Abstract
Essential thrombocythemia (ET) is heterogeneous with respect to natural history, X-chromosome inactivation patterns (XCIPs), and presence of the V617F mutation in Janus kinase 2 (JAK2). We studied 111 patients with ET; 39% were JAK2 mutant positive, and clone size (percentage mutant JAK2) was concordant with XCIP when constitutive T-cell patterns were taken into account. JAK2 mutant clones were present in both clonal and polyclonal cases as determined by XCIP, and the former had higher mutant JAK2 levels (median 26% versus 16%; P = .001). No change was observed in serial XCIP analysis of 14 polyclonal patients over a median follow-up of 61 months. Furthermore, 18 of 19 mutant-positive patients showed no significant change in mutant JAK2 level over a median follow-up of 47 months. These results suggest that, in many cases of ET, a small stable clone containing a JAK2 mutation can be maintained as a subpopulation for many years.
Introduction
Patients with essential thrombocythemia (ET) show biologic heterogeneity. Although initially considered to be a clonal disease, studies using X-chromosome inactivation patterns (XCIPs) in females have demonstrated that, once constitutional and age-acquired skewing have been taken into account, a significant proportion of patients have polyclonal myelopoiesis,1-5 although this does not exclude the presence of minor clones. More recently, the V617F mutation in Janus kinase 2 (JAK2) has been reported in a variable proportion of patients with ET (23%-72%, depending on method used),6-13 and is present in patients with both clonal and polyclonal XCIPs.11-13 The latter observation suggests that minor clones could arise on a polyclonal background and then gradually be selected over time to lead to evident clonality. We have used longitudinal studies to address this issue.
Patients, materials, and methods
Patients and samples
Approval for these studies was provided by the London Multi-center Research Ethics Committee. Patient consent was obtained according to the Declaration of Helsinki. Peripheral blood samples were obtained from 111 patients with ET (92 female, 19 male). Neutrophils and CD3+ cells were purified and DNA prepared as previously described.2
XCIPs
The human androgen receptor assay (HUMARA) was performed as previously described,2 except using a fluorescent-labeled primer, and analysis was done on the Beckman Coulter CEQ8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA). Results are given as the mean relative percentage of the smaller and larger alleles (A%:B%), respectively. There are several caveats to interpretation of an XCIP14 and a pattern was only considered to be truly clonal if (1) the myeloid cell XCIP showed more than 75% of one of the X-alleles, (2) there was more than 20% difference between the XCIP of T cells (reflecting the constitutional hemopoietic stem cell pattern) and myeloid cells, and (3) the individual was younger than 65 years of age.
JAK2 mutation analysis
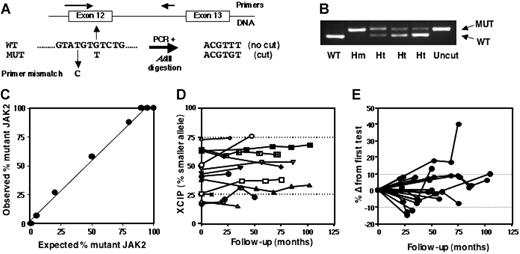
DNA was screened for presence of the G-to-T JAK2 mutation using polymerase chain reaction (PCR) with a mismatch primer and restriction endonuclease digestion to discriminate between wild-type (WT) and mutant alleles (Figure 1A-B). The relative level of mutant (percentage of total JAK2 alleles) was quantified using the same procedure with a fluorescent-labeled reverse primer and analysis of digested products was done on the Beckman Coulter CEQ8000 Genetic Analysis System. Linearity of the assay was established using DNA mixtures from homozygous WT and mutant cell lines (HL60 and HEL cells, respectively; Figure 1C).
XCIP and V617F JAK2 mutation analysis in patients with ET. (A) Principle of method using a mismatch PCR primer and AflIII digestion of PCR products. (B) Gel showing digested products from a WT control, mutant-positive homozygote (Hm), and 3 heterozygotes (Ht) with differing levels of mutant. (C) Validation of mutant JAK2 quantification assay using mixes of DNA from homozygous WT (HL60) and mutant (HEL) cell lines. (D) Serial analysis of neutrophil XCIPs in 14 patients with polyclonal myelopoiesis. Dotted lines represent the threshold for clonality (> 75% expression of one allele) provided there was more than 20% difference from the T-cell XCIP and the patient was younger than 65 years of age. (E) Serial analysis of mutant JAK2 level in neutrophil samples from 20 mutant-positive patients. Results are expressed as the percentage difference from the first sample tested. Dotted lines represent the limits of technical variation (mean ± 1 standard deviation for percentage difference in mutant level in 46 duplicate analyses).
XCIP and V617F JAK2 mutation analysis in patients with ET. (A) Principle of method using a mismatch PCR primer and AflIII digestion of PCR products. (B) Gel showing digested products from a WT control, mutant-positive homozygote (Hm), and 3 heterozygotes (Ht) with differing levels of mutant. (C) Validation of mutant JAK2 quantification assay using mixes of DNA from homozygous WT (HL60) and mutant (HEL) cell lines. (D) Serial analysis of neutrophil XCIPs in 14 patients with polyclonal myelopoiesis. Dotted lines represent the threshold for clonality (> 75% expression of one allele) provided there was more than 20% difference from the T-cell XCIP and the patient was younger than 65 years of age. (E) Serial analysis of mutant JAK2 level in neutrophil samples from 20 mutant-positive patients. Results are expressed as the percentage difference from the first sample tested. Dotted lines represent the limits of technical variation (mean ± 1 standard deviation for percentage difference in mutant level in 46 duplicate analyses).
Influence of mutant JAK2 clone size on XCIP
The observed neutrophil XCIP in JAK2 mutant–positive patients will be dependent on the X-chromosome allele expressed in the mutant-carrying clone. The expected XCIP was therefore calculated from the percentage of mutant-positive cells (x), the percentage of WT cells (y), and the percentage constitutive expression of the A and B alleles (ie, the T-cell XCIP), assuming that all mutant-positive cells were heterozygous for the mutation and that all clonal cells were mutant positive. If the A allele is expressed in the clone, the expected XCIP (A%:B%) = [(x/100) × 100 ] + [(y/100) × %A ] : [(y/100) × %B ]. Conversely, if the B allele is expressed in the clone, the expected XCIP = [(y/100) × %A ] : [(x/100) × 100 ] + [(y/100) × %B ].
Results and discussion
The JAK2 mutation was detected in 43 (39%) of 111 patients, which is in accord with the incidence reported by others.6-11,13 Twenty of the 43 mutant-positive cases had an interpretable XCIP: 5 were clonal and 15 polyclonal, 15 were not interpretable due to constitutional or age-related skewing, 2 were uninformative, 6 were male. The 5 cases with clonal myelopoiesis had a higher mutant level (median, 26%; range, 22%-47%) than the 15 with apparently polyclonal myelopoiesis (median, 16%; range, 5%-26%; P = .001) (Table 1). Only one patient (no. 5), who had a monoclonal XCIP, had a mutant level that was consistent with heterozygous mutant status in nearly all (94%) of her cells. In all other patients, heterozygous mutant status was limited to 10% to 54% of cells.
XCIP results and mutant JAK2 levels in 5 clonal and 15 polyclonal patients with ET
| Patient no. . | Clonality status . | Neut XCIP, A%:B% . | T-cell XCIP, A%:B% . | % JAK2 mutant . | % mutant+ cells* . | Expected neut XCIP if A clone, A%:B%† . | Expected neut XCIP if B clone, A%:B%† . | % Δ (O − E) . |
|---|---|---|---|---|---|---|---|---|
| 1 | C | 19:81 | 41:59 | 22 | 44 | 67:33 | 23:77 | 4 |
| 2 | C | 84:16 | 62:38 | 25 | 50 | 81:19 | 31:69 | 3 |
| 3 | C | 87:13 | 61:39 | 26 | 52 | 81:19 | 30:70 | 6 |
| 4 | C | 21:79 | 66:34 | 27 | 54 | 84:16 | 30:70 | 9 |
| 5 | C | 97:3 | 70:30 | 47 | 94 | 98:2 | 4:96 | 1 |
| 6 | P | 51:49 | 65:35 | 5 | 10 | 69:31 | 59:41 | 8 |
| 7 | P | 38:62 | 39:61 | 5 | 10 | 45:55 | 35:65 | 3 |
| 8 | P | 64:36 | 58:42 | 7 | 14 | 64:36 | 50:50 | 0 |
| 9 | P | 49:51 | 37:63 | 8 | 16 | 47:53 | 31:69 | 2 |
| 10 | P | 54:46 | 56:44 | 11 | 22 | 66:34 | 44:56 | 10 |
| 11 | P | 46:54 | 53:47 | 14 | 28 | 66:34 | 38:62 | 8 |
| 12 | P | 33:67 | 34:66 | 14 | 28 | 52:48 | 24:76 | 9 |
| 13 | P | 37:63 | 40:60 | 15 | 30 | 58:42 | 28:72 | 9 |
| 14 | P | 61:39 | 74:26 | 18 | 36 | 83:17 | 47:53 | 14 |
| 15 | P | 43:57 | 35:65 | 19 | 38 | 60:40 | 22:78 | 17 |
| 16 | P | 19:81 | 25:75 | 19 | 38 | 54:46 | 16:84 | 3 |
| 18 | P | 73:27 | 65:35 | 21 | 42 | 80:20 | 38:62 | 7 |
| 17 | P | 21:79 | 36:64 | 22 | 44 | 64:36 | 20:80 | 1 |
| 19 | P | 73:27 | 69:31 | 22 | 44 | 83:17 | 39:61 | 10 |
| 20 | P | 73:27 | 72:28 | 26 | 52 | 87:13 | 35:65 | 14 |
| Patient no. . | Clonality status . | Neut XCIP, A%:B% . | T-cell XCIP, A%:B% . | % JAK2 mutant . | % mutant+ cells* . | Expected neut XCIP if A clone, A%:B%† . | Expected neut XCIP if B clone, A%:B%† . | % Δ (O − E) . |
|---|---|---|---|---|---|---|---|---|
| 1 | C | 19:81 | 41:59 | 22 | 44 | 67:33 | 23:77 | 4 |
| 2 | C | 84:16 | 62:38 | 25 | 50 | 81:19 | 31:69 | 3 |
| 3 | C | 87:13 | 61:39 | 26 | 52 | 81:19 | 30:70 | 6 |
| 4 | C | 21:79 | 66:34 | 27 | 54 | 84:16 | 30:70 | 9 |
| 5 | C | 97:3 | 70:30 | 47 | 94 | 98:2 | 4:96 | 1 |
| 6 | P | 51:49 | 65:35 | 5 | 10 | 69:31 | 59:41 | 8 |
| 7 | P | 38:62 | 39:61 | 5 | 10 | 45:55 | 35:65 | 3 |
| 8 | P | 64:36 | 58:42 | 7 | 14 | 64:36 | 50:50 | 0 |
| 9 | P | 49:51 | 37:63 | 8 | 16 | 47:53 | 31:69 | 2 |
| 10 | P | 54:46 | 56:44 | 11 | 22 | 66:34 | 44:56 | 10 |
| 11 | P | 46:54 | 53:47 | 14 | 28 | 66:34 | 38:62 | 8 |
| 12 | P | 33:67 | 34:66 | 14 | 28 | 52:48 | 24:76 | 9 |
| 13 | P | 37:63 | 40:60 | 15 | 30 | 58:42 | 28:72 | 9 |
| 14 | P | 61:39 | 74:26 | 18 | 36 | 83:17 | 47:53 | 14 |
| 15 | P | 43:57 | 35:65 | 19 | 38 | 60:40 | 22:78 | 17 |
| 16 | P | 19:81 | 25:75 | 19 | 38 | 54:46 | 16:84 | 3 |
| 18 | P | 73:27 | 65:35 | 21 | 42 | 80:20 | 38:62 | 7 |
| 17 | P | 21:79 | 36:64 | 22 | 44 | 64:36 | 20:80 | 1 |
| 19 | P | 73:27 | 69:31 | 22 | 44 | 83:17 | 39:61 | 10 |
| 20 | P | 73:27 | 72:28 | 26 | 52 | 87:13 | 35:65 | 14 |
C indicates clonal; P, polyclonal; neut, neutrophil; Δ, difference; O − E, observed minus expected.
Assumes heterozygous mutant status in all cells.
Calculated using the equation given in “Influence of mutant JAK2 clone size on XCIP.” For example, for case 1, the expected XCIP (A%:B%) if the clone expresses the A allele = [(44/100) × 100] + [(56/100) × 41] : [(56/100) × 59] and if it expresses the B allele = [(56/100) × 41] : [(44/100) × 100] + [(56/100) × 59].
Although the presence of a clonal population would be expected to alter the XCIP, the ability to detect this difference will depend not only on the relative size of the clone but also on the constitutive XCIP of the individual and the active X-allele of the clone. We used the quantified JAK2 mutant level to reflect relative clone size (assuming all clonal cells are mutant positive and heterozygous for the mutant) and the T-cell XCIP to reflect the individuals' constitutive pattern in order to calculate the expected impact on the neutrophil XCIP according to which X-allele was expressed in the mutant-carrying cell (Table 1, allele A or B). An example of these calculations is given in the footnote to Table 1. In each of the 20 cases, the observed pattern was very close to one of the calculated XCIPs (Table 1). The percentage difference between observed and expected value of one allele ranged between 0% and 17%; in only one patient (no. 15) was the difference close to our previously determined limit of 20% for technical variation or the difference between neutrophil and T-cell XCIPs in young hematologically normal individuals.15,16 Two papers have previously suggested that in ET there is a discrepancy between the proportion of JAK2 mutant–positive cells and the clonality status by XCIP,12,17 but stringent quantitative analysis was not performed.
These results demonstrate that the relative size of the JAK2 mutant–positive population is often small and its size will influence apparent clonality status. To determine whether these clones expand due to the growth/survival advantage provided by the mutation(s) and, with time, accumulate to lead to a predominant clonal population, we performed serial analyses of neutrophil XCIPs in 14 polyclonal patients, 10 of them JAK2 mutant–positive, over a follow-up period of a median of 61 months (range, 10-102 months). We found no evidence for the emergence of a slowly growing clone (Figure 1D). Five of these patients received no therapy other than aspirin for between 10 and 87 months during the course of the study; 2 were subsequently treated with hydroxyurea but there was no change in their XCIP. Seven had received hydroxyurea throughout the study, 1 had intermittently received interferon-alpha then anagrelide. At the time of their first test, 3 of the patients were 5 to 7 years after diagnosis and therefore had had stable polyclonal disease for 12 to 16 years.
The XCIP results were corroborated by sequential analysis of JAK2 mutant levels in neutrophils from 19 mutant-positive cases over a median of 47 months (range, 22-104 months). Two patients had clonal and 10 had polyclonal XCIPs, 4 were not interpretable, 3 were male. Eighteen of the cases had remarkably stable mutant levels, and median difference between any 2 samples was 8% (range, 0%-18%; Figure 1E), including 2 patients with a consistent mutant level of 5% to 10% for 4 years. Although 13 of these patients had received cytoreductive therapy during the study (9 hydroxyurea, 2 busulphan, 1 interferon-alpha, 1 anagrelide), 5 received only aspirin for between 20 and 87 months, indicating that stability of JAK2 mutant level was not due to suppression of mutant-positive clones by antiproliferative agents. One patient showed a significant increase in JAK2 mutant level; she was elderly with a skewed XCIP and it was not possible to correlate this change with XCIP analysis. Neutrophils from 20 JAK2 WT patients also were followed over a median of 38 months (range, 22-104 months); 6 had clonal myelopoiesis, 7 were polyclonal, 4 were not interpretable, and 3 were male. None became mutant positive. At least 13 were 6 years or more from diagnosis, suggesting that it is uncommon for the mutation to occur in late-stage, stable disease.
The evidence that JAK2 mutant clones can be maintained as a subpopulation for many years poses intriguing biologic questions about the regulation of such clones. They must have a growth/survival advantage that enables them to expand to a detectable level, but then be stabilized at this level for many years without further amplification. This scenario bears a number of similarities to that observed in paroxysmal nocturnal hemoglobinuria where differences in XCIP status reflect the degree of chimerism for cells either expressing or deficient in glycosylphosphatidylinositol-anchored proteins,18 and the clonal cell populations can remain stable for significant periods of time or even regress.19
Authorship
Contribution: R.E.G. and A.J.R.A. carried out experimental analysis; M.J.N. was responsible for sample collection; R.E.G. and D.C.L. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, Royal Free and University College Medical School, 98 Chenies Mews, London WC1E 6HX United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank all patients and clinicians who provided blood samples for this study. This work was supported by a grant from the UK Medical Research Council.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal