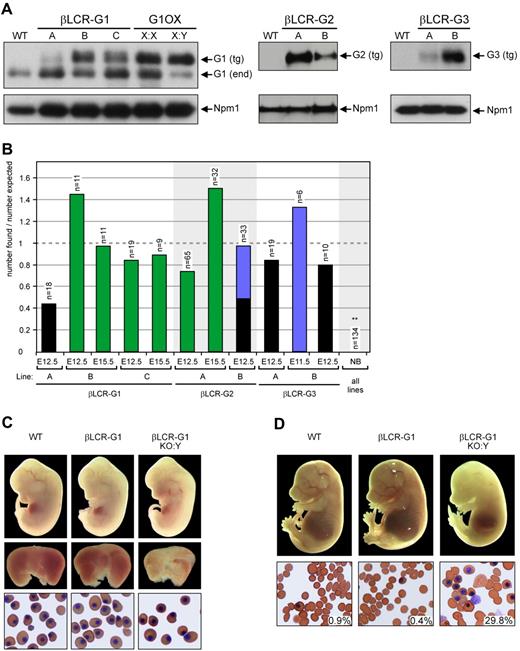
Germ line Gata1 mutants and transgenic lines used in this study. (A) Schematic representation of the Gata1 germ line mutants and (B) the constructs used for the generation of βLCR and HRD transgenic mice used in this study. Chromosomal localization of mutants and transgenes is indicated on the left; X = X chromosome; A = autosome. References to the original description of mutant or construct are given. The HRD constructs are based on the mouse Gata1 locus. IT and IE: testis- and erythroid-specific first exon, respectively; G1HE = Gata1 hematopoietic element; HS = DNaseI hypersensitive site; SA = splice acceptor; GFP = green fluorescent protein; Neo = neomycin resistance gene. The βLCR constructs are based on the human LCR (βLCR) and human β-globin gene (HBB). Abbreviations used to refer to the germ line mutants and transgenes are shown between brackets.
Germ line Gata1 mutants and transgenic lines used in this study. (A) Schematic representation of the Gata1 germ line mutants and (B) the constructs used for the generation of βLCR and HRD transgenic mice used in this study. Chromosomal localization of mutants and transgenes is indicated on the left; X = X chromosome; A = autosome. References to the original description of mutant or construct are given. The HRD constructs are based on the mouse Gata1 locus. IT and IE: testis- and erythroid-specific first exon, respectively; G1HE = Gata1 hematopoietic element; HS = DNaseI hypersensitive site; SA = splice acceptor; GFP = green fluorescent protein; Neo = neomycin resistance gene. The βLCR constructs are based on the human LCR (βLCR) and human β-globin gene (HBB). Abbreviations used to refer to the germ line mutants and transgenes are shown between brackets.
To drive expression at late stages of erythroid differentiation, we used the β-globin promoter and Locus Control Region (βLCR-G).20 Previously, we used a genomic construct containing the mouse Gata1 gene linked to the βLCR. This yielded ∼6-fold overexpression of Gata1 protein, resulting in a block in terminal erythroid differentiation (Figure 1, G1OX10 ). To attenuate the expression levels, we prepared cDNA expression constructs and used these to generate additional transgenic lines. We cloned the Gata1, Gata2, and Gata3 cDNAs in the βLCR expression vector (Figure 122,31 ). Three βLCR-Gata1 lines (βLCR-G1 A to C), 2 βLCR-Gata2 lines (βLCR-G2 A and B), and 2 βLCR-Gata3 lines (βLCR-G3 A and B) were obtained. Heterozygous animals from all transgenic lines were born at Mendelian ratios and appeared normal, including their hematologic indices (data not shown). The transgenic Gata proteins in the βLCR-G mice are expressed at different levels (Figure 2A). Furthermore, the expression levels of Gata1 in the βLCR-G1 transgenics were always lower than the ∼6-fold overexpression obtained with the genomic Gata1 construct (Figure 2A10 ). We concluded that the lower expression levels obtained with the βLCR-G cDNA constructs allow for terminal erythroid differentiation to occur.
Rescue of the Gata1 knockout by βLCR-driven Gata transgenes. (A) Characterization of βLCR-G transgenic mice. Western blot analysis of E12.5 fetal liver cells from βLCR-G1, βLCR-G2, and βLCR-G3 transgenic lines showing different expression levels of the transgene-derived Gata factors. Gata1 expression of the G1OX line10 is shown for comparison with the βLCR-G1 lines; X:X = heterozygous female; X:Y = hemizygous male. Staining of the same blots with an antibody against nucleophosmin (Npm1) was used as a loading control. (B) Rescue of the Gata1 knockout by the βLCR-G transgenes. Fetuses were isolated at the time points indicated; the bars represent the number of βLCR-G::G1KO:Y fetuses divided by the number expected according to Mendelian inheritance. n = total number of fetuses isolated. NB = newborn. Green: alive fetuses of normal size; blue: growth retarded fetuses; black: dead fetuses. **: number observed lower than expected; P < 0.01. (C) Rescue of G1KO fetuses by the βLCR-G1 transgene (line B) at E12.5. Top panel: E12.5 fetuses of genotypes indicated. Middle panel: their fetal livers. Bottom panel: erythroid cells from the circulation. (D) The βLCR-G1 transgene (line B) can rescue G1KO fetuses until E15.5. Top panel: E15.5 fetuses of genotypes indicated. Bottom panel: erythroid cells from the circulation; the percentage of nucleated cells is indicated; original magnification, 40×.
Rescue of the Gata1 knockout by βLCR-driven Gata transgenes. (A) Characterization of βLCR-G transgenic mice. Western blot analysis of E12.5 fetal liver cells from βLCR-G1, βLCR-G2, and βLCR-G3 transgenic lines showing different expression levels of the transgene-derived Gata factors. Gata1 expression of the G1OX line10 is shown for comparison with the βLCR-G1 lines; X:X = heterozygous female; X:Y = hemizygous male. Staining of the same blots with an antibody against nucleophosmin (Npm1) was used as a loading control. (B) Rescue of the Gata1 knockout by the βLCR-G transgenes. Fetuses were isolated at the time points indicated; the bars represent the number of βLCR-G::G1KO:Y fetuses divided by the number expected according to Mendelian inheritance. n = total number of fetuses isolated. NB = newborn. Green: alive fetuses of normal size; blue: growth retarded fetuses; black: dead fetuses. **: number observed lower than expected; P < 0.01. (C) Rescue of G1KO fetuses by the βLCR-G1 transgene (line B) at E12.5. Top panel: E12.5 fetuses of genotypes indicated. Middle panel: their fetal livers. Bottom panel: erythroid cells from the circulation. (D) The βLCR-G1 transgene (line B) can rescue G1KO fetuses until E15.5. Top panel: E15.5 fetuses of genotypes indicated. Bottom panel: erythroid cells from the circulation; the percentage of nucleated cells is indicated; original magnification, 40×.
Potential of βLCR-G1, -G2, and -G3 transgenes to rescue the Gata1-null mutation
We first investigated whether the Gata1-null mutation could be rescued by the βLCR-G1, βLCR-G2, and βLCR-G3 transgenes. G1KO:X female mice were bred to male mice from the different βLCR-G transgenic lines. We were surprised that no newborn βLCR-G1::G1KO:Y, βLCR-G2::G1KO:Y, or βLCR-G3::G1KO:Y animals were obtained from any of these breedings (Figure 2B).
To examine to what extent the βLCR-G1, βLCR-G2, and βLCR-G3 transgenes could rescue the Gata1-null mutation during embryonic development, we analyzed timed pregnancies. Live βLCR-G1::G1KO:Y fetuses were present at E12.5 and E15.5 (Figure 2B—D) in lines B and C, which expressed higher levels of the transgene (Figure 2A). βLCR-G1 transgenic line A, expressing low levels of the transgene, failed to prolong embryonic viability of the Gata1-null mutation, supporting previous results.10,18,30 Likewise, βLCR-G2::G1KO:Y fetuses were present at both E12.5 and E15.5 in litters from crosses between G1KO:X animals and the βLCR-G2 line with the highest transgene expression (line A; Figure 2A,B). In progeny from crosses between G1KO:X female animals and the lower expressing βLCR-G2 line B, βLCRG2::G1KO:Y fetuses were only detected at E12.5 (Figure 2B). These fetuses showed severe growth retardation or were already dead. This suggests a correlation between the rescue potential of the βLCR-G2 transgene and the expression level of the protein. No live βLCR-G3::G1KO:Y fetuses were detected at E12.5 (Figure 2B). One live βLCR-G3::G1KO:Y embryo was identified in an E11.5 litter, but this embryo showed severe growth retardation. These data are consistent with previous results indicating that, compared with Gata1 and Gata2, Gata3 is less effective in replacing Gata1 function.16
At E12.5, βLCR-G1::G1KO:Y male mice showed normal gross morphology (Figure 2C). In addition, no difference was observed between the embryonic blood of E12.5 βLCR-G1::G1KO:Y male mice and littermates, indicating that primitive, yolk sac-derived erythropoiesis had been fully rescued by the βLCR-G1 transgenes. However, βLCR-G1::G1KO:Y fetuses had small and pale livers compared with wild-type littermates, suggesting a defect in definitive, fetal liver-derived erythropoiesis. By E15.5, βLCR-G1::G1KO:Y males were very pale compared with littermates (Figure 2D). Analysis of the blood revealed that βLCR-G1::G1KO:Y male mice had relatively high numbers of nucleated erythrocytes and apparent immature cells in the circulation. A very similar phenotype was seen in βLCR-G2::G1KO:Y fetuses (data not shown). These phenotypic hallmarks were indicative of defective definitive erythropoiesis.
Taken together, these results showed that the βLCR-G1 and βLCR-G2 transgenes could rescue primitive erythropoiesis but were unable to rescue definitive erythropoiesis in a complete Gata1-null background.
Rescue of G1.05 knockdown mice by the βLCR-G1 transgene
Because transgenic mice expressing the hematopoietic Gata factors under the control of Gata1 regulatory sequences (HRD-G transgenes; Figure 1) are able to rescue erythropoiesis in G1.05 hypomorphic mice,16 we tested the rescue potential of the βLCR-G1 transgene in such mice.
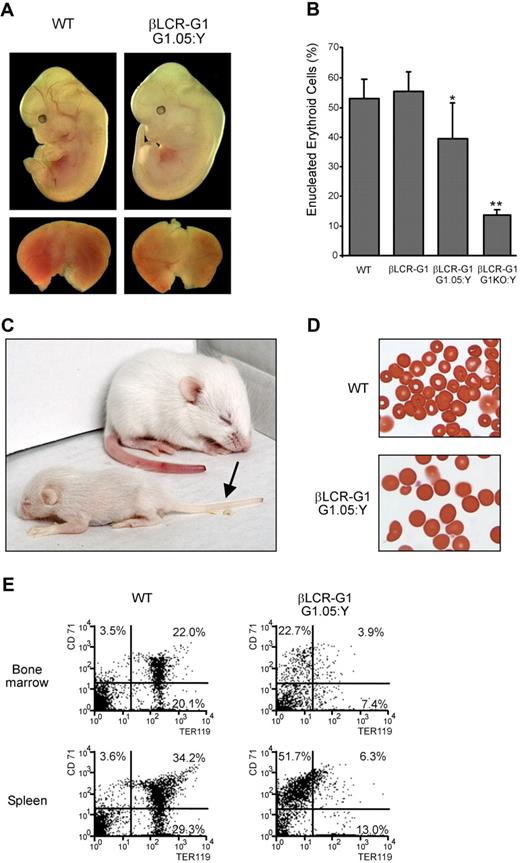
To obtain βLCR-G1::G1.05:Y male mice, we bred βLCR-G1 line B male mice to G1.05:X female mice. First, we analyzed erythropoiesis in βLCR-G1::G1.05:Y male mice during embryonic development. We found that these fetuses showed normal gross morphology, although their livers were slightly paler compared with those of wild-type littermates (Figure 3A). The differentiation potential of the fetal liver progenitors was analyzed in hanging drop cultures.24 The percentage of enucleated erythrocytes was mildly reduced by approximately 30% in βLCR-G1::G1.05:Y fetal liver cells compared with wild-type fetal liver cells. This indicates that differentiation of βLCR-G1::G1.05:Y erythroid progenitors was relatively normal, as opposed to the defective differentiation observed in βLCR-G1::G1KO:Y erythroid progenitors (Figure 3B). This defective differentiation was not due to increased apoptosis, because Annexin V staining showed that there is no increase in the number of apoptotic cells in the βLCR-G1::G1KO:Y cultures (data not shown), consistent with the notion that low levels of Gata1 prevent apoptosis of proerythroblasts.17
Rescue of the Gata1.05 mutant by the βLCR-G1 transgene. (A) Rescue of G1.05 fetuses by the βLCR-G1 transgene (line B) at E12.5. Top panel: E12.5 fetuses of genotypes indicated; bottom panel: their fetal livers. (B) Flow cytometric analysis of the percentage of enucleated (FSClow TER119+) erythroid cells in WT, βLCR-G1, βLCR-G1::G1.05:Y, and βLCR-G1::G1KO:Y E12.5 fetal livers cells after 2 days in hanging drop cultures. Enucleation was determined by cell size (FSClow). Significant difference: *, P < .05; **, P < .002, compared with WT (unpaired t test). (C) βLCR-G1::G1.05:Y pup and wild-type littermate 11 days after birth; anemia is evident as pallor of the tail (arrow). (D) Peripheral blood of βLCR-G1::G1.05:Y pup and wild-type littermate; original magnification, 40×. (E) Flow cytometric analysis of bone marrow (top panel) and spleen (bottom panel) of βLCR-G1::G1.05:Y pup and wild-type littermate. Percentages of cells expressing CD71 and/or TER119 are indicated.
Rescue of the Gata1.05 mutant by the βLCR-G1 transgene. (A) Rescue of G1.05 fetuses by the βLCR-G1 transgene (line B) at E12.5. Top panel: E12.5 fetuses of genotypes indicated; bottom panel: their fetal livers. (B) Flow cytometric analysis of the percentage of enucleated (FSClow TER119+) erythroid cells in WT, βLCR-G1, βLCR-G1::G1.05:Y, and βLCR-G1::G1KO:Y E12.5 fetal livers cells after 2 days in hanging drop cultures. Enucleation was determined by cell size (FSClow). Significant difference: *, P < .05; **, P < .002, compared with WT (unpaired t test). (C) βLCR-G1::G1.05:Y pup and wild-type littermate 11 days after birth; anemia is evident as pallor of the tail (arrow). (D) Peripheral blood of βLCR-G1::G1.05:Y pup and wild-type littermate; original magnification, 40×. (E) Flow cytometric analysis of bone marrow (top panel) and spleen (bottom panel) of βLCR-G1::G1.05:Y pup and wild-type littermate. Percentages of cells expressing CD71 and/or TER119 are indicated.
We then assessed whether βLCR-G1::G1.05:Y male mice were viable. Of 15 pups, 2 live βLCR-G1::G1.05:Y animals were obtained, demonstrating that definitive erythropoiesis had been restored sufficiently to allow perinatal survival. They were very frail, displayed growth retardation, suffered from anemia (Figure 3C), and died within 3 weeks. Analysis of the peripheral blood of a βLCR-G1::G1.05:Y animal showed abnormal morphology of the erythroid cells (Figure 3D). The bone marrow and spleen showed a severe reduction in TER119+ erythroid cells (Figure 3E).
Together, these data demonstrate that the βLCR-G1 transgene facilitates the partial rescue of the definitive erythropoiesis defect exhibited by the G1.05 hypomorphic allele and suggested that the normal expression profile of Gata1 might be mimicked by the combined actions of the βLCR-G1 transgene and the G1.05 allele, allowing terminal differentiation to occur, albeit at limited levels.
Rescue of Gata1-null mice by HRD-G transgenes
Because the G1.05 allele can contribute to the rescue phenotype by the βLCR-G1 transgene and may have contributed to the previously reported rescue by HRD-G transgenes,16 we tested the potential of the HRD-G transgenes to rescue the Gata1-null mutation.
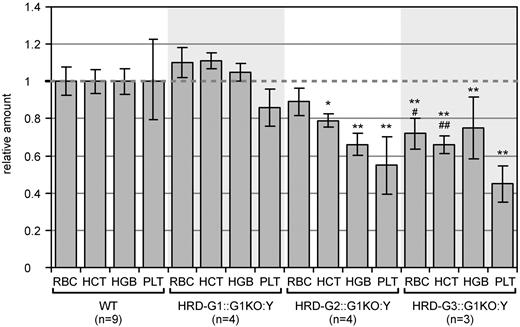
To investigate this, G1KO:X female mice were mated with HRD-G1 transgenic male mice.16 Seven rescued males (HRD-G1::G1KO:Y) were identified of a total of 41 newborn pups. This is close to the expected Mendelian ratio of 1 of 8. All rescued mice developed normally, were fertile, and showed no signs of anemia or thrombocytopenia (Figure 4). This demonstrates that Gata1, when expressed under the control of the HRD, rescues the Gata1-null phenotype in the erythroid and megakaryocytic lineages.
Hematologic parameters of Gata1 knockout mice rescued by HRD-driven Gata transgenes. Mice of the genotypes indicated were bled at an age of 8 to 12 weeks, and hematologic parameters were determined. For all parameters, values for the WT animals were normalized to 1. RBC = red blood cells; HCT = hematocrit; HGB = hemoglobin; PLT = platelets; error bars indicate ± SD. Significant difference: *, P < .05; **, P < .01, compared with WT; #, P < .05; ##, P < .01, compared with HRD-G2::G1KO:Y (unpaired t test).
Hematologic parameters of Gata1 knockout mice rescued by HRD-driven Gata transgenes. Mice of the genotypes indicated were bled at an age of 8 to 12 weeks, and hematologic parameters were determined. For all parameters, values for the WT animals were normalized to 1. RBC = red blood cells; HCT = hematocrit; HGB = hemoglobin; PLT = platelets; error bars indicate ± SD. Significant difference: *, P < .05; **, P < .01, compared with WT; #, P < .05; ##, P < .01, compared with HRD-G2::G1KO:Y (unpaired t test).
Next, we tested the potential of the other hematopoietic Gata transcription factors to rescue the Gata1-null mutation. We crossed G1KO:X female mice with HRD-Gata2(HRD-G2) and HRD-Gata3(HRD-G3) transgenic male mice16 and analyzed the offspring. These breedings gave rise to progeny with the genotype HRD-G2::G1KO:Y and HRD-G3::G1KO:Y at expected Mendelian ratios. These animals developed normally and showed hematologic parameters close to the normal range but had a mild to moderate reduction in red blood cell count, hemoglobin content, hematocrit level, and platelet number (Figure 4). As observed previously with the rescue of the G1.05 mutation,16 these values are lower in HRD-G3::G1KO:Y than in HRD-G2::G1KO:Y animals. Together, these results show unequivocally that the embryonic lethality of the Gata1-null phenotype can be rescued by Gata1, Gata2, and Gata3 transgenes driven by Gata1 regulatory sequences.
Comparison between HRD- and βLCR-driven transgene expression
The distinction in rescue potential between the HRD- and βLCR-driven Gata transgenes suggests that dynamic spatiotemporal regulation of Gata factor levels is essential for definitive erythropoiesis. We therefore compared the expression profiles of the transgenes under the control of the 2 distinct regulatory elements. To compare expression of transgene-derived Gata factor directly with that of endogenous Gata1, we used the HRD-G2 and β-LCR-G2 (line A) transgenic mice. E12.5 fetal livers of HRD-G2 and βLCR-G2 transgenic fetuses were collected and erythroblasts expanded in liquid cultures containing Dex, Epo, and SCF for 3-4 days. The cells were then allowed to terminally differentiate by removing Dex and SCF, increasing the Epo concentration, and adding iron-saturated transferrin to the medium.25,26 Samples were collected at several time points during differentiation until just before enucleation. Nuclear extracts were prepared for Western blot analysis of the expression of endogenous Gata1 and transgene-derived Gata2 proteins (Figure 5A). The expression of nucleophosmin (Npm1), a nucleolar protein,32 was used to normalize for the amount of protein loaded.
HRD- and βLCR-driven transgenes have distinct expression patterns during terminal erythroid differentiation. (A) Fetal liver cells were grown and switched to differentiation conditions. Nuclear proteins were isolated at the times indicated, and analyzed by Western blot to compare the expression patterns of endogenous Gata1 and the Gata2 protein under the control of HRD and βLCR (line A) transgenes. Top panel: staining of the same blot for Npm1 was used as a loading control. Bottom panel: expression of Gata2 and Gata1 proteins. A higher exposure of the area indicated is shown on the right (B) Graphical representation of the expression levels of the Gata2 and Gata1 proteins after normalization for Npm1 expression. The expression level of HRD-derived Gata2 was determined relative to the Npm1 expression level, and the observed ratio was normalized to 1 at 24 hours of differentiation. The normalization factor was applied to the values obtained for the Gata2/Npm1 ratios at the other time points, for both the HRD-G2 and βLCR-G2 samples. For differentiation time 0 hours and 36 hours, the difference between βLCR-driven Gata factor expression is significantly different from that of endogenous Gata1 (lower, P < .01 and higher, P < .04, respectively [4 independent experiments]). HRD-driven Gata factor expression is not significantly different from endogenous Gata1 at any time point analyzed (unpaired t test).
HRD- and βLCR-driven transgenes have distinct expression patterns during terminal erythroid differentiation. (A) Fetal liver cells were grown and switched to differentiation conditions. Nuclear proteins were isolated at the times indicated, and analyzed by Western blot to compare the expression patterns of endogenous Gata1 and the Gata2 protein under the control of HRD and βLCR (line A) transgenes. Top panel: staining of the same blot for Npm1 was used as a loading control. Bottom panel: expression of Gata2 and Gata1 proteins. A higher exposure of the area indicated is shown on the right (B) Graphical representation of the expression levels of the Gata2 and Gata1 proteins after normalization for Npm1 expression. The expression level of HRD-derived Gata2 was determined relative to the Npm1 expression level, and the observed ratio was normalized to 1 at 24 hours of differentiation. The normalization factor was applied to the values obtained for the Gata2/Npm1 ratios at the other time points, for both the HRD-G2 and βLCR-G2 samples. For differentiation time 0 hours and 36 hours, the difference between βLCR-driven Gata factor expression is significantly different from that of endogenous Gata1 (lower, P < .01 and higher, P < .04, respectively [4 independent experiments]). HRD-driven Gata factor expression is not significantly different from endogenous Gata1 at any time point analyzed (unpaired t test).
The endogenous Gata1 protein was detected before induction of differentiation, it increased during differentiation, and it declined at the later stages (Figure 5A,B). Expression of the HRD-G2-derived transgenic protein followed a similar pattern: it was expressed in undifferentiated erythroblasts and expression increased significantly during differentiation, leveling out at later stages (Figure 5A,B). The Gata2 transgene under the control of the βLCR showed a temporal expression pattern very distinct from that of the endogenous Gata1 gene. The transgenic protein was detectable in undifferentiated erythroblasts but was greatly upregulated during differentiation, still increasing at the latest time point analyzed (Figure 5A,B). Similar results were obtained with the HRD and βLCR transgenes driving Gata1 and Gata3 expression (data not shown). To determine the impact of protein stability on the expression levels of Gata1 and Gata2, we cultured HRD-G2 erythroblasts in the presence of the protein translation inhibitor cycloheximide. We observed that the half-life of Gata2 was approximately 30 minutes, in agreement with previous data.33 In addition, we found that the half-life of Gata1 is similar to that observed for Gata2. Finally, the half-lives of Gata1 and Gata2 did not change 24 hours after the induction of differentiation (data not shown).
We conclude that HRD-driven expression mimicked the endogenous Gata1 expression profile during terminal erythroid differentiation, whereas βLCR-driven expression resulted in a very distinct pattern; expression sharply increased during terminal differentiation. These distinct spatiotemporal expression patterns provide an explanation for the difference in rescue potential of the HRD-versus the βLCR-driven transgenes.
Molecular analysis of Gata2 expression in βLCR-G2::G1KO:Y fetal liver cells
The rescue of the Gata1-null mutation by the HRD-G2 and HRD-G3 transgenes demonstrates conclusively that Gata2 and Gata3 could functionally replace Gata1 in the erythroid lineage. This is surprising because endogenous Gata2 mRNA expression is highly elevated in Gata1-null erythroid progenitors but does not provide rescue.6 Because the βLCR-G2 transgene rescues the apoptosis of the Gata1-null mutation but not terminal erythroid differentiation (Figure 6A), we analyzed the erythroid differentiation defect in βLCR-G2::G1KO:Y fetal livers at the molecular level. We examined the expression levels of genes that are normally either repressed or activated by Gata1 during erythroid differentiation.34 Consistent with a block in differentiation, we found that Myb expression was aberrantly high, whereas expression of globins (Hbb-b and Hbb-a), heme synthesis enzymes (Alas2 and Alad), and the erythroid structural protein Epb4.9 was reduced in βLCR-G2::G1KO:Y fetal liver cells (Figure 6B). In addition, the expression of endogenous Gata2 mRNA was significantly increased in βLCR-G2::G1KO:Y fetal liver cells. The endogenous mouse Gata2 gene had 2 alternative promoters; the distal promoter was specifically active in hematopoietic progenitor cells.29 We examined which promoter was responsible for the transcriptional up-regulation of Gata2 in βLCR-G2::G1KO:Y cells. RT-PCR reactions strongly suggested exclusive use of the proximal promoter. Transcription from the distal promoter was undetectable in βLCR-G2::G1KO:Y fetal liver cells (Figure 6C).
Gata2 expression during terminal differentiation of βLCR-G2::G1KO:Y and HRD-G2::G1KO:Y fetal liver cells. βLCR-G2 line A was used. (A) Size distribution of TER119+ wild-type, βLCR-G2::G1KO:Y, and HRD-G2::G1KO:Y cells after 2 days in hanging drop cultures. The percentage of small, enucleated cells is indicated. Significant difference: *, P < 0.05; **, P < 0.01; compared with WT (unpaired t test). (B) RQ-PCR analysis of gene expression. RNA was isolated from βLCR-G2::G1KO:Y fetal liver cells, and gene expression was determined with real-time quantitative RT-PCR as described previously.28 Bar graphs depict higher (white area) or lower (gray area) expression levels relative to WT fetal liver cells; error bars indicate ± SD (n ≥ 3 for each gene measured). Significant difference: *, P < 0.05; **, P < 0.01 compared with WT (unpaired t test). (C) RT-PCR detecting the distal and proximal exon 1 of endogenous Gata2 mRNA; RNA from dendritic cells is a positive control for detection of distal exon 1. (D) Top panel: Western blot of βLCR-G2-and HRD-G2-derived Gata2 in Gata1-null cells during erythroid differentiation; expression of Gata1 and Gata2 in HRD-G2 cells wild type for endogenous Gata1 is shown for comparison. Bottom panel: detection of Npm1 serving as a loading control.
Gata2 expression during terminal differentiation of βLCR-G2::G1KO:Y and HRD-G2::G1KO:Y fetal liver cells. βLCR-G2 line A was used. (A) Size distribution of TER119+ wild-type, βLCR-G2::G1KO:Y, and HRD-G2::G1KO:Y cells after 2 days in hanging drop cultures. The percentage of small, enucleated cells is indicated. Significant difference: *, P < 0.05; **, P < 0.01; compared with WT (unpaired t test). (B) RQ-PCR analysis of gene expression. RNA was isolated from βLCR-G2::G1KO:Y fetal liver cells, and gene expression was determined with real-time quantitative RT-PCR as described previously.28 Bar graphs depict higher (white area) or lower (gray area) expression levels relative to WT fetal liver cells; error bars indicate ± SD (n ≥ 3 for each gene measured). Significant difference: *, P < 0.05; **, P < 0.01 compared with WT (unpaired t test). (C) RT-PCR detecting the distal and proximal exon 1 of endogenous Gata2 mRNA; RNA from dendritic cells is a positive control for detection of distal exon 1. (D) Top panel: Western blot of βLCR-G2-and HRD-G2-derived Gata2 in Gata1-null cells during erythroid differentiation; expression of Gata1 and Gata2 in HRD-G2 cells wild type for endogenous Gata1 is shown for comparison. Bottom panel: detection of Npm1 serving as a loading control.
We then analyzed the expression of Gata2 protein in βLCR-G2::G1KO:Y erythroid progenitors, using HRD-G2::G1KO:Y cells as a control. Fetal liver erythroblasts were cultured and the nuclear proteins analyzed before and 24 hours after induction of differentiation. Western blot analyses revealed that Gata2 protein was expressed in HRD-G2::G1KO:Y cells before differentiation and increased when differentiation was initiated, similar to the protein expression pattern observed in HRD-G2::WT erythroid cells (Figures 5 and 6D). Expression of Gata2 protein was low but detectable in βLCR-G2::G1KO:Y cells before differentiation, but this was not upregulated after induction of differentiation (Figure 6D), in contrast to the Gata2 protein expression pattern observed in βLCR-G2::WT cells (Figure 5). Moreover, expression of endogenous Gata2 protein was negligible in βLCR-G2::G1KO:Y erythroblasts, despite the presence of high levels of Gata2 mRNA (Figure 6B—D). These results indicate that βLCR-G2::G1KO:Y erythroblasts were unable to differentiate into erythroid progenitors that up-regulate the β-globin promoter driving the βLCR-G2 transgene, and hence the βLCR-driven transgenes are incapable of rescuing the erythroid differentiation phenotype. The negligible expression of endogenous Gata2 protein in these cells further explained why, despite dramatically increased levels of endogenous Gata2 mRNA, the Gata1-null phenotype was not rescued.
Discussion
Here we have used transgenic mice to probe the impact of Gata factor levels on the development of cells within a single hematopoietic lineage (Figure 7). Gata1 is essential for the expansion and differentiation of several cell types in the hematopoietic system; indeed, lineage-specific “knockdown” mutations introduced in the Gata1 locus have revealed important roles in development of eosinophils,35,36 mast cells,37 and megakaryocytes,38 in addition to the absolute requirement of Gata1 for erythroid development.8,39 Reduced expression of Gata1 severely affects the erythroid lineage,18,30 indicating that erythropoiesis is critically dependent on threshold levels of Gata1.
Dynamic regulation of Gata factor levels is essential for definitive erythroid development. Development of the definitive erythroid lineage is shown from progenitor cells to the terminally differentiated enucleated erythrocyte. (A) Gata 1 expression in the germline mutants used in this study. (a) Overexpression of Gata1 at late stages of development blocks terminal differentiation including enucleation. (b) In wild-type cells, Gata1 expression increases around the CFU-e/proerythroblast stage.61 (c) Low expression of Gata1 in the G1.05 mutant allows erythroid cells to develop until the proerythroblast stage. Prolonged survival of progenitors can result in a leukemic condition. (d) Gata1-null cells succumb to apoptosis at the proerythroblast stage. (B) (a) HRD-G transgenes reproduce the expression pattern of endogenous Gata1. (b and c) HRD-G transgenes rescue the G1.05 and G1KO germ line mutations, respectively. (C) (a) βLCR-G cDNA transgenes are expressed at low levels in progenitors and are sharply upregulated at late stages of development. (b) combination of the βLCR-G1 transgene with the G1.05 mutation results in partial rescue of erythroid development. (c) βLCR-G transgenes are expressed but fail to be upregulated in G1KO erythroid progenitors. Development is blocked but apoptosis is rescued. The intensity of black shading indicates protein expression levels; sloped lines indicate overexpression.
Dynamic regulation of Gata factor levels is essential for definitive erythroid development. Development of the definitive erythroid lineage is shown from progenitor cells to the terminally differentiated enucleated erythrocyte. (A) Gata 1 expression in the germline mutants used in this study. (a) Overexpression of Gata1 at late stages of development blocks terminal differentiation including enucleation. (b) In wild-type cells, Gata1 expression increases around the CFU-e/proerythroblast stage.61 (c) Low expression of Gata1 in the G1.05 mutant allows erythroid cells to develop until the proerythroblast stage. Prolonged survival of progenitors can result in a leukemic condition. (d) Gata1-null cells succumb to apoptosis at the proerythroblast stage. (B) (a) HRD-G transgenes reproduce the expression pattern of endogenous Gata1. (b and c) HRD-G transgenes rescue the G1.05 and G1KO germ line mutations, respectively. (C) (a) βLCR-G cDNA transgenes are expressed at low levels in progenitors and are sharply upregulated at late stages of development. (b) combination of the βLCR-G1 transgene with the G1.05 mutation results in partial rescue of erythroid development. (c) βLCR-G transgenes are expressed but fail to be upregulated in G1KO erythroid progenitors. Development is blocked but apoptosis is rescued. The intensity of black shading indicates protein expression levels; sloped lines indicate overexpression.
Some of us have shown previously that the hematopoietic Gata transcription factors Gata1, Gata2, and Gata3, when expressed under the control of Gata1 regulatory sequences, are able to rescue the lethal anemia of the G1.05 knockdown mutation.16 We now demonstrate that these transgenes can also rescue the phenotype of the complete Gata1-null mutation. This unequivocally excludes the possibility that the remaining 5% of endogenous Gata1 expression in the G1.05 mouse was required for the rescue of erythropoiesis by the HRD-driven Gata factors. It is noteworthy that the knockin of Gata3 cDNA in the Gata1 locus, which resulted in partial rescue of erythropoiesis, was attributed to insufficient accumulation of Gata3 protein.15 This is consistent with the importance of Gata factor levels reported here. Unlike the knockin experiment, the transgenic approach enables the use of independent lines displaying a range of expression levels, resolving the apparent discrepancy between the results reported on rescue of the Gata1-null mutation by Gata3. However, our results also indicate some factor-specific functions, because Gata3 was the least effective of the 3 Gata factors tested in rescuing the Gata1-null mutation. Nevertheless, the question that arises is which are the common essential features of the hematopoietic Gata factors that allow the observed interchangeability. It is likely that the zinc fingers are the most important single determinant of this property. These motifs are highly homologous between Gata1, -2, and -3, whereas little amino acid sequence homology is found outside these domains.40 Furthermore, the zinc fingers are not only responsible for DNA binding, but are also the most important protein-protein interaction domain, mediating interactions with the essential hematopoietic transcription factors Fog1 41 , Gfi-1B,42 and Tal1/Ldb1,43 and with chromatin modifying complexes MeCP1, ACF/WCRF,42 and p300/CBP.44,45 The importance of the zinc finger domains of Gata1 has also been demonstrated by transgenic mapping experiments.46 Finally, some of us have recently demonstrated that the endodermal Gata4 factor cannot substitute for Gata1 in erythropoiesis; a partial rescue was observed when the C-terminal region of Gata4 was replaced with the C-terminal region of Gata1, containing the C-terminal zinc finger.47 Together with the data reported here, these results set the stage for future experiments aimed at the functional dissection of the subdomains of the hematopoietic GATA factors in vivo.
Cell lineage commitment and determination
Several studies have assigned lineage determination and commitment properties to Gata factors. It has been suggested that Gata1 plays an integral role in directing myelo-erythroid lineage fate decisions during embryogenesis in the zebrafish.48 Primary mouse hematopoietic progenitors can be reprogrammed by the instructive action of Gata1.49 Enforced expression of Gata2 induces megakaryocytic differentiation of ES cell-derived hematopoietic cells, which is thought to be an effect on lineage commitment.50 Consistent with this notion, ectopic Gata2 inhibits erythroid development.51,52 Collectively, the sequential expression of Gata2 and Gata1 most likely plays an important role in the outcome of these lineage commitment decisions. Our data show that, in the case of Gata1, factor-specific properties play a relatively minor role in comparison to the dynamic regulation of the spatiotemporal expression pattern. It will be interesting to determine whether this model also applies to the other functions of the Gata factors in the hematopoietic system, and in other tissues, such as the skin, where Gata3 is a regulator of cell fate determination.53
Interplay of Gata1 and Gata2 in erythropoiesis
Because the hematopoietic expression profiles of Gata1 and Gata2 overlap partially, functional redundancy may exist between these factors. The increased severity of the erythroid phenotype observed in Gata1/Gata2 compound knockout mice is consistent with this notion.54 These observations raise an interesting conundrum, because the expression levels of endogenous Gata2 mRNA are increased considerably in Gata1-deficient erythroid progenitors,6 yet this does not result in phenotypic rescue of these cells. We also observed raised levels of endogenous Gata2 mRNA expression in βLCR-G2::G1KO:Y progenitors, to ∼20% of the mRNA levels obtained with the HRD-G2 transgene (data not shown), but this is not accompanied by an increase in Gata2 protein levels suggesting an additional layer of regulation. Protein stability is an unlikely mechanism, because Gata2 derived from the βLCR-G2 transgene is detectable, and the rescue of the Gata1 knockout by the HRD-G2 transgene would also not be possible. Alternatively, endogenous Gata2 mRNA might be subject to translational control in these cells. The Gata2 gene has 2 alternative noncoding first exons, the distal 1S and the proximal 1G exon. The 1S exon is used preferentially in immature hematopoietic cells.29 In contrast, the 1G exon is used in βLCR-G2::G1KO:Y erythroid progenitors. The 1G exon of Gata2 contains several potential translational control elements, including 2 upstream AUG codons embedded in strong translation initiation motifs (Supplemental Figure 1A). These elements may attenuate Gata2 protein expression similar to the mechanism that controls expression of Tal1 and C/EBP transcription factors.55,56 In addition, computer modeling using Mfold57 predicts that the 1G exon may adopt a Y-shaped secondary structure (Supplemental Figure 1B). Such a structure could function as an internal ribosomal entry site (IRES) directing Gata2 expression under specific conditions only. In contrast, such features are not present in the 1S exon of Gata2, the 1E exon of Gata1 used in the HRD rescue constructs, or the first exon of the βLCR rescue constructs. We suggest that exclusive use of the 1G promoter renders Gata2 mRNA subject to translational control in erythroid progenitors. This, in combination with the short half life of the factor and the rising levels of Gata1 that normally occur at this stage of development, will aid in the rapid exchange of Gata2 for Gata1 and repression of the Gata2 gene by Gata1.42,58,59
Rescue of the Gata1-null mutation: the devil is in the regulation of the level
We found that, when expressed under the control of β-globin regulatory sequences, the hematopoietic Gata proteins were unable to rescue Gata1-null definitive erythroid cells. However, these transgenes can rescue primitive erythropoiesis. This is consistent with previous results indicating that primitive erythropoiesis is less sensitive to perturbations in Gata activity.10,15,46 We show that βLCR transgene-driven expression is low in definitive erythroid precursors and rises sharply during terminal differentiation. In contrast, the HRD-driven transgenes display significant expression already in erythroid precursors, which rises during erythropoiesis and is attenuated during terminal differentiation. From these data, we conclude that the correct spatiotemporal regulation of the expression level is a critical determinant for the appropriate execution of erythropoiesis (Figure 7). Supporting this conclusion, we show that the βLCR-G1 transgene can partially rescue blocked development of G1.05 erythroid precursors. The residual Gata1 expression from the G1.05 allele activates the βLCR-G1 transgene sufficiently to facilitate further progression through differentiation. βLCR transgenes are apparently unable to establish a positive feedback loop on their expression in Gata1-null erythroid cells, even though low levels of Gata factor are detectable. Gata1-null erythroid precursors undergo apoptosis.6 Similar to the observation that apoptosis is rescued in G1.05 erythroid progenitors,17 we find that the low levels of Gata factor in βLCR-G::G1KO:Y cells are sufficient to prevent apoptosis. The failure of these cells to up-regulate expression of the βLCR transgenes beyond basal levels indicates that different Gata1 target genes respond to different threshold levels of Gata factor. We suggest that target genes activated late during erythroid differentiation, such as the globins, are dependent on high levels of Gata activity, whereas target genes activated early, such as those mediating cell survival, are already responding to low levels.34 The basal expression of the βLCR transgenes in the absence of endogenous Gata1 may have its roots in the promiscuous expression of hematopoietic genes that is thought to be a hallmark of undifferentiated hematopoietic cells.60
In conclusion, our data demonstrate the importance of dynamic regulation of the expression levels of key regulatory factors by cis-regulatory elements for the orchestration of a lineage-specific differentiation program. This provides a paradigm for other lineage-specific differentiation programs, where fine-tuning of spatiotemporal regulation of key developmental factors may also be hard-wired in cis-acting elements.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the animal caretakers for animal husbandry and Elaine Dzierzak for carefully reading the manuscript.
This work was supported by EUR breedtestrategie (M.vL., S.P.), the Dutch Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek) 901-08-092, DN 82-286, 050-10-051 (M.vL., F.G., S.P.), the Dutch Cancer Foundation NKB EUR 2000-2273 (F.G., S.P.) and the EU fp6 program MRTN-CT-2004-005499 (M.vL., F.G., S.P.).
Authorship
Contribution: R.F., M.vL., K.O., F.G., M.Y., and S.P. designed research; R.F., A.W., R.S., N.G., and R.R. performed research; R.F., K.O., M.Y., and S.P. analyzed data; and R.F., M.vL., K.O., F.G., M.Y., and S.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sjaak Philipsen, Erasmus MC, Department of Cell Biology, P.O. Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: j.philipsen@erasmusmc.nl.


![Figure 5. HRD- and βLCR-driven transgenes have distinct expression patterns during terminal erythroid differentiation. (A) Fetal liver cells were grown and switched to differentiation conditions. Nuclear proteins were isolated at the times indicated, and analyzed by Western blot to compare the expression patterns of endogenous Gata1 and the Gata2 protein under the control of HRD and βLCR (line A) transgenes. Top panel: staining of the same blot for Npm1 was used as a loading control. Bottom panel: expression of Gata2 and Gata1 proteins. A higher exposure of the area indicated is shown on the right (B) Graphical representation of the expression levels of the Gata2 and Gata1 proteins after normalization for Npm1 expression. The expression level of HRD-derived Gata2 was determined relative to the Npm1 expression level, and the observed ratio was normalized to 1 at 24 hours of differentiation. The normalization factor was applied to the values obtained for the Gata2/Npm1 ratios at the other time points, for both the HRD-G2 and βLCR-G2 samples. For differentiation time 0 hours and 36 hours, the difference between βLCR-driven Gata factor expression is significantly different from that of endogenous Gata1 (lower, P < .01 and higher, P < .04, respectively [4 independent experiments]). HRD-driven Gata factor expression is not significantly different from endogenous Gata1 at any time point analyzed (unpaired t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/12/10.1182_blood-2006-11-060491/4/m_zh80120701940005.jpeg?Expires=1765886466&Signature=GcPXuuQjKqLIJorQT2NUwhYfUP0HitUbNCp9I5Xk8jzz0KDSyUti20NWXPavam5VFzjlZifeLoR6ccTGvGwQ~YCfnhrIsQdngwQTEq-aNIO9MqR~ZCZGVPgV5rQfI0Q5LQ~sTTc4DyqNm-q8J2HiBd-r4MmdtHpa2xpW1xrxfw8PimuU9fcSbPzLRlTwyOLp6CnPoROVdno3xabLTTJ5CvrImRMl02SVQ1exfPPPqYIS3cr9-DV3Buh4AqoIhT8wh~nwEcr1iidL5MY4lu70NEHbLi3rkWvV9Bfupmo48lhyokOsI3gS4wF6wEF0eudb2WMW5wUn9p1QIrWRXBMyTA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

