Abstract
The broad and often contrasting effects of type I interferons (IFNs) in innate and adaptive immunity are belied by the signaling via a single receptor, IFN-α receptor (IFNAR). Here, we show that IFN-α/β induces opposing effects on the immunologic outcome of antigen cross-presentation depending on dendritic cell (DC) maturation status. Despite equivalent IFNAR expression, immature conventional DCs (cDCs) activate STAT1 in response to IFN-α/β, whereas exposure of mature DCs to IFN-α/β results in signaling via STAT4. Microarray analysis revealed numerous transcriptional changes resulting from the altered signaling. Importantly, STAT1 signaling resulted in significant inhibition of CD40L-induced IL-12 production, accounting for the inhibition of CD8+ T-cell activation. These data provide evidence for a molecular switch in signaling pathways concomitant with DC maturation that offers a novel mechanism by which DCs modulate the integration of signals from the surrounding environment.
Introduction
Type I interferons (IFNs) play an important role in direct antiviral defense as well as linking the innate and adaptive immune responses. The mechanism by which IFN-α/β acts in coordinating these responses cannot be clearly categorized into proinflammatory versus anti-inflammatory.1 In fact, the complexity of its biology may be revealed by the suggested clinical effects of IFN-α/β–suppressing autoimmune responses in patients with multiple sclerosis, yet potentiating T-cell priming in patients with tumors and chronic hepatitis C.2-4 These opposing clinical effects are consistent with the pleiotropic effects of IFN-α/β at the cellular level. For example, exposure of natural killer (NK) cells and naive T cells to IFN-α/β during viral infection inhibits their production of IFN-γ5 ; however, IFN-α/β has also been shown to trigger NK-cell cytotoxicity and stimulate proliferation of certain T-cell subsets.6,7
This complexity of IFN biology extends to conventional dendritic cells (cDCs). Considered the “sentinels” of the immune system, DCs are responsible for integrating immune responses. Tissue DCs, which exist in an immature state of differentiation, are responsible for capturing and processing antigen, as well as transmitting proinflammatory and immune regulatory signals to cells within draining lymph nodes.8 DCs also possess a unique ability to cross-present exogenous antigen, offering a mechanism for priming CD8+ T cells specific for viruses that do not directly infect DCs. Recent data suggest that type I IFNs may serve as a “danger” signal during this process, facilitating DC activation9 and licensing DCs to cross-prime10 ; however IFN-α/β has also been shown to inhibit the production of IL-12 by DCs,11 and may also inhibit DCs from stimulating TH1-cell differentiation.12
Using an in vitro system, we evaluated the effects of IFN-α/β on the cross-presentation of antigen. Importantly, our consideration of the distinct maturation stages and unique biologic activities of cDCs led to a surprising result. Immature DCs (iDCs) exposed to IFN-α/β (referred to herein as IFN-early), reflecting the microenvironment of a tissue DC in the presence of IFN-α/β, were impaired in their ability to activate CD8+ T cells via the cross-presentation pathway. In contrast, matured DCs that were then exposed to IFN-α/β (referred to herein as IFN-late), mimicking the microenvironment of DC/T engagement within the lymph node in the presence of IFN-α/β, demonstrated enhanced T-cell activation. Investigation into the regulation of these opposing effects revealed a molecular switch in IFN-α/β–signaling networks that is a consequence of DC maturation. Our findings offer insight into how IFN-α/β shapes the adaptive immune responses to cross-presented antigen and uncover a novel mechanism by which DCs modulate their response to cytokine signals from the surrounding environment.
Materials and methods
Isolation and preparation of cells
Peripheral-blood mononuclear cells (PBMCs), DCs, and T cells were prepared as previously described.13 PBMCs were isolated from whole blood by sedimentation over Ficoll-Hypaque (Amersham Pharmacia Biotech, Piscataway, NJ). iDCs were prepared from the T-cell–depleted fraction by culturing cells in the presence of 1000 U/mL GM-CSF (Berlex, Seattle, WA) and 500 to 1000 U/mL IL-4 (R&D Systems, Minneapolis, MN) for 6 days.14,15 To generate mature DCs, cultures were stimulated on day 6 with 50 ng/mL TNF-α (Alexis Biochemicals, Lausen, Switzerland) and 0.1 μM PGE2 (Sigma, St Louis, MO) or 10 ng/mL LPS (Sigma) for 36 to 48 hours.16 At days 6 to 7, more than 95% of the cells were CD14−CD83− LA-DRlo DCs. After maturation, on days 8 to 9, 70% to 95% of the cells were of the mature CD14−CD83+HLA-DRhi phenotype. CD4+ and CD8+ T cells were purified to more than 99% purity by positive selection with the magnetic-activated cell sorting (MACS) column purification system (Miltenyi Biotech, Auburn, CA). pDCs were also purified by MACS with BDCA-4 microbeads. pDCs were stimulate with CpG (2216: 5′-GGGGGACGATCGTCGGGGGG-3′) in 1% single-donor plasma. After 24 hours, cultures were harvested and supernatants purified for centrifugation.
Induction of apoptotic death
The mouse lymphoma cell line EL4 (number TIB-39, American Type Culture Collection, Rockville, MD) was used as a source of apoptotic cells because they can be efficiently infected with influenza virus and do not induce marked background T-cell activation to mouse antigens. The EL4 cells were infected with influenza and apoptosis was triggered by exposure to a 60UVB lamp for 120 seconds (calibrated to provide 2 mJ/cm2/s).
Endogenous and exogenous loading of DCs with antigen
For endogenous loading, mDCs were infected with 0.002 HAU influenza A/PR8/1976 (Charles River, Spafas, CT) per cell. Virus was quenched and removed with 2 washes of media with 5% pooled AB human serum (Labquip, Niagara Falls, NY). For loading DCs with exogenous antigen, iDCs were cocultured with influenza-infected EL4s that had been exposed to UVB.17 Cocultures were incubated in the presence of TNF-α/PGE2 to allow for phagocytosis of the apoptotic EL4 cells, antigen processing, and DC maturation to occur. mDCs were then used for functional assays.
Detection of antigen-specific T cells: ELISPOT for IFN-γ release
Antigen-loaded DCs were collected, counted, and added to purified T-cell populations in plates that had been coated with 10 μg/mL anti-IFNγ monoclonal antibody (mAb; clone Mab-1-D1K, Mabtech, Cincinnati, OH). In all experiments, 2 × 105 T cells were added to 6.6 × 103 DCs to give a 30:1 T-cell/DC ratio. The cultures were incubated for 40 to 44 hours at 37°C. Cells were washed out with mild detergent and the enzyme-linked immunospot assay (ELISPOT) plate was incubated with 1 μg/mL of a biotin-conjugated IFN-γ mAb (clone Mab 7BG-1, Mabtech) and developed using the Vectastain Elite kit (Vector Laboratories, Burlingame, CA). Colored spots indicated the cells that had released IFN-γ and data are reported as spot-forming cells/106 CD8+ T cells. Data from triplicate wells were averaged and mean data are reported.
Quantitative RT-PCR for analysis of STAT mRNA expression
RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and cDNA was synthesized from 1 to 2 μg RNA using oligo-dT (Roche, Indianapolis, IN) and Superscript reverse transcriptase (Invitrogen) according to manufacturer's instruction. STAT-specific mRNA is quantified relative to TATA box-binding protein (TBP) using FAM/MGB coupled Taq Man probes from Applied Biosystems (Weiterstadt, Germany; STAT1: Hs00234829_m1; STAT2: Hs00237139_m1; STAT4: Hs00231372_m1). Quantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed using the Taq Man Universal PCR Master mix (Applied Biosystems) according to the manufacturer's instructions. The probe 4326322E for TBP was used as an internal control in each reaction. The reactions were run on a PTC200 equipped with a Chromo4 detector (MJ Research, Watertown, MA). The analyses were performed with Opticon Monitor software version 2.03. All the measures were performed in duplicate and validated when the difference in Ct between the 2 measures was less than 0.3. The ratio gene of interest to housekeeping genes was calculated according to the formula: ratio = 2−ΔCt (ΔCT = mean Ct gene − mean Ct housekeeping).
Analysis of STAT and IKB-α phosphorylation
DCs were harvested, washed with PBS, resuspended in 10% FBS containing media and stimulated with indicated concentration of rIFN-α, purified IFN-α/β, IFN-γ (Sigma), or CD40L for 30 minutes at 37°C. For Western blot analysis, lysates were prepared in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8, 1% NP40, 0.5% DOC, 0.03% SDS, 2 mM EDTA). Total protein was determined and 20 μg was loaded on to 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to PVDF membrane and blotted with indicated antibodies (Cell Signaling Technologies, Beverly, MA). Blots were probed first with phospho-specific STAT antibodies, stripped with low pH, and reprobed with total STAT antibodies to ensure p-STAT is monitored in the context of total STAT. For intracellular fluorescence-activated cell sorting (FACS) analysis, samples were fixed, permeabilized, and labeled as described.18 Cells were stained with STAT1-PE, STAT3-PE, and STAT4-APC (PharMingen, San Diego, CA) as indicated.
Affymetrix microarray U133A
Biotinylated cRNA fragments were prepared from monocyte-derived DCs as per Affymetrix protocol (http://www.affymetrix.com). Cells (107) were washed in PBS and lysed using Qiashredder (Qiagen, Valencia, CA). Total RNA was isolated using RNeasy Mini Kit (Qiagen). cDNA and fragmented biotinylated labeled cRNA were subsequently prepared as per manufacturer's instructions (Affymetrix, Santa Clara, CA and Enzo Life Sciences, Farmingdale, NY) and 20 μg fragmented cRNA was loaded onto Affymetrix U133A. Data were analyzed using Microarray Suite 5.0 (Affymetrix) and compiled using a custom database (adapted from a database that was generously provided by Dr Nir Hacohen, Harvard University, Cambridge, MA).
Detection of cytokines
IL-12p70 levels were confirmed in triplicate using IL-12 enzyme-linked immunosorbent assay (ELISA; R&D Systems). IFN-α ELISA (PBL Biomedical, Piscataway, NJ) was performed in triplicate and data from 2 donors were averaged.
Analysis of IFN-α/β and IL-12 production in MCMV-challenged mice
C57BL/6 were obtained from Charles River (France); STAT1−/−(129SVE) from Taconic Farms (Germantown, NY); and STAT4−/−(Balb/c) from Jackson Labs, Bar Harbor, ME). IFNAR−/− mice were backcrossed 12 times to C57BL/6 mice. RB6-8C5 (anti-GR-1) bioreactor supernatant was obtained from monoclonal antibody facility (Memorial Sloan-Kettering Cancer Center [MSKCC], New York, NY) and mice were depleted with 500 μg at 36 hours and 12 hours before infection. Mice were infected with 4 × 105 PFU/mL salivary gland-prepared MCMV. At 36 hours after infection, serum samples were taken and spleens were harvested, incubated at a 1:50 dilution of anti-FcγR (BD PharMingen, San Diego, CA) to block nonspecific antibody interaction in mouse FACS wash buffer. Cells were surface stained with CD8 FITC (1:150 dilution, BD PharMingen) and CD11c PE (1:150 dilution, BD PharMingen), fixed, and permeabilized with Cytoperm/Cytofix (BD PharMingen), and stained with IL-12 APC (1:150, BD PharMingen).
Results
DC maturation determines the effect of IFN-α/β on antigen cross-presentation
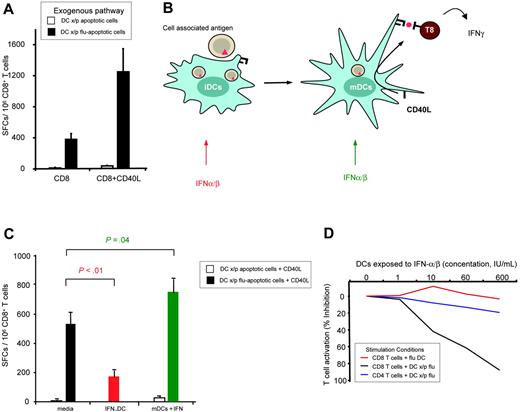
To explore the effects of IFN-α/β on antigen cross-presentation, we used an experimental system based on the activation of primary human memory CD8+ T cells. DCs cross-presenting antigen were generated by coculturing iDCs with apoptotic, influenza (flu)–infected MHC-mismatched cells in the presence of TNF-α/PGE2, used as a potent maturation stimulus.17 Antigen-loaded mDCs were then cultured with purified CD8+ T cells and the activation of influenza-specific IFN-γ–producing cells was monitored using ELISPOT. Maximal CD8+ T-cell activation by cross-presentation requires CD40L engagement, mimicking the effect of CD4+ T-cell/DC interactions (Figure 1A and Albert et al17 ). Of note, to carefully evaluate the effect of adding IFN to cDCs, it was important to use TNF-α because it triggers DC maturation without inducing endogenous IFN-α/β; this was demonstrated both directly by measuring IFNs present in the culture wells and indirectly by the absence of IFN-induced genes in the transcriptional profile of TNF-α–matured DCs (data not shown).19 Using this experimental system, we were able to selectively evaluate effects of exogenous IFN-α/β on immature and mature DCs. Specifically, leukocyte-derived IFN-α/β or recombinant IFN-α was added to iDCs during antigen capture and maturation (Figure 1B). Alternatively, to model the production of IFN-α/β in the lymph node, IFN-α/β was added to mDCs before T-cell engagement (Figure 1B). In both cases, IFN-α/β was washed out before T-cell coculture to avoid direct effects on T cells.20 Surprisingly, the effect of IFN-α/β on the regulation of T-cell activation depended on the maturation state of the DCs. IFN-α/β inhibited CD8+ T-cell activation when exposed to iDCs (Figure 1C, IFN-DC); in contrast, no inhibition was observed when adding the IFN-α/β directly to the mDCs (Figure 1C, mDC + IFN). In fact, this latter condition resulted in statistically significant enhancement of T-cell activation (P = .04).
Type I IFNs have a dual effect on cross-presenting conventional DCs. (A) Cross-presenting DCs require CD40L for maximal T-cell activation. DCs loaded with exogenous antigen were prepared by cross-presentation of influenza infected apoptotic cells (DCs x/p flu-apoptotic cells) and compared to uninfected apoptotic cells (DCs x/p apoptotic cells). DC groups were cultured with syngeneic purified CD8+ T cells. T-cell activation was monitored by IFN-γ ELISPOT and displayed as spot-forming cells (SFCs)/106 CD8+ T cells. Recombinant CD40L was added to cultures as indicated. (B) The effects of IFN-α/β were tested on immature and mature DCs, respectively. As illustrated, IFN-α/β was added to iDC during antigen capture and maturation or to mDCs during CD40 cross-linking and CD8+ T-cell engagement. (C) DCs cross-presenting uninfected (unfilled bars) or influenza loaded (filled bars) apoptotic cells were treated with media alone (black bars), exposed to 60 IU/mL IFN-α/β during antigen capture (red bars), or 60 IU/mL IFNα/β following maturation and antigen presentation (green bars). Data are representative of 10 experiments with statistical P indicated. Similar results were obtained when using recombinant IFN-α (data not shown). (D) iDCs were exposed to a dose range of IFN-α/β (concentration 0-600 IU/mL). In the case of direct presentation of antigen, mDCs were directly infected with influenza virus (flu DC). For cross-presentation, the iDCs with cocultured with apoptotic influenza-expressing apoptotic cells (DCs x/p flu-AC) during the maturation process and interferon stimulation. In each condition, the CD8+ T-cell and CD4+ T-cell activation were monitored by IFN-γ ELISPOT. Percent inhibition of T-cell activation is displayed. Maximal stimulation (no IFN-α/β) in the experiment shown is equivalent to 400 spot forming cells/million T cells.
Type I IFNs have a dual effect on cross-presenting conventional DCs. (A) Cross-presenting DCs require CD40L for maximal T-cell activation. DCs loaded with exogenous antigen were prepared by cross-presentation of influenza infected apoptotic cells (DCs x/p flu-apoptotic cells) and compared to uninfected apoptotic cells (DCs x/p apoptotic cells). DC groups were cultured with syngeneic purified CD8+ T cells. T-cell activation was monitored by IFN-γ ELISPOT and displayed as spot-forming cells (SFCs)/106 CD8+ T cells. Recombinant CD40L was added to cultures as indicated. (B) The effects of IFN-α/β were tested on immature and mature DCs, respectively. As illustrated, IFN-α/β was added to iDC during antigen capture and maturation or to mDCs during CD40 cross-linking and CD8+ T-cell engagement. (C) DCs cross-presenting uninfected (unfilled bars) or influenza loaded (filled bars) apoptotic cells were treated with media alone (black bars), exposed to 60 IU/mL IFN-α/β during antigen capture (red bars), or 60 IU/mL IFNα/β following maturation and antigen presentation (green bars). Data are representative of 10 experiments with statistical P indicated. Similar results were obtained when using recombinant IFN-α (data not shown). (D) iDCs were exposed to a dose range of IFN-α/β (concentration 0-600 IU/mL). In the case of direct presentation of antigen, mDCs were directly infected with influenza virus (flu DC). For cross-presentation, the iDCs with cocultured with apoptotic influenza-expressing apoptotic cells (DCs x/p flu-AC) during the maturation process and interferon stimulation. In each condition, the CD8+ T-cell and CD4+ T-cell activation were monitored by IFN-γ ELISPOT. Percent inhibition of T-cell activation is displayed. Maximal stimulation (no IFN-α/β) in the experiment shown is equivalent to 400 spot forming cells/million T cells.
The inhibitory effect of IFN-α/β is selective for CD40-dependent cross-presentation
To evaluate if the inhibitory effect of IFN-α/β on iDCs is specific for cross-presentation, we compared DCs cross-presenting antigen to those directly infected with influenza virus. iDCs were treated with increasing doses of IFN-α/β (10-60 IU/mL) during their maturation. These IFN-DCs were washed extensively and exposed to live influenza virus. Again, IFN-γ production by influenza-specific CD8+ T cells was used as a measure of the capacity of DCs to stimulate T cells. Whereas the cross-presentation pathway was sensitive to IFN (black line in Figure 1D black line), no inhibition of CD8+ T-cell activation was observed when IFN-DCs were directly infected (red line in Figure 1D). Further, the IFN-mDCs loaded with exogenous antigen showed no defect in stimulating influenza-reactive CD4+ T cells (blue line in Figure 1D), thus providing an internal control that antigen had been captured and that the DCs were capable of engaging T cells. From these experiments, we conclude that IFN-α/β has a selective effect on cross-presenting DCs, in turn altering their ability to activate T cells.
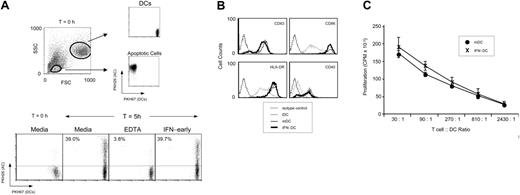
This selective effect may be due to decreased antigen capture, inefficient DC maturation, or the skewing of activation signals specific for T-cell activation by cross-presentation (eg, CD40 engagement). To directly evaluate antigen capture, apoptotic cells and iDCs were labeled with fluorescent lipophilic dyes as described.21 Capture of the red-labeled apoptotic cells by the green-labeled DCs was monitored by flow cytometry (Figure 2A). After 5 hours, about 40% of the iDCs had captured an apoptotic-cell body/bleb. Addition of IFN-α/β to the cocultures did not inhibit antigen capture nor did it alter the kinetics of uptake. We next evaluated the effect of IFN-α/β on phenotypic and functional maturation of the DCs. Importantly, the addition of IFN-α/β did not affect TNF-α–mediated maturation as monitored by expression of CD40, CD83, HLA-DR, and CD86 (Figure 2B). Moreover, the allostimulatory capacity of IFN-DCs was equivalent to mDCs, as measured using a standard allogeneic mixed lymphocyte reaction (Figure 2C). These findings suggested that exposure of iDCs to IFN-α/β during the maturation process may be affecting the ability of mDCs to be “licensed” by CD40, a critical downstream requirement for the activation of antigen-specific CD8+ T cells when antigen is captured from an exogenous source.17
IFN-α/β does not alter phagocytosis or phenotypic or functional maturation of DCs. (A) Phagocytosis of PKH26 (red dye)–labeled apoptotic cells by PKH67 (green dye)–labeled iDCs was monitored by flow cytometry. iDCs and apoptotic T cells were distinguished based on forward scatter (FSC) and side scatter (SSC) properties, as shown. Gating on iDCs using scatter and FL-1 criteria permitted monitoring of cells that engulfed a red-labeled dying cell, as based on their becoming double positive (lower FACS plots). Cocultures were incubated in the presence of media, EDTA, or IFN-α as indicated. Percent phagocytosis is indicated. (B) FACS analysis of CD83, CD86, HLA-DR, and CD40 expression levels on iDCs, mDCs, or DCs matured in the presence of IFN-α/β (IFN-DC). (C) Allostimulatory potential of mDCs or DCs matured in the presence of IFN-α/β (IFN-DC) was monitored by stimulation of T-cell proliferation after 5 days of culture. Proliferation was monitored by incorporation of 3H-thymidine. Triplicate wells are averaged and SEM is represented by error bars.
IFN-α/β does not alter phagocytosis or phenotypic or functional maturation of DCs. (A) Phagocytosis of PKH26 (red dye)–labeled apoptotic cells by PKH67 (green dye)–labeled iDCs was monitored by flow cytometry. iDCs and apoptotic T cells were distinguished based on forward scatter (FSC) and side scatter (SSC) properties, as shown. Gating on iDCs using scatter and FL-1 criteria permitted monitoring of cells that engulfed a red-labeled dying cell, as based on their becoming double positive (lower FACS plots). Cocultures were incubated in the presence of media, EDTA, or IFN-α as indicated. Percent phagocytosis is indicated. (B) FACS analysis of CD83, CD86, HLA-DR, and CD40 expression levels on iDCs, mDCs, or DCs matured in the presence of IFN-α/β (IFN-DC). (C) Allostimulatory potential of mDCs or DCs matured in the presence of IFN-α/β (IFN-DC) was monitored by stimulation of T-cell proliferation after 5 days of culture. Proliferation was monitored by incorporation of 3H-thymidine. Triplicate wells are averaged and SEM is represented by error bars.
DC maturation alters IFN-α/β signaling by switching STAT use
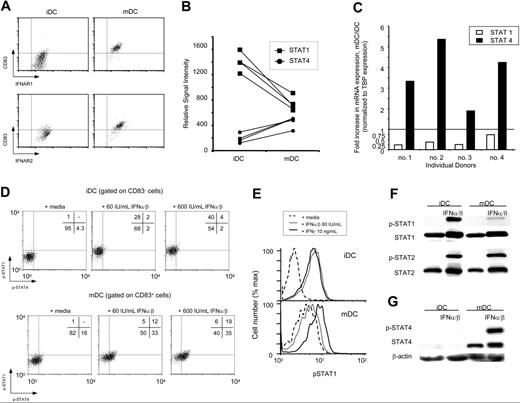
In light of the correlation of distinct DC maturation states with the paradoxical effects of IFN, we considered the possibility that intrinsic differences between iDC and mDC IFN-α/β responsiveness could account for the opposing effect on T-cell activation. Three possibilities were evaluated to directly test this hypothesis: (1) differences at the level of IFN-α/β receptor (IFNAR) expression; (2) use of distinct signaling pathways; and (3) altered CD40-mediated transcriptional events, resulting from differences in IFN-α/β signaling. First, expression of IFNAR on iDCs and mDCs was evaluated by FACS. Surface expression of both subunits, IFNAR1 and IFNAR2, were equivalent in iDCs and mDCs (Figure 3A). Immunoblot analysis of IFNAR1 and IFNAR2 confirmed that there are equivalent levels of both subunits in iDCs and mDCs (data not shown). Based on these data, we conclude that differential receptor expression does not account for the distinct effects of IFN-α/β on iDCs and mDCs.
DC maturation alters IFN-α/β signaling by modulating STAT expression. (A) iDCs and mDCs were stained with anti-CD83-Cy5 and mAbs specific for IFNAR1 (AA3, Biogen, Cambridge, MA) or IFNAR2 (MMHAR-2, Calbiochem, San Diego, CA). Anti–mouse IgG PE (Jackson Immunochemicals, Raritan, NJ) was used to visualize anti-IFNAR binding. Dot plots representative of a single donor are shown. Three individuals were evaluated and revealed similar results. (B) Transcriptional analysis of iDCs and mDCs was performed using Affymetrix microarray U133A. Data were analyzed and normalized using MAS5.0. The relative signal intensity of 4 biologic replicates is displayed for STAT1 and STAT4 expression in iDCs and mDCs. (C) Quantitative PCR was performed as described in “Materials and methods.” Fold increase in mRNA expression of mDCs/iDCs is displayed graphically for STAT1 and STAT4 as indicated. (D) iDCs and mDCs were stimulated with media alone or indicated concentrations of IFN-α/β for 30 minutes. Cells were stained with anti-CD83–FITC to identify CD83− iDCs and CD83+ mDCs. Cells were fixed and permeabilized as described in “Materials and methods” and stained for p-STAT1-PE and p-STAT4-APC. Percentage of events in each quadrant is indicated on the respective plot. Data are representative of 3 independent experiments. (E) iDCs and mDCs were stimulated with media alone, 60 IU/mL IFN-α/β, or 10 ng/mL IFN-γ for 30 minutes as indicated. Cells were fixed, permeabilized, and stained for p-STAT1, as described. (F-G) iDCs and mDCs were exposed to 60 IU/mL IFN-α/β for 30 minutes. Protein (20 μg) from the total-cell lysates were run on an 8% SDS-PAGE, transferred to PVDF membranes, and blotted with indicated antibodies. Blots were stripped and probed for total STAT1/2 or β-actin as a control for loading the gel.
DC maturation alters IFN-α/β signaling by modulating STAT expression. (A) iDCs and mDCs were stained with anti-CD83-Cy5 and mAbs specific for IFNAR1 (AA3, Biogen, Cambridge, MA) or IFNAR2 (MMHAR-2, Calbiochem, San Diego, CA). Anti–mouse IgG PE (Jackson Immunochemicals, Raritan, NJ) was used to visualize anti-IFNAR binding. Dot plots representative of a single donor are shown. Three individuals were evaluated and revealed similar results. (B) Transcriptional analysis of iDCs and mDCs was performed using Affymetrix microarray U133A. Data were analyzed and normalized using MAS5.0. The relative signal intensity of 4 biologic replicates is displayed for STAT1 and STAT4 expression in iDCs and mDCs. (C) Quantitative PCR was performed as described in “Materials and methods.” Fold increase in mRNA expression of mDCs/iDCs is displayed graphically for STAT1 and STAT4 as indicated. (D) iDCs and mDCs were stimulated with media alone or indicated concentrations of IFN-α/β for 30 minutes. Cells were stained with anti-CD83–FITC to identify CD83− iDCs and CD83+ mDCs. Cells were fixed and permeabilized as described in “Materials and methods” and stained for p-STAT1-PE and p-STAT4-APC. Percentage of events in each quadrant is indicated on the respective plot. Data are representative of 3 independent experiments. (E) iDCs and mDCs were stimulated with media alone, 60 IU/mL IFN-α/β, or 10 ng/mL IFN-γ for 30 minutes as indicated. Cells were fixed, permeabilized, and stained for p-STAT1, as described. (F-G) iDCs and mDCs were exposed to 60 IU/mL IFN-α/β for 30 minutes. Protein (20 μg) from the total-cell lysates were run on an 8% SDS-PAGE, transferred to PVDF membranes, and blotted with indicated antibodies. Blots were stripped and probed for total STAT1/2 or β-actin as a control for loading the gel.
We next considered the possibility of differential signaling downstream of IFNAR. To screen for potential points of divergent regulation, we analyzed transcriptional profiles of iDCs and mDCs from 4 individuals using Affymetrix U133A microarrays. Analysis of genes known to be involved in IFN-α/β signaling revealed possible differences in IFN response (Table 1). Specifically, microarray analysis identified changes in the relative signal intensity of STAT1 and STAT4 between iDCs and mDCs (Table 1; Figure 3B). Classically, STAT1 and STAT2 are phosphorylated in response IFN-α/β, forming a heterotrimer with IRF-9, which binds to IFN-stimulated response element (ISRE)–containing genes.22 In certain cell types, STAT4 may be phosphorylated in response to IFN-α/β, leading to transcription of distinct sets of genes.23 These changes in absolute levels of STAT1 and STAT4 mRNA were further evaluated by quantitative PCR in 4 donors, demonstrating that STAT1 is down-regulated by 1.5- to 4-fold and STAT4 is up-regulated by 2- to 5-fold on maturation (Figure 3C). Although mRNA expression correlated with protein expression, it should be noted that donor variability exists (data not shown).
Analysis of genes involved in IFN-α/β signaling and possible differences in IFN response
| Gene name . | Affymetrix ID . | iDC avg* . | A/P† . | P . | mDC avg . | A/P . | P . | Fold change (mDC/iDC)‡ . |
|---|---|---|---|---|---|---|---|---|
| IFNAR1 | 204191_at | 124 (55–188) | A/P | .165 | 139 (73–150) | A/P | .133 | 1.1 |
| IFNAR2 | 204786_s_at | 126 (51–279) | P | .004 | 122 (57–208) | P | .003 | 1.0 |
| JAK1 | 201648_at | 1377 (1227–1821) | P | <.001 | 3782 (2799–4632) | P | <.001 | 2.7 |
| TYK2 | 205546_s_at | 491 (315–880) | P | .011 | 963 (605–1229) | P | .005 | 2.0 |
| STAT1 | 200887_s_at | 1306 (892–1597) | P | <.001 | 730 (641–908) | P | <.001 | 0.6 |
| STAT1 | 97935_3_at | 720 (370–924) | P | <.001 | 474 (310–616) | P | <.001 | 0.7 |
| STAT2 | 205170_at | 164 (132–185) | P | .003 | 156 (109–189) | P | .009 | 0.9 |
| STAT3 | 208991_at | 928 (670–1179) | P | <.001 | 2084 (1458–2467) | P | <.001 | 2.2 |
| STAT4 | 206118_at | 206 (126–297) | A/P | .092 | 430 (319–507) | P | .024 | 2.1 |
| STAT5A | 203010_at | 492 (343–675) | P | .037 | 994 (772–1143) | P | .006 | 2.0 |
| STAT5B | 205026_at | 110 (87–139) | P | .008 | 239 (205–285) | P | .002 | 2.2 |
| STAT6 | 201331_s_at | 1975 (1773–2188) | P | <.001 | 1424 (1252–1681) | P | <.001 | 0.7 |
| STAT6 | 201332_s_at | 368 (306–428) | P | .007 | 269 (183–302) | A/P | .044 | 0.7 |
| Gene name . | Affymetrix ID . | iDC avg* . | A/P† . | P . | mDC avg . | A/P . | P . | Fold change (mDC/iDC)‡ . |
|---|---|---|---|---|---|---|---|---|
| IFNAR1 | 204191_at | 124 (55–188) | A/P | .165 | 139 (73–150) | A/P | .133 | 1.1 |
| IFNAR2 | 204786_s_at | 126 (51–279) | P | .004 | 122 (57–208) | P | .003 | 1.0 |
| JAK1 | 201648_at | 1377 (1227–1821) | P | <.001 | 3782 (2799–4632) | P | <.001 | 2.7 |
| TYK2 | 205546_s_at | 491 (315–880) | P | .011 | 963 (605–1229) | P | .005 | 2.0 |
| STAT1 | 200887_s_at | 1306 (892–1597) | P | <.001 | 730 (641–908) | P | <.001 | 0.6 |
| STAT1 | 97935_3_at | 720 (370–924) | P | <.001 | 474 (310–616) | P | <.001 | 0.7 |
| STAT2 | 205170_at | 164 (132–185) | P | .003 | 156 (109–189) | P | .009 | 0.9 |
| STAT3 | 208991_at | 928 (670–1179) | P | <.001 | 2084 (1458–2467) | P | <.001 | 2.2 |
| STAT4 | 206118_at | 206 (126–297) | A/P | .092 | 430 (319–507) | P | .024 | 2.1 |
| STAT5A | 203010_at | 492 (343–675) | P | .037 | 994 (772–1143) | P | .006 | 2.0 |
| STAT5B | 205026_at | 110 (87–139) | P | .008 | 239 (205–285) | P | .002 | 2.2 |
| STAT6 | 201331_s_at | 1975 (1773–2188) | P | <.001 | 1424 (1252–1681) | P | <.001 | 0.7 |
| STAT6 | 201332_s_at | 368 (306–428) | P | .007 | 269 (183–302) | A/P | .044 | 0.7 |
RNA from iDCs and mDCs was analyzed by Affymetrix microarray U133A. Affymetrix data were compiled from 4 individual donors. Average relative mRNA expression for iDCs (iDC avg) and mDCs (mDC avg) are indicated with the range indicated in parentheses.
Determination of the absence or presence (A/P) of a gene transcript as well as the P values were calculated using Affymetrix statistical software MAS5.0
Fold change of average expression levels are expressed as a ratio of mDC avg/iDC avg.
To evaluate STAT activation in DCs following IFN-α/β stimulation, we analyzed phosphorylation of STAT proteins by intracellular flow cytometry and Western blot. FACS analysis offers the advantage of per-cell analysis, thus allowing evaluation of differential IFN-α/β–induced STAT phosphorylation while simultaneously determining the state of DC maturation. DC populations were stained and gated for their expression of CD83 (a DC maturation marker), and intracellular antibodies specific for p-STAT allowed determination of how the respective cell types had responded to IFN-α/β stimulation. The CD83− iDCs phosphorylated STAT1, and not STAT4, in response to IFN-α/β, whereas CD83+ mDCs preferentially showed phosphorylation of STAT4 and a significantly reduced phosphorylation of STAT1 (Figure 3D; P < .003, n = 6). One concern in comparing 2 distinct cell types by cytometry is potential differences in their relative autofluorescence. Because this is indeed the case for iDCs and mDCs (Figure 3D-E), we generated lysates from populations of IFN-α/β–stimulated DCs (> 95% phenotypically similar based on cDC maturation markers) and monitored STAT phosphorylation by immunoblot. iDCs exposed to IFN-α/β resulted in phosphorylation of STAT1 and STAT2 (Figure 3F). In contrast, despite the presence of total STAT1 and IFN-α/β–induced STAT2 phosphorylation, mDCs showed reduced levels of STAT1 phosphorylation (Figure 3F). Instead, IFN-α/β signaling resulted in activation of STAT4 (Figure 3G).
To test if the inhibition of STAT1 phosphorylation in mDCs is intrinsically impaired, we evaluated the ability of IFN-γ to trigger STAT1 activation. Notably, IFN-γ treatment of mDCs resulted in efficient phosphorylation of STAT1, indicating that inhibition of pSTAT1 is in fact specific to IFN-α/β signaling (Figure 3E). Together, these results suggest that DC maturation alters IFN-α/β signaling through the modulation of STAT utilization.
IFN-α/β signaling results in differential regulation of CD40L-induced IL-12
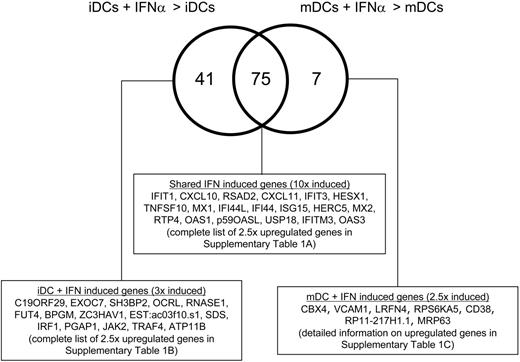
To evaluate the effect of this differential signaling on gene expression following IFNAR engagement, we again used Affymetrix U133A microarrays. As expected, a shared IFN genetic signature could be observed in both iDCs and mDCs (Figure 4; Table S1A, available on the Blood website; see the supplemental Materials link at the top of the online article). In addition, it was possible to identify differences in IFN-α/β–inducible genes, comparing the genes up-regulated in iDCs exposed to exogenous IFN-α versus mDCs exposed to exogenous IFN-α (Figure 4; Table S1B-C). Using stringent cut-off points for gene expression, we find that 41 genes are uniquely induced in iDCs and 7 genes are uniquely up-regulated in mDCs at 2 hours following IFN stimulation.
Differential induction of IFN-α–inducible genes in iDCs and mDCs. Affymetrix U133A data sets were queried to identify genes up-regulated 2 hours following exposure to IFN-α. Only genes with an absolute signal intensity more than 100 and P < .05 were included. Shared genes up-regulated by more than 10-fold are indicated and a complete list of shared 2.5-fold up-regulated genes is included in Table S1A. Genes up-regulated uniquely by IFN-α treatment of iDCs are listed and a complete list of genes uniquely 2.5-fold up-regulated are detailed in Table S1B. Finally, genes up-regulated uniquely by IFN-α treatment of mDCs are listed and a complete annotation of these changes is offered in Table S1C.
Differential induction of IFN-α–inducible genes in iDCs and mDCs. Affymetrix U133A data sets were queried to identify genes up-regulated 2 hours following exposure to IFN-α. Only genes with an absolute signal intensity more than 100 and P < .05 were included. Shared genes up-regulated by more than 10-fold are indicated and a complete list of shared 2.5-fold up-regulated genes is included in Table S1A. Genes up-regulated uniquely by IFN-α treatment of iDCs are listed and a complete list of genes uniquely 2.5-fold up-regulated are detailed in Table S1B. Finally, genes up-regulated uniquely by IFN-α treatment of mDCs are listed and a complete annotation of these changes is offered in Table S1C.
In light of our functional data suggesting that IFN-α treatment of iDCs impaired their ability to be licensed, we also defined the gene set that distinguished DCs matured in the presence of IFN-α/TNF-α/PGE2 from mDCs that had been matured simply in the presence of TNF-α/PGE2. Our interest in this comparison is that the differentially regulated genes in iDCs matured in the presence of IFN-α may result in an altered response to CD40 engagement. We find 71 genes that are 2.5 × higher in DCs matured in the presence of IFN-α suggesting that early exposure to IFN-α results in a lasting genetic program with potential effects on DC licensing (data not shown). To evaluate how this shift in gene expression altered the response to CD40 engagement, we queried for gene changes in which: mDCs + CD40L ≥ 2 × mDCs; and mDCs + CD40L ≥ 2 × IFN-α-mDCs + CD40L (n = 2). With such a Boolean search, we exclude gene changes that are simply a result of IFN treatment of the DCs. From the list generated, genes were excluded if they did not meet a “p” call in mDCs or mDCs + CD40L in either or both of the experiments, as per the Affymetrix MAS5.0 statistical package. Data on iDCs, mDCs, IFNα-mDCs, mDCs + CD40L, and IFN-α-mDCs + CD40L from the 2 individual donors were averaged, and means are graphically represented to reveal CD40L-induced transcription changes that distinguish treatment of mDCs versus IFN-mDCs. This analysis yielded 9 candidate genes that were inhibited by IFN-early (Figure S1).
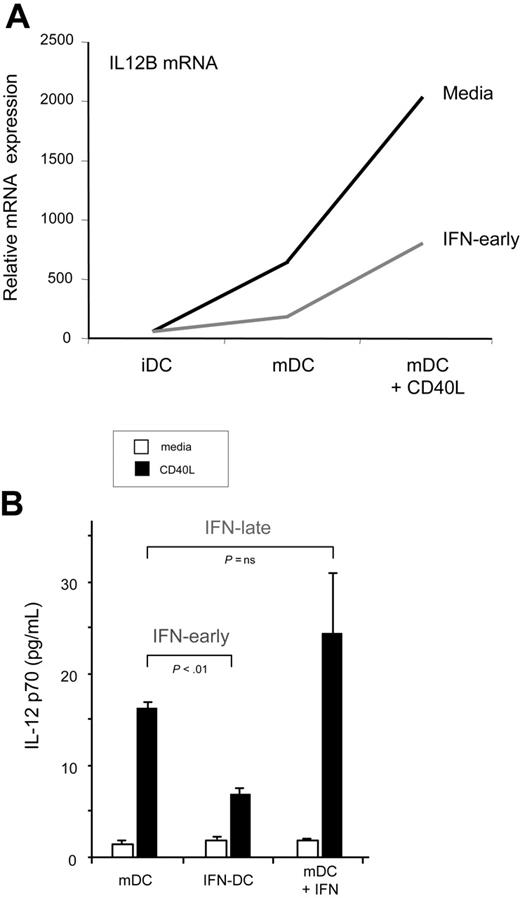
In light of the critical role that IL-12 plays in cross-priming, we further explored the observation of differential IL-12p40 mRNA expression (Figure 5A). Functional IL-12 exists as a heterodimer of IL-12p35 and IL-12p40. Interestingly, no differential IL-12p35 mRNA expression was observed with IFN-α treatment (data not shown). To evaluate the regulation of functional IL-12p70 in the respective cell types, we used an IL-12p70 ELISA (no reported cross-reactivity with IL-12p40; R&D Systems). Suggesting that IL-12p40 is limiting in mDCs, we demonstrated that, similar to IL-12p40 mRNA, CD40L-induced production of IL-12p70 was inhibited by IFN-early (Figure 5B). In contrast, no inhibition of CD40L-induced IL-12 production is seen following IFN-α/β treatment of mDCs (Figure 5B). Although others have reported the synergistic effect of IFN-α/β and CD40L in enhancing IL-12p70 production, the synergy we observed is not statistically significant at the doses of IFN-α used in these experiments.
IFN-early, not IFN-late, inhibits CD40L-triggered IL-12. (A) Affymetrix analysis shows relative expression of IL-12B subunit in iDCs, mDCs, and mDCs + CD40L. Untreated, black line. IFN-α/β–treated iDCs, gray line. (B) Supernatants from DCs matured alone (mDCs), in the presence of IFN-α/β (IFN-DC), or DCs exposed to IFN-α/β after maturation (mDC + IFN) were treated with CD40L as indicated for 2 hours. IL-12p70 was measure by ELISA and the average of triplicate wells are reported. P calculated by paired t test are shown.
IFN-early, not IFN-late, inhibits CD40L-triggered IL-12. (A) Affymetrix analysis shows relative expression of IL-12B subunit in iDCs, mDCs, and mDCs + CD40L. Untreated, black line. IFN-α/β–treated iDCs, gray line. (B) Supernatants from DCs matured alone (mDCs), in the presence of IFN-α/β (IFN-DC), or DCs exposed to IFN-α/β after maturation (mDC + IFN) were treated with CD40L as indicated for 2 hours. IL-12p70 was measure by ELISA and the average of triplicate wells are reported. P calculated by paired t test are shown.
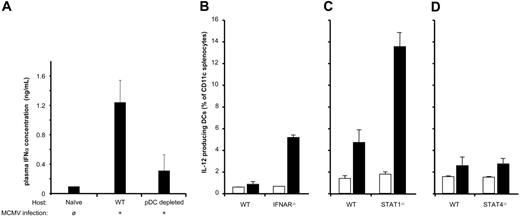
To evaluate the role of the STAT signaling switch in this dual regulation of IL-12 production, we used an in vivo model and monitored the effects of IFN-α/β signaling on IL-12p70 production. IFN-α/β produced by pDCs stimulated during MCMV infection inhibits IL-12p70 production by CD11c+ DCs.24 Using intracellular cytokine staining and FACS analysis we determined IL-12p70 production in CD11c+ splenocytes 36 hours after challenge with MCMV. We confirmed that MCMV triggers a strong IFN response that is responsible for a weak IL-12 signal in wild-type mice, but in the absence of the IFNAR, there is a significant increase in IL-12p70 production by cDCs (Figure 6A-B). Using this experimental system, we evaluated the STAT-dependence of this IFNAR-dependent IL-12 inhibition in DCs. STAT1 and STAT4 KO mice, along with the respective 129SVE and Balb/c controls, were challenged with MCMV. Consistent with our findings in human DCs, we observed that STAT1−/− mice failed to down-regulate IL-12p70 production whereas STAT4−/− behaved like their respective control (Figure 6C-D). Of note, the distinct strain backgrounds used each have different baseline production of IFN and DC-derived IL-12p70, in part due to the number of pDCs responding to the MCMV infection.25 Based on these mouse studies, we conclude that the inhibitory effects of IFN-α/β are mediated by STAT1 and not STAT4 signaling. The mechanism by which the STAT1 signaling pathways impinge on CD40-mediated signaling, however, remains unclear. Analysis of NF-κB activation in the respective cell types following CD40L stimulation suggested no differential phosphorylation or degradation of IκBα (Figure S2).
STAT1, but not STAT4, is required for IFN-α/β inhibition of IL-12p70 production by DCs. (A) Wild-type C57BL/6 or pDC-depleted mice were challenged with 4 × 105 PFU/mL salivary-gland prepared MCMV Smith strain. pDC-depleted mice were generated using anti–Gr-1 antibody (clone RB6-8C5) as described in “Material and methods.” pDC depletion was confirmed by FACS analysis on splenocytes of treated mice (data not shown). Serum samples at 36 hours after MCMV challenge were analyzed for IFN-α production by ELISA. (B-D) IFNAR−/−, STAT1−/−, or STAT4−/− mice and their respective controls (C57BL/6, 129SVE, and Balb/c) were challenged with MCMV. At 36 hours after infection, spleens were harvested and stained with CD8-FITC, CD11c-PE, and anti–IL-12–APC as described in “Materials and methods.” The percentage of CD11c+ DCs that stained positively for IL-12 is expressed as bar graph.
STAT1, but not STAT4, is required for IFN-α/β inhibition of IL-12p70 production by DCs. (A) Wild-type C57BL/6 or pDC-depleted mice were challenged with 4 × 105 PFU/mL salivary-gland prepared MCMV Smith strain. pDC-depleted mice were generated using anti–Gr-1 antibody (clone RB6-8C5) as described in “Material and methods.” pDC depletion was confirmed by FACS analysis on splenocytes of treated mice (data not shown). Serum samples at 36 hours after MCMV challenge were analyzed for IFN-α production by ELISA. (B-D) IFNAR−/−, STAT1−/−, or STAT4−/− mice and their respective controls (C57BL/6, 129SVE, and Balb/c) were challenged with MCMV. At 36 hours after infection, spleens were harvested and stained with CD8-FITC, CD11c-PE, and anti–IL-12–APC as described in “Materials and methods.” The percentage of CD11c+ DCs that stained positively for IL-12 is expressed as bar graph.
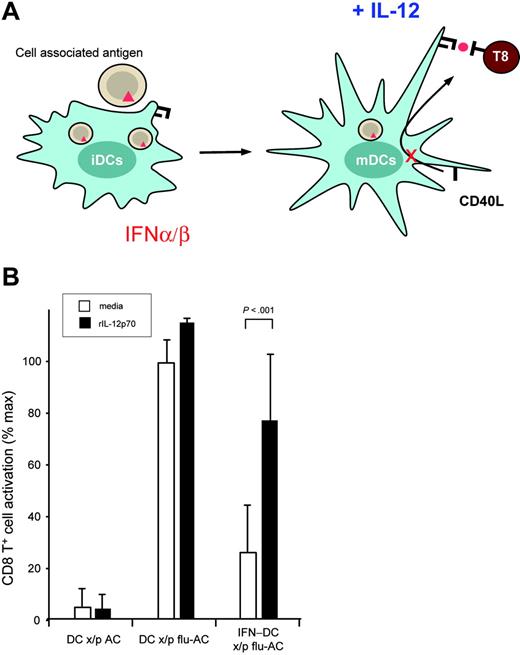
To determine the relevance of IL-12p70 inhibition, we evaluated the ability to overcome the IFN-early block by adding rIL-12p70 to the IFN-DCs cross-presenting antigen. ELISPOT results from 3 donors were normalized to the donor-specific maximal T-cell response and the percent maximal T-cell response is shown (Figure 7). Strikingly, IL-12p70 rescued the ability to stimulate antigen-specific CD8+ T cells in 3 of 3 individual donors (P = .005, n = 3). Although other factors may be involved, the decrease in CD40L-induced IL-12 seems to be a critical component of the inhibitory effects of IFN-α/β. These data also suggest that cross-presentation per se remains intact in IFN-DCs and that the dominant mechanism of regulation is via inhibition of CD40-dependent licensing of DCs. Thus, differential responsiveness of iDCs and mDCs to IFN-α/β regulates the production of cytokines that determine the immunologic outcome of cross-presentation.
Exogenous IL-12 overcomes IFN-α/β inhibition of CD40-dependent cross-presentation. (A) The schematic illustrates that IFN-α/β treatment (red) of iDCs during antigen capture and maturation inhibits CD40-induced IL-12 production. Exogenous IL-12p70 was added to DC/T-cell cultures and the effect on cross-presentation was assessed by ELISPOT. (B) T-cell activation by DCs cross-presenting apoptotic cells (DC x/p AC), DCs cross-presenting apoptotic cells loaded with influenza antigen (DC x/p flu-AC), or DCs matured in the presence of IFN-α/β cross-presenting influenza antigen (IFN DC x/p flu-AC) was monitored by IFN-γ ELISPOT. In addition to CD40L alone (media), recombinant human IL-12 (rIL-12p70) was added to the DC/T-cell cultures. Data from 3 donors was normalized to the donor-specific maximal T-cell response and the percent maximal T-cell response in the presence or absence of IFN-α/β and IL-12p70 is shown.
Exogenous IL-12 overcomes IFN-α/β inhibition of CD40-dependent cross-presentation. (A) The schematic illustrates that IFN-α/β treatment (red) of iDCs during antigen capture and maturation inhibits CD40-induced IL-12 production. Exogenous IL-12p70 was added to DC/T-cell cultures and the effect on cross-presentation was assessed by ELISPOT. (B) T-cell activation by DCs cross-presenting apoptotic cells (DC x/p AC), DCs cross-presenting apoptotic cells loaded with influenza antigen (DC x/p flu-AC), or DCs matured in the presence of IFN-α/β cross-presenting influenza antigen (IFN DC x/p flu-AC) was monitored by IFN-γ ELISPOT. In addition to CD40L alone (media), recombinant human IL-12 (rIL-12p70) was added to the DC/T-cell cultures. Data from 3 donors was normalized to the donor-specific maximal T-cell response and the percent maximal T-cell response in the presence or absence of IFN-α/β and IL-12p70 is shown.
Discussion
Although it is widely accepted that type I IFNs offer an important link between innate and adaptive immune responses,26 there exist contrasting data regarding their immunoregulatory effects. With respect to DC biology, there is support for IFN-α/β as a “danger” signal, triggering DC maturation and enhancing CD8+ T-cell cross-priming.9,10,27 These findings, however, must be contrasted with evidence for IFN-α/β as a negative regulator of IL-12 production and its promoting TH2 skewing by DCs.12 Our study offers new insight into these paradoxical effects and suggests a mechanism by which a particular cell lineage may respond differently to IFN and execute distinct immunologic programs. Here, we demonstrated that the maturation state of the DC determines the immunoregulatory effect of type I IFNs. When mDCs were exposed to IFN-α/β, a proinflammatory response ensued, as suggested by enhanced DC-mediated CD8+ T-cell activation (Figure 1). This observation is consistent with the role of IFN-α/β in enhancing TLR-mediated cytokine production28,29 and the “licensing” of DCs for cross-priming.10 In contrast, we discovered that iDCs exposed to IFN-α/β during antigen capture and maturation are impaired in their ability to activate CD8+ T cells (Figure 1). This inhibitory effect of IFN-early is specific for CD40-dependent CD8+ T-cell activation (Figures 1 and 3), implicating the modulation of a “third signal” produced by the DCs.30
How can IFN-α/β achieve opposite effects on iDCs versus mDCs when a single receptor exists? We show that a molecular switch in signaling networks is coupled to DC maturation and regulates the differential effects of IFN-α/β on antigen cross-presentation. Specifically, iDCs phosphorylate STAT1 and STAT2 on IFN-α/β stimulation and seem to use the canonical type I IFN-signaling pathway, whereas mDCs phosphorylate STAT4 and STAT2 (Figure 3). This switch is reflective of similar intracellular regulation in NK and T cells.5,31
The molecular switch in signaling pathways selectively targets an IFN-α/β signaling network. This is supported by mDCs responding to IFN-γ by signaling via STAT1 (Figure 3E). Notably, IFNAR signaling requires STAT2 for activation of both STAT132 and STAT4,33 whereas IFN-γ receptor binds and directly activates STAT1. These differences may account for the selective regulation of pSTAT1 downstream of IFNAR. Although the mechanism of STAT1 regulation in mDCs remains unknown, up-regulation of STAT4 may be directly responsible for inhibiting STAT1 activation. Microarray screening did not reveal notable changes in the expression of candidate genes such as suppressor of cytokine signaling SOCS1, or SOCS3. Interestingly, mDCs also show impaired phosphorylation of STAT3 in response to IFN-α/β stimulation (data not shown). In light of the ability of STAT3 to heterodimerize with STAT1 in the absence of STAT2,22 our findings demonstrate that the switch extends to redundant pathways to offer a functional skewing of the IFN-α/β signaling network.
CD4+ T cells, acting via CD40L/CD40 engagement, serves as a “licensing” signal for mature DCs17,34 by triggering IL-12 production, a critical component of what has been termed signal 3.30 The contrasting effects of IFN-α/β on cross-presentation reside at this point of regulation, with type I IFNs serving to overcome CD4+ T-cell help in some systems,10 but acting as a potent inhibitor of IL-12 in others.12 We propose that the switch in STAT utilization may account for this alternate regulation. IFN-α/β stimulation of iDCs activates STAT1-dependent gene transcription, which blunts CD40-induced IL-12 production. Taking advantage of the in vivo MCMV mouse model and the observation that virally induced IFN-α/β inhibits IL-12p70 production in CD11c+ DCs,24 we were able to test the role for STAT1 versus STAT4. Strikingly, the IFN produced in STAT1−/− mice failed to inhibit IL-12 production. We are careful not to overinterpret the STAT4−/− data due to differences between mice and humans in how the IFNAR recruits STAT4, but we are confident of the importance of the STAT1-mediated inhibitory pathway. This STAT1-mediated inhibition may result from both direct transcriptional inhibition of the IL-12p40 promoter35 as well as indirect transcriptional inhibition of IL-12p40 and p35 secondary to autocrine IL-10 stimulation.36 It has also been reported that p-STAT1 may act directly to inhibit TNF-α–mediated IL-12 production via inhibition of NF-κB activation37 ; however, our analysis suggests no direct modulation of IκB activation (Figure S2).
Regarding the physiologic and pathologic relevance for our findings, we suggest that spatial compartmentalization of IFN-α/β production may offer a resolution for its paradoxical immunoregulatory effects on conventional DCs. Immature and mature myeloid DCs reside in different physiologic locations, with the former residing within peripheral tissue and the latter homing to the T-cell area of draining lymph organs.8 During acute viral infection, TLR engagement on pDCs would induce trafficking to the lymph node and production of robust amounts of type I IFNs. In this situation, IFN-α/β would act on mature myeloid DCs that had migrated from the peripheral tissue with captured antigen. The exposure of mDCs to IFN-α/β may act directly on the DCs to synergize with other inflammatory stimuli and enhance IL-12 production. Our model does not rule out an additional, direct effect of IFN-α or other pDC-derived cytokines on CD8+ T-cell activation.20 For example, recent data suggest that activation of STAT4 in CD8+ T cells may potentiate IFN-γ production.38
In situations of chronic infection, type I IFNs in the tissue (produced by parenchymal cells, pDCs that have moved into sites of chronic inflammation39 or by immature myeloid DCs themselves40 ) may exert a counter-inflammatory effect on iDCs as they differentiate into mDCs. Specifically, our data predict that IFN-α/β, produced in the tissue during antigen capture, inhibits subsequent T-cell activation by cross-presenting DCs. The restriction of this inhibition to cross-presentation is likely due to the CD40 dependence in our model system and may be more broadly applicable to other CD4-dependent responses.41 The effects of IFN-early may explain how some viruses evade adaptive immune responses, subverting the host immune response by triggering IFN-α/β production in the infected tissue without stimulating pDCs.24 For example, hepatitis C virus induces robust levels of IFN-α/β production soon after infection.42 Although it remains unclear whether DCs can be infected, the high levels of IFN-α/β in the liver may play an important role in preventing effective CD8+ T-cell response in chronically infected patients.43 In addition, the inhibitory effect of IFN on tissue DCs may account for the recent observation of an inverse correlation between length of remission and pDC infiltration in early-stage breast cancer.44 Finally, the results presented here may offer a more detailed mechanism for the therapeutic effects of type I IFNs in both multiple sclerosis2 and HCV treatment.
These findings show that IFN-α/β signaling is coupled to DC maturation, enabling distinct effects on the immunologic outcome of cross-presentation. This plasticity illustrates a novel mechanism by which DCs modulate the integration of signals from the surrounding environment. These distinct signaling pathways correspond to distinct genetic signatures in iDCs and mDCs and may offer important insight into the biologic impact of IFN-α/β in distinct tissue compartments. These findings suggest a rationale for spatial compartmentalization of pDCs during viral pathogenesis and offer insight into more effective therapeutic delivery of type I IFNs.
Authorship
Contribution: R.L. conceptualized, designed and performed experiments; was responsible for the acquisition, analysis, and interpretation of data; was responsible for drafting and revising the manuscript; and assisted in creating all figures and tables. D.B. conceptualized, designed, and performed selected experiments involving quantitative PCR and assisted in acquisition and analysis of data. S.P. contributed technical expertise to experiments, provided key technology, contributed to experimental design, and was responsible for revising the manuscript. C.R. conceptualized and designed experiments, supervised studies, and revised the manuscript. R.D. conceptualized and designed experiments, supervised studies, and revised the manuscript. M.A. conceptualized and designed experiments, supervised analysis and interpretation of data, was responsible for creation of figures and tables, and was responsible for drafting and revising the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew L. Albert, Institut Pasteur, 25 Rue du Dr Roux, Paris, France 75724; e-mail. albertm@pasteur.fr.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported in part by grants from the National Institutes of Health (NIH) Medical Scientist Training Program (MSTP) grant GM07739, W. M. Keck Foundation Medical Scientist Fellowship (R.S.L.), L'Association pour la Recherche sur le Cancer (S.P.), NIH, the Greenberg Medical Research Institute and the Ellison Medical Foundation Senior Scholar's award in Global Infectious Disease (C.M.R.), Howard Hughes Medical Institute, NIH R01 CA85784 and General Clinical Research Center Grant M01-RR00102 (R.B.D.), La Ligue Nationale Contre le Cancer, Doris Duke Charitable Foundation, and EURYI Scheme, European Science Foundation (M.L.A.).
The authors would like to thank Dr Nir Hacohen for generously providing a customized database for the analysis of our Affymetrix data set. We also thank Josiane Ragimbeau for her technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal