Abstract
Bone morphogenetic protein (BMP) signaling is an important regulator of hematovascular development. However, the progenitor population that responds to BMP signaling is undefined, and the relative role of downstream mediators including Smad1 is unclear. We find that Smad1 shows a distinctive expression profile as embryonic stem (ES) cells undergo differentiation in the embryoid body (EB) system, with peak levels in cell populations enriched for the hemangioblast. To test the functional relevance of this observation, we generated an ES cell line that allows temporal control of ectopic Smad1 expression. Continuous expression of Smad1 from day 2 of EB culture does not disturb hematopoiesis, according to colony assays. In contrast, a pulse of Smad1 expression exclusively between day 2 and day 2.25 expands the population of progenitors for primitive erythroblasts and other hematopoietic lineages. This effect correlates with increased levels of transcripts encoding markers for the hemangioblast, including Runx1, Scl, and Gata2. Indeed, the pulse of Smad1 induction also expands the blast colony-forming cell (BL-CFC) population at a level that is fully sufficient to explain subsequent increases in hematopoiesis. Our data demonstrate that Smad1 expression is sufficient to expand the number of cells that commit to hemangioblast fate.
Introduction
The progenitors that generate the early hematovascular system are derived from ventral mesoderm, which in mammals contributes to the yolk sac primitive blood lineage and associated vasculature.1 The bone morphogenetic protein (BMP) signaling pathway is an essential regulator of the process by which mesoderm acquires ventral character.2 Thus, ectopic activation of BMP signaling expands ventral mesoderm derivatives including increased numbers of embryonic hematopoietic cells, whereas inhibition of the pathway causes a corresponding loss in hematopoiesis. Genetic loss-of-function experiments in mice and fish have confirmed the conserved requirement for BMPs and downstream signaling components for the generation of blood cells.3-5 However, because the pathway is essential first for the formation of normal ventral mesoderm, it has been more difficult to define regulatory functions that act on the subsequent commitment steps of mesoderm toward a hematopoietic fate. BMPs can influence hematopoietic progenitors in vitro,6,7 but whether this reflects the normal process of stem cell commitment during embryogenesis is less clear.
The Smads comprise a family of transcriptional cofactors that specifically transduce activated TGF-β/BMP signals.8 Smads 1, 5, and 8 constitute the receptor-activated Smads (R-Smads) that on phosphorylation by a type I BMP receptor bind the co-Smad4, common to all TGF-β/BMP pathways, to form heteromeric protein complexes. These R-Smad/co-Smad complexes translocate to the nucleus and cooperate with other transcription factors to modulate target gene expression. Mice deficient for Smad1 or Smad5 display varying degrees of defects in hematopoietic and vascular development. Smad1 knockout mice exhibit a defect in chorioallantoic fusion and die between E9.5 and E10.5, with some embryos displaying disruption of yolk sac angiogenesis.9,10 Loss of Smad5 is also embryonically lethal.11,12 Smad5−/− yolk sacs exhibit an increased number of high-proliferative potential colony-forming cells (HPP-CFCs), and embryonic stem (ES) cells deficient in Smad5 produce an elevated number of blast colony-forming cells (BL-CFCs).13 Again, the multitude of defects and early lethality in these knockout models preclude a detailed characterization of early events regulating specification of the hematopoietic system. The data are, however, consistent with functions for BMP signaling in the development of both hematopoietic and endothelial lineages, which are thought to be derived from an earlier common precursor, the hemangioblast.
Characterizing the molecular determinants of hemangioblast development in vivo is challenging, although the presence of the hemangioblast in early mouse embryos was confirmed at the posterior region of the primitive streak.14 This observation followed on studies using the ES cell/embryoid body (EB) system that identified the BL-CFC as the in vitro equivalent of the hemangioblast.15-17 The BL-CFC is generated in the EB as mesoderm commits to hematovascular fates, preceding the ability to detect committed primitive erythroblasts. In response to vascular endothelial growth factor (VEGF), BL-CFCs exhibit both hematopoietic and endothelial potential. Within blast colonies, genes are expressed that are common to hematopoietic and vascular development, including Scl, Flk1, and Runx1.18,19 The ES/EB system effectively demonstrates the progressive commitment of progenitors, including the hemangioblast, to adopt a hematopoietic fate. Here we report the ability of Smad1 to expand hematopoiesis by enhancing hemangioblast development during EB differentiation. We find that Smad1 can only affect this expansion within a defined developmental window, after which continued expression of Smad1 is refractory to the expansion.
Materials and methods
Gene expression profiling
Semiquantitative RNA expression analysis was performed using the 3′-end poly(A)+ reverse transcription-polymerase chain reaction (RT-PCR) method.20 RNA was reverse transcribed, 3′-end poly(A) tailed, and amplified by PCR using primers with the sequence: 5′-GTTAACTCGAGAATTC(T)24-3′. Products were subjected to brief electrophoresis through a 1.8% agarose gel, blotted onto a BrightStar-Plus nylon membrane (Ambion, Austin, TX), and hybridized with 32P-radiolabeled probes generated by random priming (Roche, Indianapolis, IN). The murine Smad1 probe consisted of a 1.8-kb fragment of 3′-UTR sequences generated by EcoR1 digestion of a pBS-mSmad1 cDNA clone (provided by Brigid Hogan, Duke University, Durham, NC). The Smad5 3′-UTR sequence was identified from a search of the EST database. The EST designated mto1e09.r1 (Image Clone no. 619816) was isolated by an EcoR1/Not1 digestion from the pT7T3D-Pac1 vector yielding a 700-bp Smad5 probe. The L32 control probe was a 1.6-kb Sac1 fragment generated from pGEM1-L32-4A.
Generation of Smad1-inducible ES cells
The approach for generating transgene-inducible ES cells is described.21 The Xho1/Not1 ires-EGFP fragment from pIRES2-EGFP (Clontech, Palo Alto, CA) was subcloned into the plox vector (provided by George Daley, Harvard Medical School, Boston, MA) engineered to contain a flag-tag upstream of cloning sites, generating plox-flag-iresEGFP. The murine Smad1 ORF was isolated by PCR from pBS-mSmad1 and shuttled into pCR 2.1-TOPO (Invitrogen, Carlsbad, CA). EcoRV and Age1 generated a Smad1 insert that was subcloned into EcoRV and Xma1 sites of plox-flag-iresEGFP. This targeting construct, plox-Flag-mSmad1ORF-iresEGFP (20 μg), was coelectroporated with 20 μg pSalk-CRE into the parental cell line Ainv18 (8 × 106 cells). Following selection and expansion in 400 μg/mL G418 (Cellgro, Herndon, VA), cDNA was isolated (Qiagen, Valencia, CA) and subjected to PCR analysis to confirm site-specific transgene integration.
Loxin F primer: 5′-CTAGATCTCGAAGGATCTGGAG-3′; Loxin R primer: 5′-ATACTTTCTCGGCAGGAGCA-3′.
ES cell growth and differentiation
ES cells were maintained on irradiated embryonic feeder cells in Dulbecco modified Eagle medium (DMEM) supplemented with 20% ES-qualified fetal calf serum (Gemini, West Sacramento, CA), 50 IU penicillin (Cellgro), 50 μg/mL streptomycin (Cellgro), LIF (2% conditioned medium), and 1.5 × 10−4 M monothioglycerol (MTG; Sigma, St Louis, MO). To generate day 3.75 EBs, cells were plated at a density of 35 000 cell/mL. Differentiation of day 3.75 EBs was carried out in 60-mm ethylene oxide-treated Petri grade dishes in IMDM supplemented with 15% FCS (Atlas Biologicals, Fort Collins, CO), 2 mM l-glutamine (Gibco BRL, Grand Island, NY), 200 μg/mL transferrin (Roche), 0.5 mM ascorbic acid (Sigma), and 4.5 × 10−4 M MTG. For the generation of day 5.75 EBs, ES cells were trypsinized and plated at a density of 7500 cells/mL in differentiation media. The day 5.75 EBs were grown under the same conditions described but supplemented with 5% protein-free hybridoma medium (PFHM-II; Gibco BRL). For continuous Smad1 induction, EBs were exposed to 1 μg/mL doxycycline. For EBs induced on days 0, 1, or 2, 0.5 μg/mL doxycycline was added to the culture media on days 3, 4, and 5. For transient transgene induction, EBs were induced with 1 μg/mL doxycycline for 6 hours, harvested, and gently suspended in fresh differentiation media lacking doxycycline. To examine GFP induction, iGFP and iSmad1iresEGFP ES cell lines were untreated or exposed to 1 μg/mL doxycycline for 8 hours before cells were analyzed on a fluorescence-activated cell sorting (FACS) Scan Flow Cytometer with a 488-nm blue laser (Becton Dickinson, Bedford, MA) for GFP expression (FL1 channel). Cell fractionation of iSmad1 ES cell-derived EB cells (∼ 5 × 106 cells) was performed exactly as per manufacturer's instructions using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL).
Colony assays
For the generation of blast cell colonies, day 3.75 EB cells were plated in 1% methylcellulose (Fluka, Ronkonkoma, NY) supplemented with 10% FCS (Atlas, lot no. A50224A), VEGF (5 ng/mL), Kit ligand (KL; 1% conditioned medium), IL-6 (10 ng/mL), and 25% D4T endothelial cell-conditioned medium.15 For the growth of hematopoietic progenitors (from day 5.75 EBs), colonies were generated in IMDM containing 1% methylcellulose, 10% plasma-derived serum (PDS; Antech, Tyler, TX), 5% PFHM-II, and specific cytokines as follows: primitive erythroblasts (erythropoietin [EPO] 2 U/mL); macrophages (IL-3 [1% conditioned media], M-CSF [5 ng/mL]); megakaryocytes (IL-3 [1% conditioned media], IL-11 [5 ng/mL], thrombopoietin [TPO, 5 ng/mL]); mixed colonies (KL [1% conditioned media], IL-3 [10 ng/mL], G-CSF [30 ng/mL, GM-CSF [10 ng/mL], IL-11 [5 ng/mL], IL-6 [5 ng/mL], TPO [5 ng/mL], and M-CSF [5 ng/mL]). IL-3 and Kit ligand were derived from media conditioned by Chinese hamster ovary (CHO) cells transfected with IL-3 and KL expression vectors, respectively (kindly provided by Genetics Institute). EPO (Amgen, Cambridge, MA) was obtained from the Albert Einstein hospital pharmacy. All other cytokines were purchased from R&D Systems (Minneapolis, MN).
Quantitative RT-PCR
The iSmad1 EBs were induced for 6 hours with 1 μg/mL doxycycline from day 2 to day 2.25. Cells from EBs at day 2.25, 2.5, 3, and 4 along with age matched uninduced control EBs were harvested and total RNA isolated (TRI REAGENT, Molecular Research Center, Cincinnati, OH). First-strand cDNA synthesis was performed (Superscript III First-Strand Synthesis System for RT-PCR, Invitrogen) and the cDNA subjected to quantitative RT-PCR analysis (Opticon DNA Engine 2, MJ Research, Watertown, MA).
Smad1: F5′-GTGTATGAACTCACCAAAATGTGC-3′, R5′-TAACATCCTGCCGGTGGTATTC-3′; Brachyury: F5′-CTCTGGTCTGTGAGCAATGG-3′,R5′-GAGCCTCGAAAGAACTGAGC-3′; Gata2: F5′-AAGCCCAAGCGGAGGCTGTCTG-3′, R5′-GGTCAGTGGCCTGTTAACATTG-3′; Gata1: F5′-AGGCCCTGGAAGACCAGGAAG-3′, R5′-AGAAAGGACTGGGAAAGTCAGC-3′; Runx1: F5′-GCCTCTCTGCAGAACTTTCC-3′, R5′-GACGGCAGAGTAGGGAACTG-3′; Vegf: F5′-CTCTACCTCCACCATGCCAAG-3′, R5′-GGTACTCCTGGAGGATGTCCACC-3′; Gapdh: F5′-TTCACCACCATGGAGAAGGC-3′, R5′-GGCATGGACTGTGGTCATGA-3′.
Results
Smad1 expression levels peak during stages of EB development associated with hemangioblast commitment
A major advantage of the ES/EB system is that defined cell populations can be isolated in vitro, representing early embryonic stages that are not readily accessible during mammalian embryogenesis. As a first step to investigate specificity for BMP-associated Smads, we compared their relative expression patterns at distinct stages of EB development. We took advantage of ES cells containing the GFP cDNA inserted into the Brachyury (Bra) locus, which provides a marker for the progressive commitment of ES cells to a mesendoderm fate. Shown previously,22 Bra:GFP expression combined with analysis of cell surface expression of the VEGF receptor Flk1, can be used to isolate distinct subpopulations including predominantly nonmesodermal cells (Bra−Flk1−), committed mesendoderm (Bra+, Flk1−), and mesoderm enriched for the hemangioblast, the progenitor to hematopoietic and vascular lineages (Bra+, Flk1+). We isolated these samples in addition to a variety of other EB and embryo-derived samples that represent various stages of hematopoietic and endothelial development, and compared the transcription profiles for Smad1, Smad5, and Smad8. Although Smad activity is regulated after translation, whether the genes are regulated in addition at the transcript level has not been fully evaluated. Total RNA was isolated from each sample and processed using a method that promotes quantitative amplification of cDNA, followed by Southern blotting using specific probes,23 and Smad transcript levels were normalized to those for the ribosomal protein gene L32.
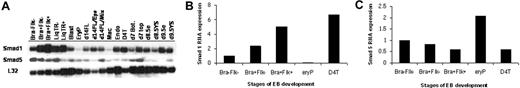
The transcript levels for Smad8 were uniformly low or undetectable and did not appear to diverge significantly (data not shown). In contrast, distinct stage-specific differences were noted for Smad1 and Smad5 as shown by a representative blot in Figure 1A. The changes in expression were particularly noteworthy for Smad1 during defined early stages of hematopoietic commitment (Figure 1B). When normalized to Bra−Flk− cells, Smad1 transcript levels are up-regulated several-fold as ES cells become committed to mesoderm (Bra+Flk−) and then further increased in the hemangioblast-enriched progenitors (Bra+Flk+). The transcript levels are markedly decreased in cells derived from primitive erythroid (EryP) colonies but maintained at a relatively high level in cells that represent endothelial fate, including D4T cells, a polyoma middle T-transformed endothelial cell line derived from EBs.15 In contrast to Smad1, Smad5 transcript levels are fairly constant during the early commitment phases, but are retained at a relatively high level in erythroid compared to endothelial derivatives (Figure 1C). The data suggest 2 possible roles for Smad1. First, Smad1 levels might influence in a positive manner the commitment of early mesoderm cells to a hemangioblast fate. Second, it might be necessary to down-regulate Smad1 levels for the appropriate development of differentiated hematopoietic cells, for example, during erythropoiesis.
Smad1 expression levels correlate with hemangioblast specification during ES/EB differentiation. (A) mSmad1 RNA expression in cells or colonies derived from the ES cell/EB system, or embryonic tissues. Ribosomal L32 expression levels serve as an internal control. Samples are as follows: Bra−Flk−, nonmesodermal early EB cells; Bra+Flk−, EB cells sorted for Brachyury (mesoderm population); Bra+Flk+, ES cells sorted for Brachyury and Flk1 (enriched for the hemangioblast population); LiqTR(− and +), transitional colonies from EB cells grown in the presence or absence of cytokines; Blast, blast (hemangioblast-derived) colonies; EryP, primitive erythroblast colonies; d14FL/EPO/mix, cells from day 14 fetal liver cultured without cytokines or with EPO (2 U/mL) only or with a mix of cytokines (EPO, 2 U/mL; kit ligand and IL-3, each 1% conditioned media; IL-11, 5 ng/mL; and TPO, 5 ng/mL); Mac, macrophage colonies; Endo, endothelial cells formed from day 6 embryoid bodies cultured with bFGF (100 ng/mL); D4T, an endothelial cell line; day 7, bottom and top, respectively, of whole day 7 mouse embryos; d8.5 (e and YS), day 8.5 mouse embryo and yolk sac, respectively; d9.5 (e and YS), day 9.5 mouse embryo and yolk sac, respectively. (B) Quantification of mSmad1 RNA expression in a subset of the samples from panel A normalized to the L32 control and graphed as fold change compared to Bra−Flk−. These are data from one representative experiment, but essentially the same results were obtained in at least 3 independent experiments. (C) The same as panel B, except analyzing relative Smad5 mRNA levels. Again, essentially the same results were obtained in at least 3 independent experiments.
Smad1 expression levels correlate with hemangioblast specification during ES/EB differentiation. (A) mSmad1 RNA expression in cells or colonies derived from the ES cell/EB system, or embryonic tissues. Ribosomal L32 expression levels serve as an internal control. Samples are as follows: Bra−Flk−, nonmesodermal early EB cells; Bra+Flk−, EB cells sorted for Brachyury (mesoderm population); Bra+Flk+, ES cells sorted for Brachyury and Flk1 (enriched for the hemangioblast population); LiqTR(− and +), transitional colonies from EB cells grown in the presence or absence of cytokines; Blast, blast (hemangioblast-derived) colonies; EryP, primitive erythroblast colonies; d14FL/EPO/mix, cells from day 14 fetal liver cultured without cytokines or with EPO (2 U/mL) only or with a mix of cytokines (EPO, 2 U/mL; kit ligand and IL-3, each 1% conditioned media; IL-11, 5 ng/mL; and TPO, 5 ng/mL); Mac, macrophage colonies; Endo, endothelial cells formed from day 6 embryoid bodies cultured with bFGF (100 ng/mL); D4T, an endothelial cell line; day 7, bottom and top, respectively, of whole day 7 mouse embryos; d8.5 (e and YS), day 8.5 mouse embryo and yolk sac, respectively; d9.5 (e and YS), day 9.5 mouse embryo and yolk sac, respectively. (B) Quantification of mSmad1 RNA expression in a subset of the samples from panel A normalized to the L32 control and graphed as fold change compared to Bra−Flk−. These are data from one representative experiment, but essentially the same results were obtained in at least 3 independent experiments. (C) The same as panel B, except analyzing relative Smad5 mRNA levels. Again, essentially the same results were obtained in at least 3 independent experiments.
Generation of inducible ES cells directing expression of Smad1
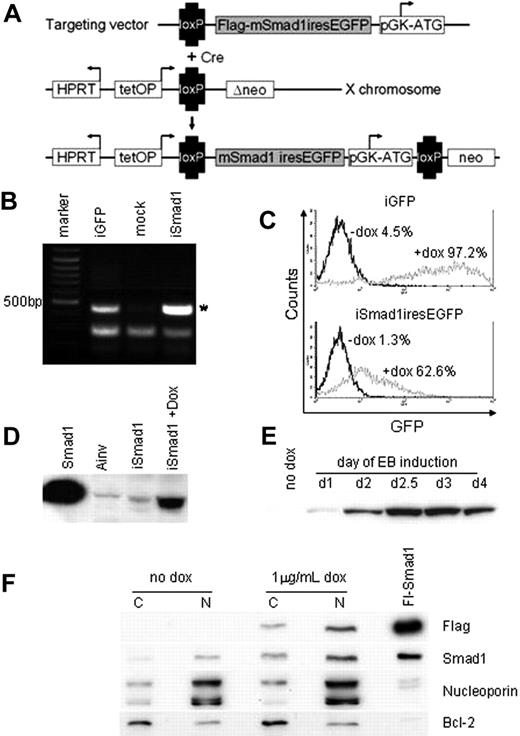
A system for conditional control of transgene expression in ES cells was described previously21 and we adapted this to direct expression of Smad1 during EB development. According to this system, a cDNA is targeted by homologous recombination downstream of the tetracycline (tet) operator. The parental Ainv18 ES cell line also contains an expression cassette for the reverse tet transactivator inserted into the constitutively expressed ROSA26 locus. We created a targeting vector with a cDNA encoding flag-tagged Smad1 upstream of an IRES element that directs expression of GFP. Following coelectroporation into Ainv18 cells with a vector expressing Cre recombinase, G418-resistant ES cell lines were generated with the Smad1-GFP cassette placed under control of the tet operator, as illustrated in Figure 2A. Site-specific transgene integration was demonstrated by PCR analysis of cDNA isolated from selected ES cell clones, designated iSmad1 (Figure 2B). The iSmad1 cells were induced with 1 μg/mL doxycycline and subsequent FACS analysis demonstrated significant levels of GFP expression (Figure 2C). Western blotting analysis of extracts from doxycycline-treated iSmad1 cells confirmed effective ectopic Smad1 expression (Figure 2D). Furthermore, sustained transgene expression is achieved throughout stages of EB development from day 1 to day 6 (Figure 2E). This demonstrates the major advantages of using inducible ES cells over retroviral approaches for evaluating the consequence of ectopic gene expression, because the retrovirus approach targets necessarily the earliest stages of development, and transgene expression is frequently silenced in host cells, thus precluding analysis at later stages.24
Generation of inducible ES cells directing Smad1 and eGFP expression. (A) iSmad1iresEGFP ES cells were generated using the approach described in Kyba et al.21 Cre-mediated recombination of the targeting vector into the lox site on the X chromosome restores G-418 (NEO) resistance leading to the isolation of transgenic cells capable of tet-on induced sustained transgene expression. (B) Genomic PCR analysis of iSmad1 ES cells demonstrates site-specific transgene integration. Ainv-GFP is an iGFP line and positive control. The asterisk indicates a 500-bp PCR product that is generated only by a correctly integrated transgene. (C) FACS analysis of iGFP and iSmad1iresEGFP ES cell lines showing responsiveness to 1 μg/mL doxycycline. (D) An anti-Flag Western blot of extracts from iSmad1iresEGFP cells untreated (iSmad1) or exposed for 8 hours to 1 μg/mL doxycycline (+dox). A Smad1+ control sample (first lane) along with extract from Ainv parental ES cells (second lane) are also included. (E) An anti-Flag Western blot of extracts made from day 6 embryoid body samples that were induced at different times of development. Once added, the induction was maintained in the EBs until day 6 as described in “Materials and methods.” In all samples EBs were cultured until day 6 when they were harvested and processed by Western blotting. (F) Western blot analysis of cytoplasmic and nuclear extracts from untreated (no dox) and iSmad1 EBs that were induced with doxycycline at day 2 and harvested at day 3 (1 μg/mL dox). For each sample, 30 μg protein lysate was analyzed with the antibodies indicated on the right side of the panel. Note that panel F is composed of 4 different panel strips from top to bottom; each independent strip represents the same samples that were analyzed with the different antisera. Nucleoporin and Bcl-2 are nuclear and cytoplasmic markers, respectively, which indicates that the enriched separation of nuclear and cytoplasmic compartments was successful. The lane marked Fl-Smad1 is a positive control whole cell extract generated by transient transfection of a flag-tagged Smad1 expression vector.
Generation of inducible ES cells directing Smad1 and eGFP expression. (A) iSmad1iresEGFP ES cells were generated using the approach described in Kyba et al.21 Cre-mediated recombination of the targeting vector into the lox site on the X chromosome restores G-418 (NEO) resistance leading to the isolation of transgenic cells capable of tet-on induced sustained transgene expression. (B) Genomic PCR analysis of iSmad1 ES cells demonstrates site-specific transgene integration. Ainv-GFP is an iGFP line and positive control. The asterisk indicates a 500-bp PCR product that is generated only by a correctly integrated transgene. (C) FACS analysis of iGFP and iSmad1iresEGFP ES cell lines showing responsiveness to 1 μg/mL doxycycline. (D) An anti-Flag Western blot of extracts from iSmad1iresEGFP cells untreated (iSmad1) or exposed for 8 hours to 1 μg/mL doxycycline (+dox). A Smad1+ control sample (first lane) along with extract from Ainv parental ES cells (second lane) are also included. (E) An anti-Flag Western blot of extracts made from day 6 embryoid body samples that were induced at different times of development. Once added, the induction was maintained in the EBs until day 6 as described in “Materials and methods.” In all samples EBs were cultured until day 6 when they were harvested and processed by Western blotting. (F) Western blot analysis of cytoplasmic and nuclear extracts from untreated (no dox) and iSmad1 EBs that were induced with doxycycline at day 2 and harvested at day 3 (1 μg/mL dox). For each sample, 30 μg protein lysate was analyzed with the antibodies indicated on the right side of the panel. Note that panel F is composed of 4 different panel strips from top to bottom; each independent strip represents the same samples that were analyzed with the different antisera. Nucleoporin and Bcl-2 are nuclear and cytoplasmic markers, respectively, which indicates that the enriched separation of nuclear and cytoplasmic compartments was successful. The lane marked Fl-Smad1 is a positive control whole cell extract generated by transient transfection of a flag-tagged Smad1 expression vector.
Finally, we tested if induced expression using this approach is sufficient to increase levels of nuclear Smad1 because expressed protein presumably requires phosphorylation for nuclear import. This is challenging because there are no antibodies specific for phospho-Smad1 (they react also to phospho-Smad5). We tested numerous antibodies purported to be specific to Smad1, but they all cross-react with Smad5. We considered that it might be possible to detect increased levels of cellular Smad1/5 and phospho-Smad1/5. Indeed, induction of Smad1 results in an apparent 2- to 3-fold increase in levels of total cellular Smad1/5 (data not shown). However, in repeated experiments, we could find no evidence for increased levels of phospho-Smad1/5 (data not shown). Yet by analyzing specifically nuclear and cytoplasmic extracts, we confirmed that induced flag-tagged Smad1 is localized predominantly in the nucleus (Figure 2F top row) and that induction of Smad1 increases levels of nuclear Smad1/5 (Figure 2F second row). Therefore, measuring steady-state phospho-Smad1/5 levels does not equate with defining the accumulation of nuclear Smad1. Total phospho-Smad levels might be tightly regulated, but not rate limiting for Smad activation, or mechanisms that normally restrict Smad to the cytoplasm in the absence of phosphorylation are limiting. Regardless, the system can be used to effectively increase the levels of nuclear (biologically active) Smad1.
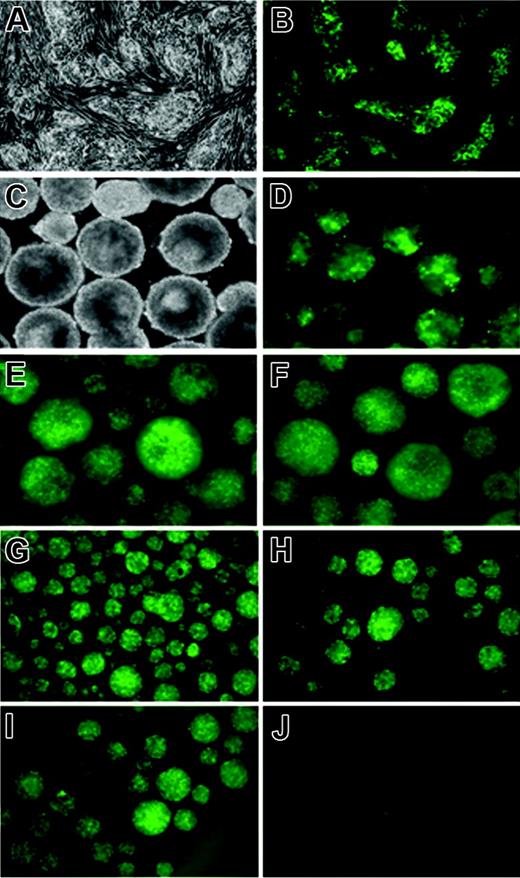
To ensure that iSmad1 ES cells express a consistent and uniform level of transgene expression, several subclones were isolated and their transgene expression profiles examined. We continued studies using one subclone line (sc5) that displayed a strong uniform level of GFP expression in response to doxycycline treatment (Figure 3A-B) and showed no background or “leaky” GFP expression in the absence of induction. The sc5 line is also capable of sustained GFP transgene expression throughout EB development (Figure 3C-F). An important and useful feature of using inducible ES cells is the ability (in principle) to support transgene expression that can subsequently be turned off. We induced EBs that were generated using sc5 ES cells with a 6-hour pulse of doxycycline from day 2 to 2.25, and GFP expression was analyzed for several days following withdrawal (washout) of the drug. High levels of transgene expression were confirmed within 6 hours after induction is initiated (Figure 3G) and GFP expression is maintained but gradually dissipates to undetectable levels by day 4 of EB development (Figure 3H-J). Although the washout is effective at reversing transgene expression, we note that this assay for GFP is not quantitative and the half-life of GFP is unlikely to reflect the corresponding turnover of Smad1. Thus, inducible ES cells directing sustained but reversible Smad1 expression were generated. These ES cells were next used to analyze quantitatively the hematopoietic potential when Smad1 expression is manipulated.
Kinetics of GFP expression in the iSmad1iresEGFP subclone 5. (A) Phase and (B) GFP fluorescence images of untreated and 1-μg/mL doxycycline-treated iSmad1iresEGFP sc5 ES cells, respectively. (C) Phase and (D-F) GFP fluorescence images of day 5.75 iSmad1iresEGFP sc5 EBs that were continuously induced starting on day 0 (D), day 2.75 (E), and day 4 (F). In panels G-J, the iSmad1iresEGFP EBs were induced from day 2 to day 2.25 according to the washout method described in “Materials and methods,” and GFP expression was monitored. (G) Day 2.25 EBs, (H) day 2.5 EBs, (I) day 3 EBs, (J) day 4 EBs. Images were visualized using an AxioCam MRm camera (Zeiss, Thornwood, NY) attached to a Zeiss Axiovert 200M microscope that was equipped with a 10×/0.30 numerical aperture Plan-Neofluar objective. Images were processed using Zeiss Axiovision software version 4.5.
Kinetics of GFP expression in the iSmad1iresEGFP subclone 5. (A) Phase and (B) GFP fluorescence images of untreated and 1-μg/mL doxycycline-treated iSmad1iresEGFP sc5 ES cells, respectively. (C) Phase and (D-F) GFP fluorescence images of day 5.75 iSmad1iresEGFP sc5 EBs that were continuously induced starting on day 0 (D), day 2.75 (E), and day 4 (F). In panels G-J, the iSmad1iresEGFP EBs were induced from day 2 to day 2.25 according to the washout method described in “Materials and methods,” and GFP expression was monitored. (G) Day 2.25 EBs, (H) day 2.5 EBs, (I) day 3 EBs, (J) day 4 EBs. Images were visualized using an AxioCam MRm camera (Zeiss, Thornwood, NY) attached to a Zeiss Axiovert 200M microscope that was equipped with a 10×/0.30 numerical aperture Plan-Neofluar objective. Images were processed using Zeiss Axiovision software version 4.5.
Forced expression of Smad1 during the hemangioblast window of EB development expands EryP colony formation
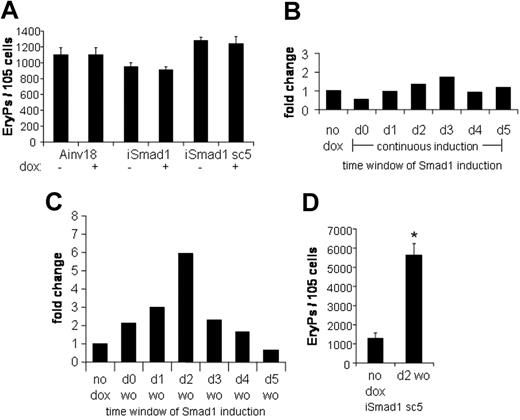
The progression of ES cells to form differentiated primitive erythroid colonies is well characterized in the EB system and correlates closely with normal mouse development. The committed erythroid progenitor, EryP-CFC, is specified by day 4 of EB development. If Smad1 regulates the development of EryP-CFC, it would be expected that induction of Smad1 at day 4 would result in altered numbers of EryP colonies after plating out progenitors at day 6 under conditions that promote erythroid development. For example, forced expression of Stat5 under these conditions leads to a 4-fold enhancement of the EryP-CFC population.25 Therefore, EBs were generated using iSmad1 cells and were induced with doxycycline from day 4 to day 6, at which point their EryP potential was examined. No effect on EryP formation was observed with this protocol in either iSmad1 mixed or iSmad1 clonal sc5 ES cell lines (Figure 4A). The EryP colony-forming potential of the Ainv18 parental ES cells was also examined and this confirmed there are no independent effects of doxycycline on the EryP program. Definitive hematopoietic development (assayed by macrophage colony formation) in iSmad1 EBs was also unchanged under these conditions (data not shown). These results indicate that overexpression of Smad1 does not affect the committed EryP-CFC population. Nor does maintenance of Smad1 expression appear to inhibit end-stage erythroid differentiation.
Activating Smad1 signaling after EryP-CFC specification has no effect on EryP numbers, but a timed pulse of Smad1 expands EryP colony formation. (A) The primitive EryP colony-forming potential of Ainv18, iSmad1 mixed, and iSmad1 sc5 EBs were examined. EBs were induced on day 4 and replated at day 6 in the presence of EPO (2 U/mL). EryP colonies were scored 5 days later. Ainv18 is the parental ES cell line and control. For each sample, n = 3. (B) Data from a single representative experiment analyzing the EryP colony-forming potential of iSmad1iresEGFP sc5 EBs examined after continuous induction initiated at different times. Developing iSmad1 EBs were untreated or treated with doxycycline on the indicated days and Smad1 expression was maintained as previously described (Figure 3C-F) until day 5.75 when the EBs were harvested and replated with EPO (2 U/mL). EryP colonies were scored 5 days after plating. Shown is the fold change compared to the untreated (no dox) control. The modest increases seen here with induction at day 2 or day 3 were not statistically significant in multiple experiments. (C) The iSmad1 EBs were untreated (no dox) or treated with the doxycycline by the washout (wo) procedure on the day indicated. EBs were harvested and replated with EPO (2 U/mL) on day 5.75. The data shown are from a single representative experiment. Shown is the fold change compared to the untreated control. (D) Statistical analysis of EryP colony formation from day 5.75 iSmad1 EBs comparing untreated and samples induced at day 2 to 2.25 with doxycycline. For each sample, n = 4. Error bars represent the SEM and the asterisk indicates P < .01 compared to the no dox samples.
Activating Smad1 signaling after EryP-CFC specification has no effect on EryP numbers, but a timed pulse of Smad1 expands EryP colony formation. (A) The primitive EryP colony-forming potential of Ainv18, iSmad1 mixed, and iSmad1 sc5 EBs were examined. EBs were induced on day 4 and replated at day 6 in the presence of EPO (2 U/mL). EryP colonies were scored 5 days later. Ainv18 is the parental ES cell line and control. For each sample, n = 3. (B) Data from a single representative experiment analyzing the EryP colony-forming potential of iSmad1iresEGFP sc5 EBs examined after continuous induction initiated at different times. Developing iSmad1 EBs were untreated or treated with doxycycline on the indicated days and Smad1 expression was maintained as previously described (Figure 3C-F) until day 5.75 when the EBs were harvested and replated with EPO (2 U/mL). EryP colonies were scored 5 days after plating. Shown is the fold change compared to the untreated (no dox) control. The modest increases seen here with induction at day 2 or day 3 were not statistically significant in multiple experiments. (C) The iSmad1 EBs were untreated (no dox) or treated with the doxycycline by the washout (wo) procedure on the day indicated. EBs were harvested and replated with EPO (2 U/mL) on day 5.75. The data shown are from a single representative experiment. Shown is the fold change compared to the untreated control. (D) Statistical analysis of EryP colony formation from day 5.75 iSmad1 EBs comparing untreated and samples induced at day 2 to 2.25 with doxycycline. For each sample, n = 4. Error bars represent the SEM and the asterisk indicates P < .01 compared to the no dox samples.
To determine if Smad1 could regulate the erythroid program by influencing an earlier hematopoietic stage, we conducted a time-course study of Smad1 induction. Initially, Smad1 was induced on each day of EB development from day 0 to day 5, and the induction was maintained until the EBs were harvested on day 5.75, at which point EryP colony-forming potential was examined. A representative example from such an experiment is shown in Figure 4B. Under these conditions, with continuous induction of Smad1, there is little if any significant change in the number of EryP-CFCs compared to control uninduced ES cells. There is a consistent reduction in EryPs derived from EBs induced on day 0, and this may reflect an effect of Smad1 to restrict ES cell development. Several experiments, as seen in Figure 4B, showed a minor increase in EryP-CFCs when induction was initiated around day 2 or day 3, but in the context of multiple experiments it was not statistically significant (not shown).
To investigate whether Smad1 has distinct influences at different stages of EB development, which might thereby mask effects under conditions of continuous induction, a time-course experiment was performed with transient induction of Smad1. In this protocol, Smad1 induction is initiated on each day, but maintained for only 6 hours, after which the doxycycline is washed out, followed by continued EB culture in fresh media until day 5.75, when EryP colony-forming potential is assessed. Under these conditions, striking effects in EryP development are observed, as shown in the data from one representative experiment in Figure 4C. The data generate a Gaussian-like distribution with a maximal increase in the number of EryP colonies derived when the induction of EBs is initiated at day 2. Quantification of data derived from multiple experiments (Figure 4D) shows that the induction of Smad1 under these conditions results in approximately a 5-fold increase in the number of EryP colonies (P < .01). Two conclusions can be drawn from these results. First, Smad1 affects erythropoiesis when it is expressed at the time of commitment to the hemangioblast. Second, sustained Smad1 expression during subsequent stages of EB development must be inhibitory to expanded hematopoietic development, which under conditions of continuous expression thereby masks the enhancement seen by transient expression restricted to day 2 to 2.25.
Smad1 expression expands the development of macrophage, megakaryocyte, and definitive mixed-lineage colonies
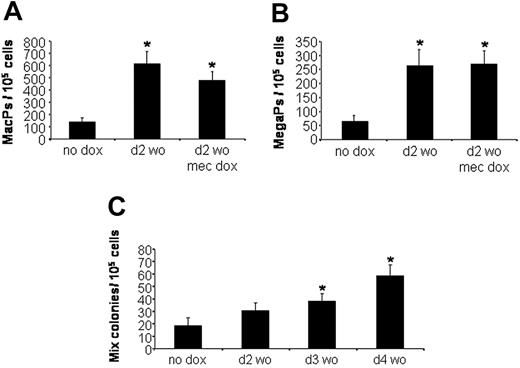
Because the changes in EryP colony formation are mediated by an early pulse of Smad1, we considered whether Smad1 is sufficient to mediate a general expansion of hematopoietic development. Using the same protocol, EBs were left uninduced or treated transiently with doxycycline for 6 hours beginning at day 2 and plated at day 5.75 under defined culture conditions that are permissive for the development of macrophage, megakaryocyte, or multilineage mixed (erythroid/megakaryocyte/macrophage/granulocyte) colonies. The macrophage and megakaryocyte progenitor populations were expanded by pulsed Smad1 approximately 5-fold, similar to the analysis of EryPs (Figure 5A and 5B, respectively). The colony numbers are unchanged if doxycycline is included in the methylcellulose plating media, again indicating that Smad1 induction does not inhibit cell differentiation. The effect on definitive mixed-lineage colonies was significant but less robust (Figure 5C), and interestingly the effect was maximized when EBs were induced at a slightly later stage (day 3 or 4) compared to the peak activity for expansion of EryPs (day 2).
Transient Smad1 expression also expands macrophage, megakaryocyte, and definitive mixed-lineage progenitors. Developing iSmad1 EBs were left untreated (no dox) or treated with doxycycline according to the day 2 washout protocol. EBs were then harvested at day 5.75 and replated to score for (A) macrophage progenitors (MacPs), (B) megakaryocyte progenitors (MegaPs), or (C) mixed-lineage progenitors (Mix). After plating, the macrophage and megakaryocyte colonies were scored on day 7, and the mixed-lineage colonies were scored on day 8. In panels A and B, the doxycycline was added for 6 hours at day 2, and a subset of EBs was also exposed to doxycycline at the time of replating in methylcellulose (mec dox) to test if this inhibits differentiation, which it does not. In panel C, the iSmad1 EBs were untreated (no dox) or treated with doxycycline for 6 hours on day 2 (d2 wo), day 3 (d3 wo), or day 4 (d4 wo). Induction at later time points did not expand the number of mixed-lineage colonies (not shown). In all cases, for each sample, n = 3. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
Transient Smad1 expression also expands macrophage, megakaryocyte, and definitive mixed-lineage progenitors. Developing iSmad1 EBs were left untreated (no dox) or treated with doxycycline according to the day 2 washout protocol. EBs were then harvested at day 5.75 and replated to score for (A) macrophage progenitors (MacPs), (B) megakaryocyte progenitors (MegaPs), or (C) mixed-lineage progenitors (Mix). After plating, the macrophage and megakaryocyte colonies were scored on day 7, and the mixed-lineage colonies were scored on day 8. In panels A and B, the doxycycline was added for 6 hours at day 2, and a subset of EBs was also exposed to doxycycline at the time of replating in methylcellulose (mec dox) to test if this inhibits differentiation, which it does not. In panel C, the iSmad1 EBs were untreated (no dox) or treated with doxycycline for 6 hours on day 2 (d2 wo), day 3 (d3 wo), or day 4 (d4 wo). Induction at later time points did not expand the number of mixed-lineage colonies (not shown). In all cases, for each sample, n = 3. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
Genes associated with hemangioblast development are affected by induced expression of Smad1
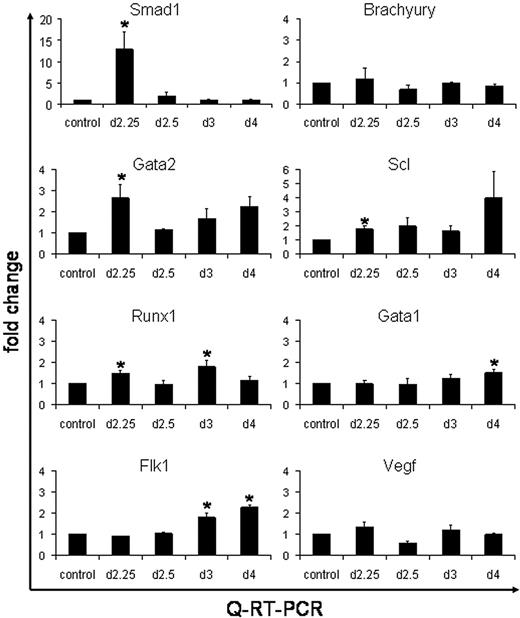
These results suggest that the enhanced levels of hematopoiesis caused by Smad1 are mediated through expansion of an early progenitor, for example, the hemangioblast. It is also plausible that the Smad1-mediated increase in the number of EryP colonies occurs from a general expansion of prehematopoietic mesoderm. To examine gene expression changes caused by induced Smad1 we used quantitative real-time PCR (QPCR) analysis. The iSmad1 ES cells were either left untreated (as control) or induced for 6 hours starting at day 2, followed by washout of the inducer. EBs were harvested on day 2.25, day 2.5, day 3, or day 4. Total RNA was isolated and converted to cDNA, which was then subjected to QPCR analysis (Figure 6). Quantification of Smad1 RNA levels confirmed that transgene expression levels are effectively induced (on average 12-fold) by 6 hours. The QPCR data demonstrate that the washout technique effectively silences the transgene with high efficiency because Smad1 transcripts are reduced essentially to baseline by the day 2.5 time point. No significant changes were observed in the levels of Brachyury transcripts, indicating that the enhancement to EryP colony formation is not caused indirectly due to a general increase in the commitment of ES cells to mesoderm. Genes were next analyzed that have been previously associated with hemangioblast development, including Gata2, Runx1, Scl, and Flk1. A statistically significant increase in the levels of Gata2, Runx1, and Scl transcripts were found by day 2.25, which is consistent with increased commitment of mesoderm to hemangioblast fate. Interestingly, transcript levels for Flk1 and for Vegf are not altered in EBs induced with Smad1, indicating that any Smad1-mediated effects on hemangioblast development are downstream of the Flk1 pathway. There is also no significant effect in the levels of Gata1 transcription at the early stages, which again suggests that Smad1 does not induce directly the commitment to an erythroblast fate, but rather that the increased numbers of EryP colonies is derived through enhanced hemangioblast development.
QPCR analysis shows activation by Smad1 of hemangioblast-associated genes. The iSmad1 EBs were left untreated or treated with doxycycline from day 2 to 2.25. Age-matched untreated and treated samples were harvested at day 2.25, day 2.5, day 3, or day 4. The cDNA was generated from isolated RNA and QPCR was performed. The data were processed using the 2-ΔΔCT method.26 The median of each sample was normalized to its respective age-matched untreated control and the median average (n = 3) was graphed as fold change in RNA expression. The QPCR analysis was performed for Smad1, Brachyury, Gata2, Scl, Runx1, Flk1, and Vegf, as indicated. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
QPCR analysis shows activation by Smad1 of hemangioblast-associated genes. The iSmad1 EBs were left untreated or treated with doxycycline from day 2 to 2.25. Age-matched untreated and treated samples were harvested at day 2.25, day 2.5, day 3, or day 4. The cDNA was generated from isolated RNA and QPCR was performed. The data were processed using the 2-ΔΔCT method.26 The median of each sample was normalized to its respective age-matched untreated control and the median average (n = 3) was graphed as fold change in RNA expression. The QPCR analysis was performed for Smad1, Brachyury, Gata2, Scl, Runx1, Flk1, and Vegf, as indicated. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
We note that the enhanced expression of hemangioblast markers is transient and that at the 2.5 day time point the levels return to baseline. However, at later time points, from day 3 to day 4 there is a second “wave” of enhanced expression, this time including Gata1, and this correlates with the commitment to hematopoietic fate. In this case the Flk1 levels are also enhanced significantly, which would be consistent with expanded hemangioblast development or the coordinate commitment of hematopoietic cells or angioblasts from the previously induced hemangioblast progenitor. Together, the data are consistent with the timed washout analysis, suggesting that the enhanced generation of EryP colonies is mediated by the expansion of an earlier progenitor, which leads subsequently to increased commitment to hematovascular development.
Smad1 enhances hemangioblast development
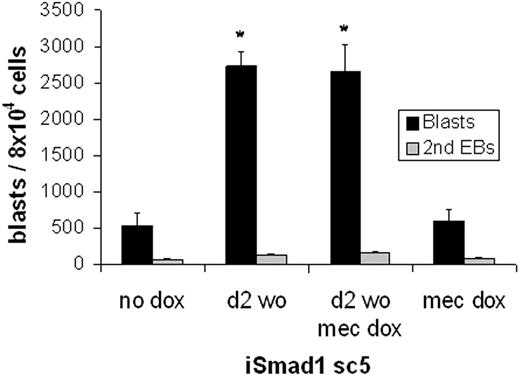
Finally, to test directly the hypothesis that Smad1-mediated expansion of the number of hematopoietic colonies results from expansion of the hemangioblast population, in vitro BL-CFC assays were performed. Again EBs were generated and then either left untreated or induced at day 2 for 6 hours with subsequent washout of the inducer. EBs were harvested at day 3 and plated under conditions established previously that support BL-CFC development (Figure 7). Compared to control ES cells, induced expression of Smad1 resulted in a significant (approximately 5-fold) increase in BL-CFC formation (P < .05). Notably, this expansion is sufficient to account fully for the increased number of hematopoietic colonies. Further induction of Smad1 at the time of replating EB cells for the blast colony formation did not augment blast development, again consistent with an effect that functions only early in EB development.
Smad1 enhances hemangioblast development. The blast-forming potential of iSmad1 EBs was examined. The iSmad1 EBs were left untreated (no dox) or treated with doxycycline from day 2 to 2.25 (d2 wo) and replated at day 3.75 for blast colony formation. In other samples, the EB-derived cells were replated with the addition of doxycycline in the methylcellulose (mec dox). Blast colonies (▪) and secondary EBs (⊡) were scored on day 4 after plating. For each sample, n = 3. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
Smad1 enhances hemangioblast development. The blast-forming potential of iSmad1 EBs was examined. The iSmad1 EBs were left untreated (no dox) or treated with doxycycline from day 2 to 2.25 (d2 wo) and replated at day 3.75 for blast colony formation. In other samples, the EB-derived cells were replated with the addition of doxycycline in the methylcellulose (mec dox). Blast colonies (▪) and secondary EBs (⊡) were scored on day 4 after plating. For each sample, n = 3. Error bars indicate the SEM and the asterisks indicate a P < .01 compared to the no dox samples.
Discussion
The hemangioblast is thought to represent the first committed hematopoietic progenitor specified from mesoderm. The transient nature of this multipotent cell makes it particularly challenging to define the transcriptional regulatory pathways that support hemangioblast development. Here we show that enforced expression of Smad1 within a defined developmental window causes a significant increase in the number of primitive erythroblasts and other hematopoietic progenitors, and that this is likely an indirect result of Smad1 enhancing hemangioblast commitment.
This interpretation is consistent with the quantification of Smad1 transcript levels during EB development, which showed a peak precisely within the cell population predicted to be most enriched for the hemangioblast (Bra+, Flk1+). We found no significant difference in the potential of ES cells expressing continuously Smad1 from day 2 to day 6 to generate erythroid progenitors, indicating that day 4 erythroblasts are not responsive to Smad1. However, 2 striking observations were made using the washout method in place of continuous induction. First, there is a significant increase in the generation of EryP colonies, dependent on a specific window of Smad1 expression, with a maximal enhancement noted using induction at day 2 followed by washout after 6 hours. Because EB cells are expected to commit to an erythroblast fate around day 4, the effect of Smad1 is consistent with expansion of an earlier progenitor. Second, continued expression of Smad1 blocks this expansion since EBs induced continuously from day 2 fail to generate the enhanced numbers of EryP colonies seen using the transient pulsed induction. The analysis of RNA levels confirmed that the Smad1 transcript levels are rapidly depleted following washout. The data suggest that Smad1 promotes the expansion of hematopoietic progenitors between days 2 and 3 but then subsequently restricts further hematopoietic expansion unless down-regulated. Because forced expression of Smad1 at day 4 does not inhibit the normal number of EryP colonies, the system is able to support baseline normal numbers of hematopoietic cells even under conditions of enhanced Smad1 levels.
In the ES/EB system, the hemangioblast is thought to develop between days 2.5 to 3.5,16 so our data are consistent with the ability of enhanced Smad1 signaling to activate target genes of the hemangioblast program. The subsequent increase in hemangioblast numbers would then expand downstream hematopoietic progenitors, including the primitive erythroblast. Indeed, the BL-CFC assay confirmed that transient induction of Smad1 enhanced hemangioblast numbers, at a level that is fully sufficient to account for the increase in numbers of EryPs and other hematopoietic progenitors. QPCR analysis on EB-derived cells following transient Smad1 induction confirmed increased levels of hemangioblast-relevant marker genes by day 2.25, including transcripts encoding Gata2, Scl, and Runx1. We note that a significant increase at day 2.25 was not seen for Flk1, which is thought to be required for the transition of mesoderm to a hemangioblast fate.27 Mouse embryos deficient for Flk1 fail to generate hematopoietic and endothelial progenitors.28 However, a more detailed analysis using Flk1−/− ES cells demonstrated that the gene is not essential for BL-CFC specification but is required for hematopoietic commitment of the hemangioblast.29 Thus, BL-CFCs develop normally in Flk1−/− EBs, but fail to respond to VEGF.29 Of course, in our system the EBs do express normal levels of Flk1; the levels are just not enhanced by Smad1 at the early stages. Because the Flk1 levels are increased by day 3, the expansion of BL-CFC development may still be dependent on Flk1, but the Flk1 gene may be a less direct target of Smad1 compared for example to Gata2 or Runx1.
In addition to Flk1, the expression levels of hematopoietic regulatory genes including Gata1 are increased at day 3 to 4 by the early pulse of Smad1, consistent with the timing of commitment to hematopoietic or angiogenic progenitors. We expect that increased commitment to hemangioblast fate should result also in expansion of other hematopoietic lineages and endothelial cells. Macrophage and megakaryocyte progenitors are expanded by Smad1 exactly as shown for EryPs. By FACS analysis we found that Smad1 induction causes a relatively modest but consistent expansion of VE-cadherin–positive cells at day 5 of EB development (average in 4 independent experiments of 1.7-fold, ± 0.2; data not shown). This modest increase might reflect the normal generation of endothelial cells independent of the hemangioblast (thus diluting the apparent fold-expansion of those that are hemangioblast-derived). Alternatively, the transient induction of Smad1 (by washout) might bias the progenitor toward hematopoietic fate, perhaps accentuated by the washout protocol, since the expression profiling data (Figure 1) indicate that Smad1 continues to be expressed at relatively high levels in the endothelial lineage. Interestingly, the expansion of the definitive mixed-lineage colonies was more efficient when induced at slightly later stages (day 3 or 4), compared to the primitive colonies (day 2), suggesting the potential for heterogeneity in the hemangioblast population for progenitors that are more committed to primitive or definitive lineages.
Smad1 is a key mediator of the BMP signaling pathway, which is implicated in regulating various aspects of hematopoiesis.30 Forced expression of BMP4 during embryogenesis expands hematopoietic mesoderm.31 Loss of BMP signaling, either by targeted mutation of the ligand3 or forced expression of dominant-negative receptors,32,33 blocks the development of hematopoietic mesoderm. Similar conclusions were derived using the mouse ES cell system, since BMP4 enhances hematopoietic mesoderm,34 and inhibition of the pathway by Smad6 expression blocks the emergence of Flk1+ mesoderm.27 We showed previously in the Xenopus system using an inducible Smad6 isoform that BMP signaling is still required for primitive erythropoiesis after the time that mesoderm is specified,35 and the results reported here support this interpretation in the mouse ES system, but in addition define the developmental window specifically at the stage of hemangioblast development.
Our results are not entirely consistent with data showing that in serum-free media EB cells respond to BMP4 to activate hematopoietic regulatory genes only after day 4 of culture.36 One possibility is that BMP4 signaling functions during early hemangioblast development through Smad1 and then stimulates expression of the erythroid differentiation program through a different mediator, for example, Smad5. This would be consistent with the distinctions we found in the relative expression patterns for Smad1 and Smad5. Loss of Smad5 results in increased numbers of HPP-CFCs in the yolk sac and increased BL-CFCs in EBs,13 which is also consistent with a role for Smad5 in directing differentiation or negatively regulating early progenitors during hematovascular development. Although Smad1 and Smad5 are closely related genes, they may play very different functions in regulating the transition states of early progenitors. Similar conclusions were made in zebrafish with regard to early dorsal-ventral patterning, based on differential activities and responsiveness to ligands, which indicates that Smad1 acts later and may be a target of Smad5.37 Indeed, differential activities may not be restricted to the BMP pathway, because ALK1 can transduce TGF-β signals through Smad1.38 Defining the differential activities of Smad1 and Smad5 with respect to progenitor biology may be essential for fully understanding how BMP signaling regulates hematovascular development.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisment” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: B.T.Z. designed the study, performed experiments, and wrote the manuscript; S.L., I.T., and L.B. performed experiments; M.L.-K., M.K., and G.K. provided expert unpublished technical advice and essential reagents; G.K. also assisted in study design; and T.E. designed the study and wrote the manuscript. Each of the authors contributed significantly to this study.
Acknowledgments
The authors are particularly grateful to George Daley (Harvard Medical School, Boston, MA) for providing the Ainv18 parental ES cells and targeting vector. Reagents were also provided by Brigid Hogan (Duke University, Durham, NC), and Genetics Institute. We thank Irene Puga (Albert Einstein College of Medicine) for providing advice and reagents for comparing nuclear and cytoplasmic lysates and Catherine Laiosa (Albert Einstein College of Medicine) for technical help with the ES/EB system. We also thank members of the Keller laboratory, including Tara Huber and Valerie Gouon-Evans, for providing important advice and members of the Evans laboratory for critical analysis of the data.
This work was supported by grants from the National Institutes of Health (HL056182; T.E. and G.K.) and the Irma T. Hirschl Trust. B.T.Z. is supported by a Medical Scientist Training Program training grant (T32 GM007288).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal