Abstract
A significant proportion of children with T-cell acute lymphoblastic leukemia (T-ALL) continue to fail therapy. Consequently, characterization of the cells that proliferate to maintain the disease should provide valuable information on the most relevant therapeutic targets. We have used in vitro suspension culture (SC) and nonobese diabetic–severe combined immune deficient (NOD/SCID) mouse assays to phenotypically characterize and purify T-ALL progenitor cells. Cells from 13 pediatric cases were maintained in vitro for at least 4 weeks and expanded in 8 cases. To characterize the progenitors, cells were sorted for expression of CD34 and CD4 or CD7 and the subfractions were evaluated in vitro and in vivo. The majority of cells capable of long-term proliferation in vitro were derived from the CD34+/CD4− and CD34+/CD7− subfractions. Moreover, the CD34+/CD4− or CD7− cells were the only subfractions capable of NOD/SCID engraftment. These T-ALL cells successfully repopulated secondary and tertiary recipients with equivalent levels of engraftment, demonstrating self-renewal ability. The immunophenotype and genotype of the original leukemia cells were preserved with serial passage in the NOD/SCID mice. These data demonstrate the long-term repopulating ability of the CD34+/CD4− and CD34+/CD7− subfractions in T-ALL and suggest that a cell with a more primitive phenotype was the target for leukemic transformation in these cases.
Introduction
Childhood acute lymphoblastic leukemia (ALL), the most common pediatric cancer, is a heterogeneous disease, composed of different genetic, clinically relevant, subtypes. Developments in flow cytometric techniques and the availability of lineage-associated monoclonal antibodies have permitted characterization of normal and leukemia cells and affirmed the immunophenotypic heterogeneity in ALL.1 The different ALL subtypes have been considered to represent clonal expansion of malignant cells that had been transformed at different maturation stages within the hemopoietic hierarchy. Therefore, the degree of maturation of the initial transformed target cell would influence the characteristics of resulting leukemic blasts.2 Furthermore, analyses of chromosomal mutations, and TcR and IgH rearrangements, provided evidence to support the hypothesis that T- and B-lineage ALL originate in cells already committed to the T- or B-cell lineages, respectively.3-7 However, these investigations were performed on bulk ALL populations, which are most likely to be the progeny of cells in the stem cell pool and hence may not be representative of the original transformed cells, from which the leukemia has arisen.
A number of recent studies have provided evidence that Philadelphia chromosome–positive (Ph+) ALL and B-cell precursor ALL may arise in cells with a primitive CD34+/CD10−/CD19−/CD33−/CD38− phenotype rather than in committed B-lymphoid cells.8-13 Therefore, B-cell precursor ALL, in common with acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), may arise as the result of a transformation event in a primitive hemopoietic cell. The transformed leukemia stem cells have self-renewal ability to perpetuate the disease, but can differentiate only to a limited extent. Consequently, these leukemia stem cells may play a critical role in disease progression and it is therefore important to increase our understanding of the biologic characteristics of this population.
T-ALL represents around 15% of pediatric ALL cases but the early stages of development of this leukemia subtype are relatively poorly understood. Difficulties in maintaining primary cultures of ALL cells have hampered investigations into the biology of this malignancy and there are only a few reports on growth of T-ALL using in vivo models.14-20 We have developed an in vitro suspension culture (SC) assay that supports the long-term proliferation of cells from childhood B-ALL, which, together with the nonobese diabetic–severe combined immune deficient (NOD/SCID) xenotransplant model, permits identification and characterization of the leukemia cells that may be responsible for maintenance of the disease in vivo.12 In the present study we have used the SC assay together with the NOD/SCID mouse model to characterize leukemic cells with long-term proliferative potential in childhood T-ALL.
Materials and methods
Patient cells
Bone marrow (BM) cells from children (median age 6 years, 9 months; range, 1-15 years) with T-ALL at presentation and/or relapse were obtained after informed consent and with approval of the Research Ethics Committee of the United Bristol Healthcare National Health Service (NHS) Trust. The characteristics of the patients included in the study are shown in Table 1. Patients were selected only on the basis of availability of material for study. Cells were separated using Ficoll-Hypaque (Sigma-Aldrich, Poole, United Kingdom) to obtain a mononuclear cell (MNC) population, then frozen in Iscove modified Dulbecco medium (IMDM; Gibco, Paisley, United Kingdom) with 50% fetal calf serum (FCS; Gibco) and 10% DMSO (Manor Park Pharmaceuticals, Bristol, United Kingdom) and stored in liquid nitrogen.
Patient characteristics
| Patient . | Karyotype . | Age at diagnosis, y . | Sex . | Disease status at biopsy . | WBC count at biopsy, × 109/L . |
|---|---|---|---|---|---|
| 1 | +10, +21 | 1 | F | Diagnosis | 105 |
| 2 | 46XY | 15 | M | Diagnosis | 406 |
| 3 | 46XX | 1 | F | Diagnosis | 233 |
| 4 | 46XY | 5 | M | Diagnosis | 44 |
| 5 | +4, +8 | 11 | M | Diagnosis | 14 |
| 6 | Complex | 1 | M | Diagnosis | 700 |
| 7 | 46XY | 14 | M | Diagnosis | 90 |
| 8 | 46XY | 5 | M | Diagnosis | 75 |
| 9 | del 9 | 9 | M | Diagnosis | 92 |
| 10 | 46XX | 6 | F | Diagnosis | 41 |
| 11 | N/A | 14 | F | Diagnosis | 118 |
| 12 | del 6q | 14 | F | Diagnosis | 362 |
| 13* | Complex | 15 | M | Diagnosis | 450 |
| 14 | t(11;14) | 6 | M | Diagnosis | 300 |
| 15 | +8,del 9p | 4 | M | Relapse 1 | 81 |
| 16 | del 9p | 9 | M | Diagnosis | 421 |
| Patient . | Karyotype . | Age at diagnosis, y . | Sex . | Disease status at biopsy . | WBC count at biopsy, × 109/L . |
|---|---|---|---|---|---|
| 1 | +10, +21 | 1 | F | Diagnosis | 105 |
| 2 | 46XY | 15 | M | Diagnosis | 406 |
| 3 | 46XX | 1 | F | Diagnosis | 233 |
| 4 | 46XY | 5 | M | Diagnosis | 44 |
| 5 | +4, +8 | 11 | M | Diagnosis | 14 |
| 6 | Complex | 1 | M | Diagnosis | 700 |
| 7 | 46XY | 14 | M | Diagnosis | 90 |
| 8 | 46XY | 5 | M | Diagnosis | 75 |
| 9 | del 9 | 9 | M | Diagnosis | 92 |
| 10 | 46XX | 6 | F | Diagnosis | 41 |
| 11 | N/A | 14 | F | Diagnosis | 118 |
| 12 | del 6q | 14 | F | Diagnosis | 362 |
| 13* | Complex | 15 | M | Diagnosis | 450 |
| 14 | t(11;14) | 6 | M | Diagnosis | 300 |
| 15 | +8,del 9p | 4 | M | Relapse 1 | 81 |
| 16 | del 9p | 9 | M | Diagnosis | 421 |
WBC indicates white blood cell.
Biopsy material at relapse was also used.
ALL cell sorting
Thawed ALL cells were suspended in Hanks balanced salt solution (HBSS; Sigma-Aldrich) plus 2% human albumin solution (HAS; BPL, Elstree, United Kingdom) at 107 cells/mL. Cells were stained with monoclonal antibodies CD34-PE and CD4 or CD7-FITC, and separate aliquots were stained with irrelevant IgG1 antibodies as isotype controls (all BD Biosciences, Oxford, United Kingdom). Cells were washed twice in HBSS with 2% HAS and maintained on ice prior to sorting. Cells were sorted using a MoFlo Cell sorter (Cytomation, Freiburg, Germany), on the basis of fluorescence intensity after gating on cells with low forward and side scatter. Sort gates were set up to exclude at least 99.9% of the cells in the isotype controls, and gate separations of at least 10 channels were used to discriminate positive from negative fractions to ensure purity. The purity of all sorted subfractions, from each patient sample, was checked during and after sorting as previously described12 ; this was always more than 98% for all populations. Cells were sorted into IMDM plus 50% FCS, maintained at 4°C, then washed, resuspended at known cell concentrations, and used in suspension culture or NOD/SCID assays.
Suspension culture assay
Suspension cultures were initiated with unsorted ALL cells or with sorted subfractions at up to 5 × 105 cells/mL, supplemented with IL-3, IL-7, and stem cell factor (SCF) and maintained as described previously.12 At weeks 1, 3, 5, and 6, the cells removed during half-media changes were analyzed by flow cytometry to determine viability and absolute cell numbers. Viability was determined by 7-amino-actinomycin D (7AAD) uptake, and absolute cell counts were established using Flow-Count Fluorospheres (Beckman Coulter, High Wycombe, United Kingdom) according to the manufacturer's instructions. Cells removed at weeks 2 and 4 were used for cytogenetic or morphologic analyses. Suspension cultures were maintained for 6 weeks then harvested; total cell content was assessed by flow cytometry and cytogenetic/morphologic analyses performed.
Transplantation of leukemic cells into NOD/SCID mice
NOD/SCID mice were bred and maintained at the University of Bristol Animal Services Unit. One hour before transplantation, 6- to 8-week-old NOD/SCID mice were irradiated with 2.5 Gy acute X-rays from a linear accelerator at a rate of 7.5 cGys−1. Unsorted ALL cells and sorted subfractions were resuspended in 0.3 mL IMDM plus 5% FCS and injected into the lateral tail vein. Animals were maintained for 8 to 10 weeks and killed electively or up until they began to exhibit clinical symptoms of the disease. At the time each mouse was killed, its gross anatomy was inspected and femoral BM samples were removed for flow cytometric and fluorescence in situ hybridization (FISH)/histologic analyses.
In some cases, MNC preparations harvested from NOD/SCID BM were used for serial transplantation experiments to evaluate the self-renewal ability of sorted subfractions. For comparison with primary transplants, equal numbers of CD45+ cells were inoculated into the serial recipients. These cells were not enriched for any particular phenotype prior to evaluation in sequential xenografts.
Evaluation of transplanted cells
Flow cytometric analysis of murine tissues.
The immunophenotype of all xenografts was examined using antibodies against CD45 and appropriate lineage markers. Cell suspensions were lysed in ammonium chloride then washed and resuspended in HBSS plus 2% HAS. In every case, cells were stained with CD45 along with 1 or 2 of the following lineage markers: CD2, CD3, CD4, CD7, CD8, CD19, CD33, CD34 (BD Biosciences), to compare the immunophenotype of the engrafted cells to that of the original patient sample at diagnosis/relapse. Separate aliquots were used as isotype controls. Cells were also stained with 7AAD as a marker of cell viability. After staining cells were washed, resuspended in HBSS plus 2% HAS and maintained on ice prior to analysis using a Coulter EPICS XL-MCL flow cytometer (Beckman Coulter). Nonviable cells were gated out based on 7AAD uptake, matched isotype controls were run for each tissue sample and used to define gate settings, which excluded at least 99.9% of the cells in the isotype control. Any positive value (ie, < 0.1%) for the isotype control was then subtracted from the percentage positive in the CD45-stained samples. Cells from noninjected NOD/SCIDs were used as additional controls to verify the gate settings. We defined human cell engraftment as expression of at least 0.1% CD45+ cells in a sample. Aliquots of cells derived from murine tissue were plated onto slides for morphologic or cytogenetic analysis.
Cytogenetic and morphologic analysis.
Cytogenetic analysis was performed on patient samples at diagnosis, on cells harvested from cultures at weeks 2, 4, and 6 and on cells harvested from transplanted NOD/SCID marrow by the Regional Cytogenetics Unit, Southmead Hospital. At least 50 nuclei per sample were scored on preparations from cultured cells and NOD/SCID marrow. If cytogenetic analysis was not possible, then cytospins of cultured cells and NOD/SCID BM were prepared. Cytospins were fixed and stained with Hema-Gurr (BDH, Poole, United Kingdom) then examined and assessed by a hematology consultant at Bristol Haematology and Oncology Centre.
Identification of clonal receptor gene rearrangements.
Diagnostic samples and cells from xenografts established with sorted ALL subfractions were screened for the presence of T-cell receptor gamma (TCRG) gene rearrangements, present in approximately 95% of patients with T-ALL. The polymerase chain reaction (PCR) primers and conditions were as described previously,21,22 with PCR products subjected to heteroduplex analysis for clonality assessment23 followed by sequencing of monoclonal products. The exact junctional rearrangement was established by comparison with all known human TCRG germ line sequences (http://imgt.cines.fr/vquest/pagesdyn_v4/index.html). Where cells harvested from NOD/SCID mice were screened, control experiments using noninjected NOD/SCID BM showed no contamination of PCR products with murine DNA.
Statistical analysis.
Statistical analyses were performed using matched paired t tests or analysis of variance with Tukey post-hoc analysis of means.
Results
Growth of T-ALL cells in suspension culture
Initially, cells from 15 patients with T-ALL were set up in suspension culture supplemented with IL-3, IL-7, and SCF. Cells from 13 of the patients were maintained for up to 4 weeks in culture, and in 11 patients, cells were maintained for up to 6 weeks. Cells from patients 9 and 11 were maintained for only 2 and 3 weeks in SC, respectively, and were not used for any further experiments. It was possible to expand the number of cultured cells in 8 of the 11 patient samples that proliferated up to week 6 (2- to 25-fold) from 5 × 105 cells at initiation of culture to 1.25 × 107 at week 6. Cytogenetic analyses by FISH were possible in 7 of the 13 patient cell samples maintained in SC and confirmed that 55% to 100% of cultured cells had the same karyotype as that seen in the patient samples at diagnosis. The numbers of abnormal cells detected by FISH were similar to the original diagnostic reports. Morphologic analyses on cultured cells from the remaining 6 patients, who had either a normal karyotype or for whom FISH probes were not available, demonstrated that most were undifferentiated blasts (60%-100%) and effete cells.
Phenotypic characterization of T-ALL cells with long-term proliferative ability in vitro
Mononuclear diagnosis cells from 8 of the 13 patient samples that proliferated in SC were labeled with antibodies against CD34 and CD4. All 4 subfractions were sorted and their proliferative ability evaluated in SC (Figure 1A). At sorting, the majority of nucleated cells were CD34−/CD4− (48% ± 5%); the CD34−/CD4+ subfraction represented 33% ± 6% whereas the CD34+/CD4+ and CD34+/CD4− subfractions represented only 4% ± 2% and 10% ± 5%, respectively. After 7 days in SC, the majority of proliferating cells (45% ± 9%) were derived from the CD34+/CD4− subfraction. Subsequently, the proportion of proliferating cells derived from the CD34+/CD4− subfraction increased throughout the course of the assay to 51% ± 10% at week 3 and 65% ± 13% at week 6. The proportion of cells derived from this subfraction was significantly higher than from any other subfraction (F = 21***, P < .001). In the cultures of CD34+CD4− cells, there was a 1.4- to 6835-fold expansion in cell numbers with an average 9.6 × 104 cells at initiation and an average 1.55 × 106 cells at week 6.
Proliferation of T-ALL cells sorted for CD34 and CD4 or CD7 in suspension culture. ALL cells from 8 patients were sorted for expression of CD34 and CD4 (A) and cells from 7 patients were sorted for expression of CD34 and CD7 (B). The proliferative capacity of each of the sorted subfractions and unsorted controls were evaluated in SC. Cultures were maintained with weekly half-media changes, and absolute cell counts derived from each sorted population and unsorted controls were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells, represented by each sorted population. The proportion of the total cells derived from each sorted subfraction is presented as the mean plus or minus the standard error (SE).
Proliferation of T-ALL cells sorted for CD34 and CD4 or CD7 in suspension culture. ALL cells from 8 patients were sorted for expression of CD34 and CD4 (A) and cells from 7 patients were sorted for expression of CD34 and CD7 (B). The proliferative capacity of each of the sorted subfractions and unsorted controls were evaluated in SC. Cultures were maintained with weekly half-media changes, and absolute cell counts derived from each sorted population and unsorted controls were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells, represented by each sorted population. The proportion of the total cells derived from each sorted subfraction is presented as the mean plus or minus the standard error (SE).
Cells from 4 of the patients described in the previous paragraph and 3 additional patients were labeled with CD34 and CD7 and the sorted subfractions were evaluated in SC (Figure 1B). The majority of cells at sorting were CD34−/CD7+ (60% ± 12%). The CD34−/CD7− subfraction represented 20% ± 5%, whereas the CD34+/CD7+ and CD34+/CD7− subfractions represented 7% ± 6% and 6% ± 4%, respectively. Similar numbers of proliferating cells were detected from each subfraction after 1 week in culture. However, after 3 weeks in culture, the majority of cells were derived from the CD34+/CD7− subfraction (53% ± 10%) and at week 6, 75% ± 2% of cells were derived from this subfraction (F = 8.1**, P < .003). In the cultures of CD34+/CD7− cells there was a 3- to 417-fold expansion in cell numbers starting with an average of 1.2 × 105 at initiation of culture to an average 3.4 × 106 at week 6.
Cytogenetic analyses, where possible, and morphologic analyses were carried out on the cultured cells at weeks 2, 4, and 6. Analyses by FISH confirmed that 52% to 100% of cultured cells derived from the CD34+/CD4+, CD34+/CD4−, CD34+/CD7+, and CD34+/CD7− subfractions contained cells with the same karyotype as the unsorted diagnostic patient samples. In the samples from cases with a normal karyotype or those for which FISH probes were not available, morphologic analyses revealed that the majority of cultured cells derived from these sorted subfractions had blast morphology, CD34+/CD4− (80%-95% blasts) and CD34+/CD7− (90%-100% blasts).
Engraftment of T-ALL cells in NOD/SCID mice
Unsorted cells from 10 patients were evaluated for their ability to engraft NOD/SCID mice (Table 2). In each case, engraftment of T-ALL cells was achieved using less than or equal to 107 cells, although the level of engraftment varied considerably among patient samples. In cases 2, 6, 12, and 14, high levels of engraftment were achieved using less than or equal to 106 primary cells. It was possible to evaluate engraftment of cells from patient 13 at diagnosis, first relapse, and second relapse. Engraftment levels were significantly higher in the relapse samples from this patient (P < .02), although there was no difference in the rate of engraftment.
Engraftment characteristics of unsorted ALL cells in NOD/SCID mice
| Patient no. . | % CD45+ in NOD/SCID*, by no. of unsorted cells injected . | % FISH+ . | |||
|---|---|---|---|---|---|
| 5 × 105 . | 1 × 106 . | 5 × 106 . | 1 × 107 . | ||
| 2 | — | 54.0 | 82.0 | 99.0 | NA |
| 4 | — | 1.0 | 2.0 | 6.0 | NA |
| 6 | 59 | 80.0 | 91.0 | 98.0 | 100 |
| 7 | — | — | — | 0.2 | NA |
| 10 | — | 0.4 | 2.6 | — | NA |
| 12 | 2 | 60.0 | 89.0 | — | — |
| 13 | — | — | 38.0 | — | 50 |
| 13† | — | — | 83.0 | — | 48 |
| 13‡ | — | — | 55.0 | 96.0 | 53 |
| 14 | — | 25.0 | 41.0 | 79.0 | 71 |
| 15† | — | 0.5 | 11.0 | — | 82 |
| 16 | — | 0.3 | 26.0 | — | 68 |
| Patient no. . | % CD45+ in NOD/SCID*, by no. of unsorted cells injected . | % FISH+ . | |||
|---|---|---|---|---|---|
| 5 × 105 . | 1 × 106 . | 5 × 106 . | 1 × 107 . | ||
| 2 | — | 54.0 | 82.0 | 99.0 | NA |
| 4 | — | 1.0 | 2.0 | 6.0 | NA |
| 6 | 59 | 80.0 | 91.0 | 98.0 | 100 |
| 7 | — | — | — | 0.2 | NA |
| 10 | — | 0.4 | 2.6 | — | NA |
| 12 | 2 | 60.0 | 89.0 | — | — |
| 13 | — | — | 38.0 | — | 50 |
| 13† | — | — | 83.0 | — | 48 |
| 13‡ | — | — | 55.0 | 96.0 | 53 |
| 14 | — | 25.0 | 41.0 | 79.0 | 71 |
| 15† | — | 0.5 | 11.0 | — | 82 |
| 16 | — | 0.3 | 26.0 | — | 68 |
— indicates not tested; NA, not applicable.
Each value represents the mean level of CD45+ cells in the femoral marrow of at least 2 NOD/SCID mice.
Sample at first relapse.
Sample at second relapse.
Serial transplantation studies were performed using BM cells harvested from NOD/SCID mice engrafted with cells from patients 2, 6, 12, 13 (at diagnosis), 14, and 16 to determine whether T-ALL could be transferred into secondary and tertiary recipients (Table 3). In each case, there were no significant differences in the levels of engraftment detected in the primary, secondary, and tertiary recipients. Passage through the NOD/SCID mice did not result in a change in phenotype (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article) or karyotype of the original leukemia.
Levels of engraftment in serial NOD/SCID transplants
| Patient no. . | No. of cells injected . | % CD45+ . | ||
|---|---|---|---|---|
| Primary . | Secondary . | Tertiary . | ||
| 2 | 1× 107 | 99.0 ± 0.3 | 95.0 ± 2.0 | 99.0 ± 0.4 |
| 6 | 1 × 107 | 98.0 ± 2.0 | 91.0 ± 8.0 | 83.0 ± 0.7 |
| 12 | 1 × 106 | 60.0 ± 8.0 | 47.0 ± 13.0 | 37.0 ± 5.0 |
| 13 | 5 × 106 | 38.0 ± 10.0 | 48.0 ± 4.0 | — |
| 14 | 1 × 107 | 79.0 ± 6.0 | 63.0 ± 11.0 | — |
| 16 | 5 × 106 | 26.0 ± 3.0 | 18.0 ± 7.0 | — |
| Patient no. . | No. of cells injected . | % CD45+ . | ||
|---|---|---|---|---|
| Primary . | Secondary . | Tertiary . | ||
| 2 | 1× 107 | 99.0 ± 0.3 | 95.0 ± 2.0 | 99.0 ± 0.4 |
| 6 | 1 × 107 | 98.0 ± 2.0 | 91.0 ± 8.0 | 83.0 ± 0.7 |
| 12 | 1 × 106 | 60.0 ± 8.0 | 47.0 ± 13.0 | 37.0 ± 5.0 |
| 13 | 5 × 106 | 38.0 ± 10.0 | 48.0 ± 4.0 | — |
| 14 | 1 × 107 | 79.0 ± 6.0 | 63.0 ± 11.0 | — |
| 16 | 5 × 106 | 26.0 ± 3.0 | 18.0 ± 7.0 | — |
— indicates not tested.
Engraftment of T-ALL subfractions in NOD/SCID mice
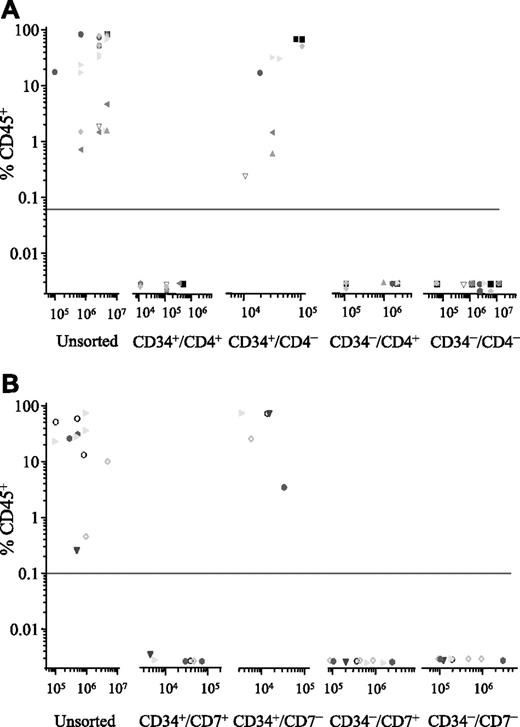
Cells from 7 patients (2, 4, 6, 7, 10, 12, and 14) were sorted for expression of CD34 and CD4, and the NOD/SCID engrafting capacity of each subfraction was investigated (Figure 2A). In each case, engraftment was detected in recipients of the CD34+/CD4− subfraction only (range, 0.5%-89% CD45+, using 104-105 cells). To achieve equivalent levels of engraftment using unsorted cells from these patients, 10- to more than 300-fold more cells were inoculated. There was no detectable engraftment in recipients of CD34+/CD4+ or the CD34− subfractions, despite injecting up to 100-fold more cells from some of these sorted subfractions.
Phenotype of NOD/SCID engrafting T-ALL cells. ALL cells were sorted for expression of CD34 and CD4 (n = 7; A) or CD34 and CD7 (n = 5; B). Both unsorted cells and the sorted subfractions were evaluated for their ability to engraft irradiated NOD/SCID recipients. Each patient is represented by a specific symbol, and each symbol depicts the engraftment obtained as measured by CD45+ cells present in the bone marrow of the NOD/SCID recipients.
Phenotype of NOD/SCID engrafting T-ALL cells. ALL cells were sorted for expression of CD34 and CD4 (n = 7; A) or CD34 and CD7 (n = 5; B). Both unsorted cells and the sorted subfractions were evaluated for their ability to engraft irradiated NOD/SCID recipients. Each patient is represented by a specific symbol, and each symbol depicts the engraftment obtained as measured by CD45+ cells present in the bone marrow of the NOD/SCID recipients.
Subsequently, cells from 2 of these patients (6 and 14) and 3 others (13, 15, and 16) were sorted for expression of CD34 and CD7, and the sorted subfractions were evaluated in the NOD/SCID assay (Figure 2B). Only the CD34+/CD7− subfraction had engrafting capacity (range, 4%-85% CD45+, using 4 × 103–8 × 104 cells). This sorting strategy enriched NOD/SCID engrafting cells 5- to more than 2000-fold compared with levels observed with unsorted subfractions. Once again there was no engraftment observed with the CD34+/CD7+, CD34−/CD7+, or the CD34−/CD7− subfractions, despite inoculating significantly higher numbers of cells.
To evaluate the self-renewal potential of these T-ALL subfractions, BM cells from NOD/SCID mice that had been engrafted with CD34+/CD4− cells from patients 2, 6, 12, and 14, and from mice engrafted with CD34+/CD7− cells from patients 6, 13, 14, and 16 were serially transplanted into secondary and, in 2 cases, tertiary recipients. The cells inoculated into serial recipients were not enriched for CD34+/CD4− or CD34+/CD7− cells, but animals were injected with an equivalent number of CD45+ cells as the primary recipients. In every case, similar levels of engraftment were observed in the secondary and tertiary transplants as had been observed in the primary transplants (Table 4)
Engraftment of sorted ALL cell populations in serial recipients
| ALL subfraction, NOD/SCID xenograft recipient . | No. of cells injected . | % CD45+ in BM . | % FISH+ . | Immunophenotype* . |
|---|---|---|---|---|
| CD34+/CD4− | ||||
| Patient 2 | ||||
| Primary recipient | 1 × 105 | 88 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Secondary recipient | 1 × 105 | 94 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Tertiary recipient | 1 × 105 | 97 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Patient 6 | ||||
| Primary recipient | 2 × 104 | 22 | 98 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Secondary recipient | 2 × 104 | 19 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Patient 12 | ||||
| Primary recipient | 1 × 105 | 59 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Secondary recipient | 1 × 105 | 47 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Tertiary recipient | 1 × 105 | 55 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Patient 14 | ||||
| Primary recipient | 3 × 104 | 47 | 72 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Secondary recipient | 3 × 104 | 43 | 70 | CD3−/CD4+/CD7+/CD19−/CD33− |
| CD34+/CD7− | ||||
| Patient 6 | ||||
| Primary recipient | 8 × 104 | 4 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Secondary recipient | 8 × 104 | 5 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Patient 13 | ||||
| Primary recipient | 1 × 104 | 82 | 64 | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Secondary recipient | 1 × 104 | 77 | 67 | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Patient 14 | ||||
| Primary recipient | 4 × 103 | 80 | 80 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Secondary recipient | 4 × 103 | 67 | 100 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Patient 16 | ||||
| Primary recipient | 2 × 104 | 85 | 74 | CD3−/CD4+/CD7+/CD19−/CD33−/CD34+ |
| Secondary recipient | 2 × 104 | 82 | 69 | CD3−/CD4+/CD7+/CD19−/CD33−/CD34+ |
| ALL subfraction, NOD/SCID xenograft recipient . | No. of cells injected . | % CD45+ in BM . | % FISH+ . | Immunophenotype* . |
|---|---|---|---|---|
| CD34+/CD4− | ||||
| Patient 2 | ||||
| Primary recipient | 1 × 105 | 88 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Secondary recipient | 1 × 105 | 94 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Tertiary recipient | 1 × 105 | 97 | — | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Patient 6 | ||||
| Primary recipient | 2 × 104 | 22 | 98 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Secondary recipient | 2 × 104 | 19 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Patient 12 | ||||
| Primary recipient | 1 × 105 | 59 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Secondary recipient | 1 × 105 | 47 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Tertiary recipient | 1 × 105 | 55 | — | CD2+/CD3−/CD4−/CD7+/CD19−/CD33−/CD34+ |
| Patient 14 | ||||
| Primary recipient | 3 × 104 | 47 | 72 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Secondary recipient | 3 × 104 | 43 | 70 | CD3−/CD4+/CD7+/CD19−/CD33− |
| CD34+/CD7− | ||||
| Patient 6 | ||||
| Primary recipient | 8 × 104 | 4 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Secondary recipient | 8 × 104 | 5 | 100 | CD2+/CD3−/CD4+/CD7+/CD8+/CD19−/CD33− |
| Patient 13 | ||||
| Primary recipient | 1 × 104 | 82 | 64 | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Secondary recipient | 1 × 104 | 77 | 67 | CD2+/CD3−/CD4+/CD7+/CD8−/CD19−/CD33− |
| Patient 14 | ||||
| Primary recipient | 4 × 103 | 80 | 80 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Secondary recipient | 4 × 103 | 67 | 100 | CD3−/CD4+/CD7+/CD19−/CD33− |
| Patient 16 | ||||
| Primary recipient | 2 × 104 | 85 | 74 | CD3−/CD4+/CD7+/CD19−/CD33−/CD34+ |
| Secondary recipient | 2 × 104 | 82 | 69 | CD3−/CD4+/CD7+/CD19−/CD33−/CD34+ |
— indicates not tested.
The cutoff for positivity was set at 20% of cells staining more intensely than the isotype control, in accordance with standard diagnostic immunophenotyping procedures defined by the British Committee for Standards in Haematology.24
Immunophenotypic analyses of the engrafted CD45+ cells removed from the NOD/SCID mice revealed that they had the same characteristic immunophenotype as the patient cells at diagnosis with high expression of CD2 (> 68%), low expression of CD3 (< 7%), and lacking expression of CD19 and CD33. Expression of CD4 was also high in the engrafted cells (> 47%), with the exception of cells from NOD/SCID mice that had been inoculated with cells from patient 12 (2%-4% CD4+). However, the low expression of CD4 was consistent with the immunophenotype of the diagnostic biopsy sample from this patient. The engrafted cells from all recipients were found to express CD7 (21%-56%), and CD8 expression was detected in recipients of samples with a CD8+ immunophenotype at diagnosis (Figure 3). FISH analyses, where possible, revealed that 64% to 100% of the engrafted cells had an abnormal karyotype, consistent with the number of abnormal cells detected in the diagnostic biopsy samples from these patients at diagnosis. These data confirm that cells with a leukemia karyotype were being transplanted in the serial xenografts. Taken together, these findings provide evidence for the self-renewal ability of the CD34+/CD4− and CD34+/CD7− T-ALL cells and suggest that they can differentiate to a certain extent in vivo, resulting in similar immunophenotypes to the bulk T-ALL cell population.
Phenotypic analysis of engrafted NOD/SCID bone marrow. Bone marrow cells removed from NOD/SCID mice that had been engrafted with sorted T-ALL cells were analyzed in more detail using additional lineage markers. Cells were initially gated on the basis of low forward and side scatter (A), then CD45+ cells were gated (B) and the expression of the lymphoid and myeloid antigens on these gated cells was investigated. The peaks representing specific lymphoid and myeloid antibodies are shown in black, and isotype controls are represented as translucent peaks on the overlay histograms. The figure shows the immunophenotype of cells removed from a NOD/SCID mouse inoculated with cells from patient 6. The engrafted ALL cells were found to express CD2, CD4, and CD7. CD8 cells were also detected in this sample, which is consistent with the high expression of CD8 in the diagnostic biopsy sample from this patient.
Phenotypic analysis of engrafted NOD/SCID bone marrow. Bone marrow cells removed from NOD/SCID mice that had been engrafted with sorted T-ALL cells were analyzed in more detail using additional lineage markers. Cells were initially gated on the basis of low forward and side scatter (A), then CD45+ cells were gated (B) and the expression of the lymphoid and myeloid antigens on these gated cells was investigated. The peaks representing specific lymphoid and myeloid antibodies are shown in black, and isotype controls are represented as translucent peaks on the overlay histograms. The figure shows the immunophenotype of cells removed from a NOD/SCID mouse inoculated with cells from patient 6. The engrafted ALL cells were found to express CD2, CD4, and CD7. CD8 cells were also detected in this sample, which is consistent with the high expression of CD8 in the diagnostic biopsy sample from this patient.
Analysis of clonal T-cell receptor gene rearrangements
To further characterize the engrafted CD45+ cells, it was possible to investigate the TCRG gene rearrangements in 4 cases, and the genotype was compared with matched patient cells at diagnosis (Table 5). In patients 6 and 14, the unique rearrangements detected at diagnosis were also observed following serial passage in the NOD/SCID model. However, in patients 2 and 13 changes in rearrangements were observed. Analysis of diagnostic material from patient 2 revealed 2 TCRG rearrangements and SIL-TAL. In cells from the corresponding xenograft, only 1 of these TCRG rearrangements was detected and a new TCRG rearrangement was identified. The SIL-TAL rearrangement was not present. In patient 13, a TCRG rearrangement present in the diagnostic material was not detected in the secondary transplant of CD34+/CD7− cells.
Sequence analysis of TCRG rearrangements at diagnosis and after engraftment
| Patient no. and sample source . | . | . | Clonal rearrangement sequence . | . |
|---|---|---|---|---|
| 6 | ||||
| Biopsy | (TRGV9*01) | tgtgccttgtgggaggt. | tttc | gaattattataagaaactcttt(TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV9*01) | tgtgccttgtgggaggt. | tttc | gaattattataagaaactcttt (TRGJ2*01) |
| 14 | ||||
| Biopsy | (TRGV3*01) | tgtgccacctgggacag. | cccca | gaattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 2° | (TRGV3*01) | tgtgccacctgggacag. | cccca | gaattattataagaaactcttt (TRGJ2*01) |
| Biopsy | (TRGV11*01) | tgtgcctgct........... | atgagg | ...........agaaactcttt (TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV11*01) | tgtgcctgct........... | atgagg | ...........agaaactcttt (TRGJ2*01) |
| 2 | ||||
| Biopsy | (TRGV3*01) | tgtgccacctgg...... | tgatagg | ...ttattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 1° | (TRGV3*01) | tgtgccacctgg...... | tgatagg | ...ttattataagaaactcttt (TRGJ2*01) |
| Biopsy | (TRGV9*01) | tgtgccttgtgggagg. | aga | .aattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 1° | (TRGV9*01) | tgtgccttgtgggagg. | aaaccttcctattattataagaaactctttggcagtgtaatattctcttgtggattgggg | ...............actcttt (TRGJ2*01) |
| Biopsy | (SIL Y07540) | caatttctggctcac | ccccaccc | gttggttttcatttcttc (TAL AL135960) |
| CD34+/CD4− NOD/SCID 1° | Clonal rearrangement not present | |||
| 13 | ||||
| Biopsy | (TRGV2*01) | tgtgccacctg....... | accacattataaac | gaattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV2*01) | Clonal rearrangement not present |
| Patient no. and sample source . | . | . | Clonal rearrangement sequence . | . |
|---|---|---|---|---|
| 6 | ||||
| Biopsy | (TRGV9*01) | tgtgccttgtgggaggt. | tttc | gaattattataagaaactcttt(TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV9*01) | tgtgccttgtgggaggt. | tttc | gaattattataagaaactcttt (TRGJ2*01) |
| 14 | ||||
| Biopsy | (TRGV3*01) | tgtgccacctgggacag. | cccca | gaattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 2° | (TRGV3*01) | tgtgccacctgggacag. | cccca | gaattattataagaaactcttt (TRGJ2*01) |
| Biopsy | (TRGV11*01) | tgtgcctgct........... | atgagg | ...........agaaactcttt (TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV11*01) | tgtgcctgct........... | atgagg | ...........agaaactcttt (TRGJ2*01) |
| 2 | ||||
| Biopsy | (TRGV3*01) | tgtgccacctgg...... | tgatagg | ...ttattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 1° | (TRGV3*01) | tgtgccacctgg...... | tgatagg | ...ttattataagaaactcttt (TRGJ2*01) |
| Biopsy | (TRGV9*01) | tgtgccttgtgggagg. | aga | .aattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD4− NOD/SCID 1° | (TRGV9*01) | tgtgccttgtgggagg. | aaaccttcctattattataagaaactctttggcagtgtaatattctcttgtggattgggg | ...............actcttt (TRGJ2*01) |
| Biopsy | (SIL Y07540) | caatttctggctcac | ccccaccc | gttggttttcatttcttc (TAL AL135960) |
| CD34+/CD4− NOD/SCID 1° | Clonal rearrangement not present | |||
| 13 | ||||
| Biopsy | (TRGV2*01) | tgtgccacctg....... | accacattataaac | gaattattataagaaactcttt (TRGJ2*01) |
| CD34+/CD7− NOD/SCID 2° | (TRGV2*01) | Clonal rearrangement not present |
Unique V and J segments or SIL-TAL genes identified are shown in parentheses. Randomly deleted bases are represented by full stops; randomly inserted bases (both N and P nucleotides) are shown in italics.
1° and 2° indicate primary and secondary NOD/SCID recipients, respectively.
Discussion
Whilst there have been significant improvements in outcome for children with T-ALL in recent years, a proportion of children with this malignancy continue to fail therapy. Intensification of current treatment regimens for childhood T-ALL may not be a feasible approach since the incidence of significant adverse risk effects would be likely to increase. A current challenge is to augment our knowledge of the biology and regulation of the cells that proliferate to maintain this malignancy and to evaluate experimental therapeutic strategies on these putative stem cells. In this study we investigated the characteristics of T-ALL cells that could proliferate in long-term assays both in vitro and in vivo.
The proliferative characteristics of childhood T-ALL cells have not been well defined, primarily due to lack of appropriate culture systems. Most studies have only investigated growth over short periods of time25-28 or for longer time periods, primarily to establish T-ALL cell lines.29 A NOD/SCID mouse fetal thymic organ culture (FTOC) system that supported growth of cells from 6 patients with T-ALL has been described, and proliferation was observed to peak at week 4.30 In the current study we analyzed the growth of childhood T-ALL cells from 15 patients using an SC assay. This assay is less complex than the NOD/SCID FTOC assay and we previously used it to characterize progenitor cells from both AML31-35 and B-cell precursor ALL.12 Cells from the majority of cases could be maintained and expanded during the culture period, and evaluation of sorted T-ALL populations revealed that cells capable of long-term proliferation were derived from the CD34+/CD4− and CD34+/CD7− subfractions. FISH and morphologic analyses confirmed that the cultured cells had the same karyotype as the patients at diagnosis and had blast morphology. These findings suggest that a hierarchy may exist in T-ALL with the CD34+/CD4− and CD34+/CD7− cells having greater proliferative potential than the other subpopulations investigated.
In order to provide more convincing evidence for the existence of a progenitor cell hierarchy in childhood T-ALL and to characterize these cells, it is essential to use in vivo models. There have been mixed reports as to the permissiveness of both SCID and NOD/SCID mice to support engraftment of T-cell leukemias, and most have indicated that only a proportion of T-ALL cases can engraft immune-deficient mice.14-20 However, in the current investigation engraftment was achieved with every leukemia sample evaluated, albeit to varying levels. When the sorted T-ALL subpopulations were evaluated in vivo, engraftment was observed only in recipients of the CD34+/CD4− and CD34+/CD7− subfractions. It was possible to enrich NOD/SCID engrafting cells by more than 3 logs, using CD34+/CD4− or CD7− cells, compared with using unsorted cells. It should be possible to further enrich these NOD/SCID leukemia-initiating cells by using a 3-color sorting strategy to purify the CD34+/CD4−/CD7− subfraction. However, this population represented less than 0.3% of the total T-ALL population in all but one case examined. Consequently, functional evaluation of the CD34+/CD4−/CD7− subfraction was beyond the scope of the current investigation, due to limited material available from these pediatric cases.
It is highly unlikely that the observed engraftment with the CD34+/CD4− and CD7− cells could be a result of contaminating lineage-positive cells in the inocula. Stringent sort gates were used to ensure high levels of purity in the sorted populations and there was no engraftment observed with the CD34+/CD4+ or CD34+/CD7+ subfractions in this model. Furthermore, serial transplantation studies demonstrated that CD34+/CD4− and CD34+/CD7− T-ALL cells were capable of self-renewal and expansion of the progenitor cell pool. Immunophenotypic analyses of the engrafted CD45+ cells suggested that the CD34+/CD4− and CD34+/CD7− subfractions underwent some degree of differentiation in vivo, generating a similar immunophenotype to the original diagnostic samples. Moreover, the cytogenetic analyses provided compelling evidence that the cells capable of serial NOD/SCID engraftment were of leukemia origin. To our knowledge, the data we present here is the largest cohort of serial engraftment of pediatric T-ALL cases in NOD/SCID mice reported to date and the first report of phenotypically defined leukemia-initiating cells for pediatric T-ALL.
To determine whether the genotype was preserved with passage through the NOD/SCID mice, the pattern of clonal TCRG rearrangements detected at diagnosis was compared to that of engrafted material from recipients of these putative leukemia-initiating cells. TCR rearrangements are unique and have previously been used to evaluate the fidelity of the NOD/SCID model as an accurate representation of the clinical disease.20,36 Dialynas et al20 found that TCRVB gene rearrangements were preserved in 2 cases evaluated. Similarly, Liem et al36 found that clonal rearrangements at diagnosis were largely preserved after 3 passages in NOD/SCID mice. In 1 of the 2 cases examined, an IGK-intron KDE rearrangement was lost and a new TCRG rearrangement was gained in the tertiary xenografts. Our findings are similar to those reported by Dialynas et al20 and Liem et al,36 in that 57% of rearrangements were identical between primary diagnostic material and samples from xenografts established with CD34+/CD4− and CD34+/CD7− cells. Some rearrangements were not present, or new rearrangements were identified, in the serial transplant samples. This is the first such analysis of phenotypically primitive T-ALL cells, serially passaged through the NOD/SCID model. It is possible that emergence of a new TCRG marker identifies a subpopulation present only at very low levels at diagnosis. Our future studies will attempt to determine if these leukemia-initiating cells contain rearrangements identical to the primary material or harbor unique rearrangements that can be identified as a predominant clone at relapse.
Although there has been some debate about whether different subtypes of T-ALL represent proliferation arrest at variable stages of T-cell development, most reports agree that the leukemic transformation event occurred in a cell that was committed to the T lineage.1,6,37 However, a significant proportion of T-ALL cases have been identified as being immature, with a similar phenotype to normal thymic precursors in that they were negative for most markers associated with the T lineage and expressed CD34, CD13, and CD33.38-40 Most of these immature T-ALL cases had clear evidence of both T- and myeloid cell potential rather than T- and B-lymphoid potential, suggesting they had arisen in an early hemopoietic precursor.38 There is evidence in normal hemopoietic development that the T and B lineages separate prior to loss of myeloid potential, rather than separation of a common lymphoid precursor41 from a common myeloid precursor.42,43 Therefore, a hierarchy of progenitor cells may exist in T-ALL, with the malignant transformation of a hemopoietic progenitor cell giving rise to a leukemia stem cell that retains the key properties of self-renewal and proliferation but fails to differentiate completely.5,44
In support of this theory, it has been reported that IL-7 receptor–negative (IL-7R−) T-ALL cells had a higher proliferative capacity than IL-7R+ cells.30 IL-7R is thought to be undetectable on early thymic progenitors,43 and expression increases once cells are committed to the T lineage.45-48 Ma et al30 found that IL-7R− T-ALL cells had a higher proliferative capacity than IL-7R+ cells in both primary and secondary FTOC cultures, and IL-7R+ cells were generated from the IL-7R− fraction in vitro. The authors concluded that a hierarchy of progenitors may well exist in T-ALL lymphoblasts, with IL-7R− cells representing the more primitive subfraction.30 Our findings that T-ALL cells with long-term proliferative capacity in vitro and NOD/SCID engrafting capacity had an early CD34+/CD4− and CD7− phenotype add further support to the existence of a hierarchy within the leukemic blast population.
Collectively, the data we present here suggest that in the T-ALL cases examined, the malignancy appears to have arisen in a cell that is more phenotypically primitive than was previously thought, with some differentiation occurring subsequent to the transformation event. Further studies will be necessary to determine whether pediatric T-ALL originates in a cell that is not restricted to the T-cell lineage or in a more primitive cell with both myeloid and lymphoid potential. Expanding our knowledge of the biologic characteristics of childhood T-ALL in these model systems should permit the development and evaluation of novel therapies that can specifically target the leukemic stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: C.V.C. processed samples, performed the in vitro experiments, analyzed the data, and helped write the report; H.M.M. performed the gene rearrangement analyses and contributed to the writing of the report; P.R.K. collated patient data and contributed to writing the report; P.V. performed the diagnostic immunophenotyping and helped analyze the data; R.S.E. helped perform the in vivo experiments and commented on the manuscript; and A.B. conceived and designed the study, supervised its execution, performed the in vivo experiments, and wrote the report. A.B. had full access to all the data in the study and had final responsibility for the decision to submit for publication.
The authors wish to thank the staff of the Regional Cytogenetics Unit, Southmead Hospital; the staff of the cell sorting facility, University of Wales College of Medicine; and Mr Paul Archer for excellent technical assistance. We wish to acknowledge the Sequencing Service (School of Life Sciences, University of Dundee, United Kingdom; http://www.dnaseq.co.uk/) for DNA sequencing. We also wish to thank the patients and their families, who gave permission for their cells to be used for research. This work was supported by grants from the Leukaemia Research Fund, United Kingdom, and the National Blood Authority.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal