Abstract
Caspases have demonstrated several nonapoptotic functions including a role in the differentiation of specific cell types. Here, we show that caspase-8 is the upstream enzyme in the proteolytic caspase cascade whose activation is required for the differentiation of peripheral-blood monocytes into macrophages. On macrophage colony-stimulating factor (M-CSF) exposure, caspase-8 associates with the adaptor protein Fas-associated death domain (FADD), the serine/threonine kinase receptor-interacting protein 1 (RIP1) and the long isoform of FLICE-inhibitory protein FLIP. Overexpression of FADD accelerates the differentiation process that does not involve any death receptor. Active caspase-8 cleaves RIP1, which prevents sustained NF-κB activation, and activates downstream caspases. Together these data identify a role for caspase-8 in monocytes undergoing macrophagic differentiation, that is, the enzyme activated in an atypical complex down-regulates NF-κB activity through RIP1 cleavage.
Introduction
A family of cysteine proteases, known as caspases, plays a central role in many forms of apoptosis.1 Two main pathways of caspase activation leading to apoptosis have been described. The intrinsic pathway involves the disruption of the outer mitochondrial membrane barrier function, thus permitting the release of proapoptotic molecules from the mitochondria to the cytosol. These molecules include cytochrome c that, in the presence of ATP, trigger oligomerization of a platform protein named apoptosis-activating factor 1 (Apaf-1). This protein recruits and activates caspase-9 in the apoptosome. In turn, caspase-9 cleaves and activates downstream effector enzymes such as caspase-3. The extrinsic pathway starts at the level of plasma membrane by engagement of death receptors such as Fas/CD95, tumor necrosis factor receptor 1 (TNF-R1), and TNF-related apoptosis-inducing ligand (TRAIL) receptors DR4 and DR5. In the presence of their respective ligand, death receptors recruit the adaptor molecule Fas-associated death domain (FADD) protein, which, in turn, recruits and activates an initiator enzyme, usually caspase-8, in the death-inducing signaling complex (DISC). Caspase-8 either directly activates the caspase cascade or connects the extrinsic to the intrinsic pathway through cleavage of the sentinel BH3-only protein Bid.1,2 Additional pathways of caspase activation involve either dependence receptors in the absence of their ligand,3 or interaction of Apaf-1–like molecules with the adaptor molecule ASC,4 or endoplasmic reticulum stress that activates caspase-12 in mice and caspase-4 in humans.5
Although in most cases caspase activation engages cells to die, recent evidence indicates nonapoptotic functions of these enzymes. For example, caspase-8 was involved in lymphocyte activation,6 which might account for the combined T- and B- cell, and natural killer (NK)–cell immunodeficiency in patients with mutated caspase-8.7 The enzymatic activity of caspase-8 is also required for fetal liver hematopoietic stem cell proliferation.8 Signaling through caspase-8 does not always require its enzymatic activity; for example, in cells in which signaling through Fas/CD95 promotes survival rather than death, caspase-8 mediates NF-κB activation and its enzymatic activity is dispensable for this function. A scaffolding-related function was suggested for the enzyme that recruits FADD, the 2 isoforms of FLICE-inhibitory protein (FLIP), and the serine/threonine receptor-interacting protein 1 (RIP1).8-11
Caspases were also involved in specific differentiation processes. Erythropoiesis could be regulated by a negative feedback loop in which mature erythroblasts expressing death-receptor ligands inhibit the differentiation of immature erythroblasts through caspase-8–mediated degradation of the transcription factor GATA-1,12 whereas a transient activation of caspases that does not lead to GATA-1 cleavage is requested for erythroid differentiation.13 Caspase activation was also demonstrated to play a role in the terminal differentiation of specific cell types that include lens epithelial cells, keratinocytes, skeletal muscle cells, megakaryocytes, osteoblasts, and drosophila spermatozoids (for a review, see Launay et al14 ).
We have previously reported an activation of caspase-3 and caspase-9 in human peripheral-blood monocytes that differentiate into macrophages in response to macrophage colony-stimulating factor (M-CSF). This caspase activation was not related to apoptosis nor was it observed in monocytes exposed to IL-4 and granulocyte-macrophage colony-stimulating factor (GM-CSF) that induce their differentiation into dendritic cells. By using the U937 human monocytic cell line exposed to phorbol ester as a model system, we showed that caspase activation actively contributed to the macrophagic differentiation process.15 The role of caspase-8 in this differentiation pathway was subsequently suggested by analysis of a mouse model of conditional caspase-8 gene knockout in myeloid bone marrow cells.16
The molecular pathway leading to caspase-8 activation in monocytes exposed to M-CSF, including its place in the proteolytic cascade of caspases and the functional consequences of its activation, remained unknown. In the present study, we demonstrate that caspase-8 is the apical enzyme in the caspase cascade that contributes to this differentiation pathway. In response to M-CSF, caspase-8 interacts with FADD, RIP1, and the long isoform of FLIP in the absence of any death receptor. NF-κB activation is transient along the macrophagic differentiation pathway and caspase-8–mediated RIP1 cleavage appears to prevent its sustained activation.
Materials and methods
Chemical reagents
M-CSF, GM-CSF, and IL-4 were obtained from R&D Systems (Minneapolis, MN) and TPA from Sigma-Aldrich Laboratories (St Louis, MO). TRAIL was obtained from Alexis (Lausen, Switzerland), the Fas agonistic antibody (clone CH11) from Biovalley (Rockville, MD), cycloheximide from Sigma-Aldrich, TNF-α from PeproTech (London, United Kingdom), and [γ-32P]ATP (259 TBq/mmol) from MP Biomedicals (Illkirch, France).
Antibodies
The following mouse monoclonal antibodies were used: an anti–human HSC70 (Santa Cruz Biotechnology, Santa Cruz, CA), anti–human caspase-8 (MBL), anti-TRAF2 (Stressgen, Victoria, BC, Canada), anti-FLIP (Alexis), and anti-RIP1, anti-FADD, anti-TRADD, and antiflotillin (PharMingen, San Diego, CA). Anti-p65, anti-Fas (Santa Cruz Biotechnology), anti-DR4 or anti-DR5 (Chemicon, Temecula, CA), and anti–cleaved caspase-3 (Cell Signaling Technology, Beverly, MA) rabbit polyclonal antibodies were also used. Secondary antibodies included HRP-conjugated goat antimouse or antirabbit antibodies (Jackson ImmunoResearch Laboratories, Bar Harbor, ME), and HRP-conjugated goat anti–mouse IgG1 and IgG2b (Southern Biotechnology, Birmingham, AL). For flow cytometry experiments, we used APC-conjugated anti-CD11b or anti-CD71 or anti-CD1a, together with an APC-conjugated isotype IgG1-matched control (PharMingen), a FITC-conjugated CD11b together with a FITC-conjugated mouse IgG1 isotype control (Immunotech, Marseille, France) anti-DR4, anti-DR5, anti-DcR1, anti-DcR2 or anti-TRAIL (Alexis), anti-Fas or Fas-L (PharMingen), and an anti–cleaved caspase-8 (Cell Signaling Technology) with its IgG1 mouse negative control (Dako, Glostrup, Denmark), and 488-Alexa goat antimouse or 568-Alexa goat antirabbit antibodies (Molecular Probes, Eugene, OR). A goat anti–caspase-8 (C-20) and a rabbit anti-FLIPL (H-150) polyclonal antibody (Santa Cruz Biotechnology) were used for immunoprecipitation experiments.
Cell culture and differentiation
The human leukemic cell line U937 (CRL-1593.2, mycoplasma free and virus free; American Type Culture Collection [ATCC], Manassas, VA), and U937 containing either pcDNA vector 3.1 or a caspase-8 dominant-negative form or the cowpox virus caspase-1 and caspase-8 inhibitor CrmA (pCDNA, C8DN, and CrmA, kindly provided by S. Grant, Medical College of Virginia, Richmond, VA), were grown in suspension in RPMI 1640 medium with glutamax-I (Gibco, Grand Island, NY) supplemented with 10% (vol/vol) fetal bovine serum (FBS; BioWhittaker, Verviers, Belgium) in an atmosphere of 95% air and 5% CO2 at 37°C. Human peripheral-blood monocytes were obtained from healthy donors with informed consent, provided in accordance with the Declaration of Helsinki, and purified using a monocyte isolation kit with a light-scattering (LS) column according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany) and exposed to appropriate cytokines. Macrophagic differentiation could be assessed by measuring the percentage of cells with a fibroblast-like shape (Station Cell Observer; Zeiss, LePecq, France). To identify these changes, cells were visualized using an Axiovert 200M inverted fluorescence microscope (Zeiss). Digital images were captured with a cooled charge-coupled device camera (AxioCam HRm, Zeiss) controlled with AxioVision software (Zeiss). Phase images were taken using a 200×/0.40 NA objective lens.
Flow cytometry and immunofluorescence assays
CD11b, CD71, and CD1a expression and caspase-8 cleavage were measured as previously described.15 To detect caspase activity, we used FAM-LETD-fmk (caspase-8), FAM-DEVD-fmk (caspases-3), and FAM-LEHD-fmk (caspases-9) detection kit FLICA (Serotec, Oxford, United Kingdom) according to the manufacturer's instructions. For nuclear p65 identification, cells were fixed in 2% paraformaldehyde for 10 minutes at room temperature (RT) and cytospun. Cells were then permeabilized for 20 minutes with 0.1% saponin and saturated 1 hour with 2% FBS, before incubation overnight with the p65 Ab (C-20) in PBS containing 2% FBS. After washing, cells were incubated for 30 minutes with 568-Alexa antirabbit antibody. Percentage of apoptotic cells was measured after nuclei staining with Hoechst 33342 (Sigma-Aldrich) using a fluorescence microscope (Nikon, Tokyo, Japan).
Vector constructs
Dominant-negative FADD (FADD-DN) and wild-type FADD (FADD-WT) vectors have been described previously.17 A RIP1 mutated on the caspase cleavage site (kindly provided by Olivier Micheau) amplified by polymerase chain reaction (PCR) was cloned into a ΔMCS lentiviral plasmid (ZEfir without its multicloning site) downstream the EF1α promoter and upstream the GFP cassette under the control of an internal ribosomal entry sequence (IRES).
Lentivirus vector production and transduction protocol
Vector particles were produced as previously described.17 U937 cells and primary monocytes were transduced with viral supernatants (at a multiplicity of infection [MOI] of ∼100, representing 100 ng/mL viral p24) on days 1 and 2 in RPMI containing 7.5% of BSA insulin transferrin (BIT). For primary cells cytokines were added at the beginning of infection and differentiation was analyzed at day 4. For U937 cells, EGFP+ U937 cells were selected 7 days after infection by cell sorting using a Coulter EPICS EPS (Beckman Coulter, Villepinte, France).
siRNA transfection
Human primary monocytes were transfected using the Human Monocyte Nucleofector Kit (Amaxa, Köln, Germany) according to the manufacturer's instructions. Briefly, 5 × 106 monocytes were resuspended into 100 μL nucleofector solution with 2 μg of either caspase-8 siRNA (forward: AGGGAACUUCAGACACCAGtt; reverse: CUGGUGUCUGAAGUUCCCUtt; Ambion, Austin, TX) or luciferase siRNA (Qiagen, Hilden, Germany) (forward: CUUACGCUGAGUACUUCGAtt; reverse: UCGAAGUACUCAGCGUAAGtt) before nucleofection with nucleofactor I (Amaxa). Cells were then immediately removed and incubated overnight with 1 mL prewarmed monocyte nucleofactor medium containing 2 mM glutamine and 10% FBS. Cells were then resuspended into complete RPMI medium and treated with appropriate cytokines to induce their differentiation into macrophages or dendritic cells.
Immunoprecipitation
Cells (100 × 106) were lysed in 1 mL lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 10% glycerol, complete protease inhibitor mixture [CPIM], Roche, Indianapolis, IN) for 30 minutes on ice. After centrifugation at 14 000g at 4°C for 30 minutes, supernatants were precleared during 2 hours at 4°C in the presence of 30 μL mixed Sepharose 6B (Sigma Aldrich) and protein G (Amersham, Freiburg, Germany). After centrifugation at 1000g for 3 minutes the supernatant was incubated with anti–caspase-8 antibody (0.2 μg/mL) or anti-FLIPL antibody (10 μg/mL) at 4°C for 20 hours in the presence of 40 μL mixed Sepharose. The precipitates were washed 4 times in lysis buffer and analyzed by immunoblotting.
Cell lysates and immunoblotting
Whole-cell lysates were prepared as previously described.15 Whole-cell lysates or lipid raft samples or immunoprecipitation samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and electroblotted to nitrocellulose membrane (Schleicher and Schuell, Keene, NH). After incubation for 2 hours at RT by 8% nonfat milk in Tris-buffered saline (TBS)–0.1% Tween-20, membranes were incubated overnight with the primary antibody diluted in TBS-milk-Tween, washed, incubated with the secondary antibody for 30 minutes at RT, and washed again before analysis with a chemiluminescence detection kit (Amersham).
EMSA
For electrophoretic mobility shift assay (EMSA), nuclear fractions were obtained by incubating the cells in lysis buffer (10 mM HEPES, pH 7.8, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.6% NP-40) in the presence of CPIM. Cell lysate was centrifuged at 1200g for 10 minutes and the pellet was washed once in lysis buffer and then resuspended in a buffer containing 20 mM HEPES, pH 7.8, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, in the presence of CPIM for 30 minutes on ice. Nuclear extracts were cleared by centrifugation at 20 000g for 30 minutes, then 5 μg was incubated with 100 000 cpm γ32P-end labeled NF-κB (5′-AGTTGAGGGGCTTTCCCAGGC-3′) consensus oligonucleotide (Promega, Madison, WI) in a reaction buffer containing 5 μL HNB (0.5 M sucrose, 15 mM Tris, pH 7.5, 60 mM KCl, 0.25 mM EDTA, pH 8, 0.125 mM EGTA, pH 5, 0.15 mM spermin, 0.5 mM spermidine, 1 mM DTT), 2 μL MgSp (10 mM MgCl2, 80 mM spermidine), 1.5 μL NaPi (10 mM NaPi, 1 mM EDTA), 10 mM DTT, and 0.2 μg poly(dI-dC). After 30 minutes, the DNA-protein complexes were separated from free oligonucleotides by electrophoresis in a 4% polyacrylamide gel and detected by a PhosphorImager.
Results
Caspase-8 is activated upstream of caspase-9 and caspase-3 in monocytic cells undergoing macrophagic differentiation
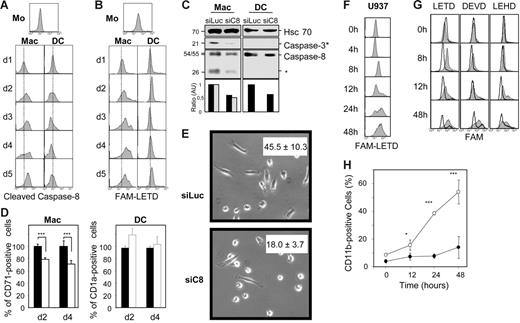
We have shown previously that several caspases were activated during M-CSF–induced differentiation of human peripheral-blood monocytes into macrophages as well as TPA-induced differentiation of U937 monocytic cells.15 The present study aimed to determine how this differentiation-associated proteolytic cascade was initiated. Because caspase-8 deletion targeted in bone marrow progenitors resulted in arrest of monocyte differentiation into macrophages,16 we focused on this enzyme. Using a flow cytometry assay, we detected the time-dependent accumulation of cleaved caspase-8 in human monocytes purified from healthy donor peripheral blood and exposed to M-CSF to trigger their differentiation into macrophages. Conversely, no accumulation of cleaved caspase-8 could be detected in peripheral-blood monocytes exposed to GM-CSF and IL-4 to induce their differentiation into dendritic cells (Figure 1A). The appearance of these active fragments in the cytoplasm was independent of any apoptotic feature15 (and data not shown). The specific activation of caspase-8 associated with macrophagic differentiation of monocytes was further suggested by using the fluorochrome inhibitor FAM-LETD-fmk (Figure 1B). siRNA-mediated down-regulation of caspase-8 expression in peripheral-blood monocytes inhibited caspase-3 cleavage (Figure 1C) and negatively interfered with their differentiation into macrophages on M-CSF exposure, as indicated by studying the expression of CD71 at the cell surface (Figure 1D) and the percentage of cells with a fibroblast-like shape (Figure 1E), without affecting their differentiation into dendritic cells on treatment with GM-CSF and IL-4, as measured by CD1a expression (Figure 1D). Similarly, the macrophagic differentiation of U937 human leukemic cells exposed to 20 nM TPA was associated with a time-dependent increase in FAM-LETD-fmk cleavage activity (Figure 1F) and cleaved caspase-8 (not shown). In this cell line, stable overexpression of a mutated caspase-8 that behaves as a dominant-negative mutant and delays the appearance of a FAM-LETD-fmk cleavage activity (Figure 1G) inhibited the TPA-induced differentiation process, as indicated by studying the time-dependent appearance of the differentiation marker CD11b (Figure 1H), morphologic changes, and cell adhesion to the culture flask (not shown). This construct also prevented the differentiation-associated activation of other caspases such as those that cleave FAM-DEVD (mainly caspase-3) and FAM-LEHD (mainly caspase-9) peptides (Figure 1G). Similar results were obtained in U937 cells expressing the cowpox virus CrmA protein that inhibits caspase-1 and caspase-8 (not shown). Together these results suggested that caspase-8 was activated upstream of caspase-9 and caspase-3 in the cascade associated with macrophagic differentiation.
Caspase-8 involvement in monocyte differentiation into macrophages. (A-B) Human monocytes (Mo) were purified from healthy donor peripheral blood and exposed for indicated times to 100 ng/mL M-CSF to trigger their macrophagic differentiation (Mac) or 100 ng/mL GM-CSF plus 10 ng/mL IL-4 and 50 μM β-mercaptoethanol for inducing their differentiation into dendritic cells (DC) before flow cytometry analysis of (A) cleaved caspase-8 expression and (B) FAM-LETD cleavage activity. (C-E) Monocytes were transfected with either Luciferase (siLuc) or caspase-8 siRNA (siC8). (C) Expression of caspase-8 and cleavage fragments and cleaved caspase-3 were analyzed by immunoblotting. Hsc70 indicates loading control. Molecular weights are indicated in kDa. The asterisk indicates cleavage fragments. Caspase-8 (▪) and cleavage fragments (⊡) were normalized to Hsc70 expression and represented as fold decreases. (D) Expression of CD71 or CD1a was studied by fluorescence-activated cell sorting (FACS) analysis. Results are normalized to values obtained in cells transfected with luciferase siRNA; ▪, siLuc; □, siC8. (E) Percentages of cells with a fibroblastic-like shape, indicating macrophagic differentiation, as observed microscopically (mean ± SD of 3 measurements). (F) U937 cells were treated with 20 nM TPA for indicated times before flow cytometry analysis of FAM-LETD cleavage activity. (G-H) U937 cells were stably transfected with an empty vector or a vector encoding a caspase-8 dominant-negative mutant (C8-DN) before flow cytometry analysis of (G) FAM-LETD, FAM-DEVD, and FAM-LEHD cleavage activities (gray curves, control vector; white curves, C8-DN) and (H) CD11b expression at the cell surface (○, control vector; •, C8-DN). One representative of at least 3 independent experiments is shown or mean ± SD of 3 independent experiments. *P < .05; ***P < .005.
Caspase-8 involvement in monocyte differentiation into macrophages. (A-B) Human monocytes (Mo) were purified from healthy donor peripheral blood and exposed for indicated times to 100 ng/mL M-CSF to trigger their macrophagic differentiation (Mac) or 100 ng/mL GM-CSF plus 10 ng/mL IL-4 and 50 μM β-mercaptoethanol for inducing their differentiation into dendritic cells (DC) before flow cytometry analysis of (A) cleaved caspase-8 expression and (B) FAM-LETD cleavage activity. (C-E) Monocytes were transfected with either Luciferase (siLuc) or caspase-8 siRNA (siC8). (C) Expression of caspase-8 and cleavage fragments and cleaved caspase-3 were analyzed by immunoblotting. Hsc70 indicates loading control. Molecular weights are indicated in kDa. The asterisk indicates cleavage fragments. Caspase-8 (▪) and cleavage fragments (⊡) were normalized to Hsc70 expression and represented as fold decreases. (D) Expression of CD71 or CD1a was studied by fluorescence-activated cell sorting (FACS) analysis. Results are normalized to values obtained in cells transfected with luciferase siRNA; ▪, siLuc; □, siC8. (E) Percentages of cells with a fibroblastic-like shape, indicating macrophagic differentiation, as observed microscopically (mean ± SD of 3 measurements). (F) U937 cells were treated with 20 nM TPA for indicated times before flow cytometry analysis of FAM-LETD cleavage activity. (G-H) U937 cells were stably transfected with an empty vector or a vector encoding a caspase-8 dominant-negative mutant (C8-DN) before flow cytometry analysis of (G) FAM-LETD, FAM-DEVD, and FAM-LEHD cleavage activities (gray curves, control vector; white curves, C8-DN) and (H) CD11b expression at the cell surface (○, control vector; •, C8-DN). One representative of at least 3 independent experiments is shown or mean ± SD of 3 independent experiments. *P < .05; ***P < .005.
Caspase-8 associates with FADD, RIP1, and FLIP isoforms along with macrophagic differentiation
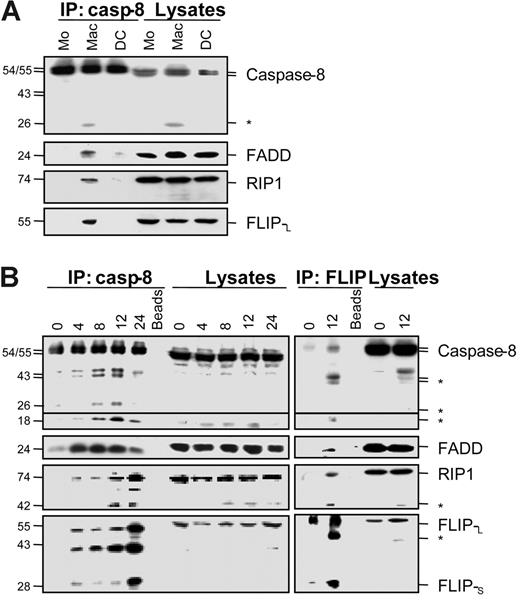
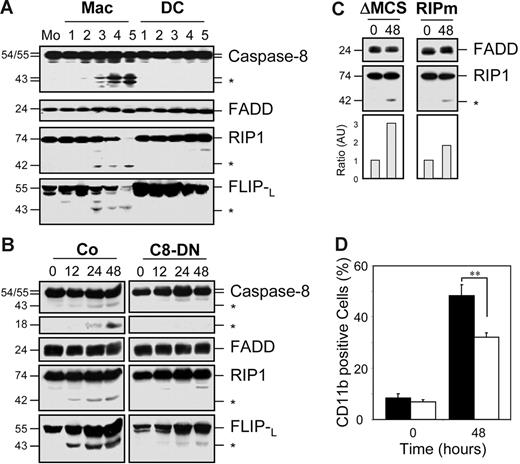
Caspase-8 activation associated with the differentiation of monocytes into macrophages was further indicated by immunoblot analyses showing the appearance of a 26-kDa caspase-8 fragment in monocytes exposed to M-CSF for 2 days. This fragment was not detected in monocytes exposed for the same time to GM-CSF and IL-4 nor in untreated cells (Figure 2A, cell lysates). Immunoprecipitation of caspase-8 in these cell extracts demonstrated that this enzyme associated with the adaptor molecule FADD, the serine/threonine kinase RIP1, and FLIPL in monocytes exposed to M-CSF (Figure 2A). These observations were further confirmed in U937 cells exposed to TPA showing the time-dependent appearance of caspase-8 cleavage fragments and the association of caspase-8 with FADD, RIP1, and the 2 isoforms of FLIP (Figure 2B). In this model, both immunoprecipitation and Western blots showed that the differentiation process was also associated with the time-dependent appearance of caspase-8 active fragments, as well as RIP1 and FLIPL fragments similar to those generated by caspase-mediated cleavage.18-20 The use of anti-FLIP antibody for immunoprecipitation confirmed the interaction of FLIP with caspase-8, FADD, and RIP1 in cells undergoing macrophagic differentiation (Figure 2B).
Caspase-8 association with FADD, RIP1, and FLIP proteins in cells undergoing macrophagic differentiation. (A) Peripheral-blood monocytes (Mo) were treated for 2 days as in Figure 1 to induce their differentiation into macrophages (Mac) or dendritic cells (DC) before lysis. These lysates were used for immunoblotting before (Lysates) or after (IP:casp-8) immunoprecipitation with an anti–caspase-8 antibody. (B) U937 cells were exposed to 20 nM TPA for indicated times (hours) before analysis as in panel A or using an anti-FLIP antibody (IP:FLIP) for immunoprecipitation. Molecular weights are in kilodaltons. The asterisk indicates cleavage products. Beads are a negative control without antibody for immunoprecipitation. One representative of at least 3 independent experiments is shown.
Caspase-8 association with FADD, RIP1, and FLIP proteins in cells undergoing macrophagic differentiation. (A) Peripheral-blood monocytes (Mo) were treated for 2 days as in Figure 1 to induce their differentiation into macrophages (Mac) or dendritic cells (DC) before lysis. These lysates were used for immunoblotting before (Lysates) or after (IP:casp-8) immunoprecipitation with an anti–caspase-8 antibody. (B) U937 cells were exposed to 20 nM TPA for indicated times (hours) before analysis as in panel A or using an anti-FLIP antibody (IP:FLIP) for immunoprecipitation. Molecular weights are in kilodaltons. The asterisk indicates cleavage products. Beads are a negative control without antibody for immunoprecipitation. One representative of at least 3 independent experiments is shown.
Death receptors are not associated with caspase-8 in cells undergoing macrophagic differentiation
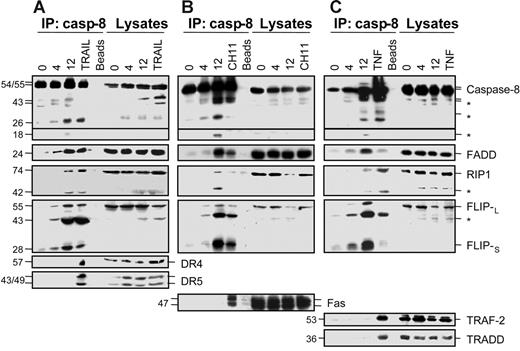
We then analyzed whether caspase-8 activation and the recruitment of FADD, RIP1, and FLIP isoforms could involve a death receptor. Blocking antibodies targeting TNF-α, TNFR1, Fas, and TRAIL did not affect TPA-induced differentiation in U937 cells15 (and data not shown). Because signaling through death receptors can occur in a ligand-independent manner,21 we checked the effect of differentiation on the expression of death receptors at the cell surface and their distribution in the plasma membrane lipid rafts. M-CSF–induced macrophagic differentiation of monocytes and TPA-induced differentiation of U937 cells (not shown) did not significantly modify the expression of Fas, TRAIL receptor DR4, and TRAIL decoy receptors DcR1 and DcR2 at the cell surface. The only change identified in death receptor expression along macrophagic differentiation was an increase in DR5 expression at the cell surface. TPA treatment did not induce any redistribution of the death receptors Fas and DR5 in plasma membrane rafts. Similarly, neither FADD nor RIP1 nor procaspase-8 was significantly redistributed in the rafts along the TPA-induced differentiation of U937 cells (see Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
When analyzed by immunoblot in cell lysates, DR5 protein level slightly increased, whereas expression of DR4, Fas, and the adaptor molecules TRAF2 and TRADD involved in TNFR1-mediated apoptosis18 remained unchanged in U937 cells undergoing TPA-induced differentiation (Figure 3). Coimmunoprecipitation experiments using an anti–caspase-8 antibody were repeated in U937 cells undergoing TPA-induced differentiation and compared to those performed in U937 cells exposed to either TRAIL (Figure 3A) or CH11 anti-Fas agonistic antibody (Figure 3B) or TNF-α (Figure 3C). All these treatments induced interaction of caspase-8 with specific receptor (TRAIL and Fas pathways) or adaptor (TNF pathway) molecules (Figure 3) as well as with FADD and FLIP isoforms. In response to TRAIL and TNF-α, caspase-8 also interacted with RIP1 (Figure 3). In U937 cells exposed to TPA, these experiments confirmed the recruitment of FADD, RIP1, and FLIP isoforms, whereas neither DR4 nor DR5 nor Fas nor TRAF2 nor TRADD was associated with caspase-8 (Figure 3A-C).
Caspase-8 does not associate with death receptors in cells undergoing macrophagic differentiation. U937 cells were exposed to 20 nM TPA for indicated times (hours) before lysis. These lysates were used for immunoblotting before (Lysates) or after (IP:casp-8) immunoprecipitation with an anti–caspase-8 antibody. As positive controls, U937 cells were treated with 500 ng/mL TRAIL for 30 minutes (A) or 100 ng/mL CH11 Fas antibody plus 0.8 μg/mL CHX for 30 minutes (B) or 500 ng/mL of TNF-α for 3 hours (C). Molecular weights are in kilodaltons. The asterisk indicates cleavage products. Beads are negative controls without antibody for immunoprecipitation. One representative experiment is shown.
Caspase-8 does not associate with death receptors in cells undergoing macrophagic differentiation. U937 cells were exposed to 20 nM TPA for indicated times (hours) before lysis. These lysates were used for immunoblotting before (Lysates) or after (IP:casp-8) immunoprecipitation with an anti–caspase-8 antibody. As positive controls, U937 cells were treated with 500 ng/mL TRAIL for 30 minutes (A) or 100 ng/mL CH11 Fas antibody plus 0.8 μg/mL CHX for 30 minutes (B) or 500 ng/mL of TNF-α for 3 hours (C). Molecular weights are in kilodaltons. The asterisk indicates cleavage products. Beads are negative controls without antibody for immunoprecipitation. One representative experiment is shown.
A role for FADD in macrophagic differentiation
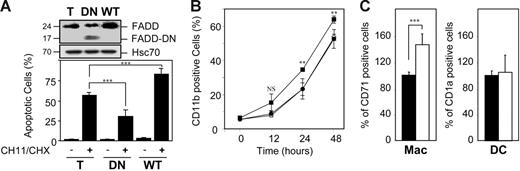
We then used a lentiviral construct to introduce either wild-type or a FADD dominant-negative mutant deleted of most of the death effector domain in U937 cells and their expression was checked by Western blotting (Figure 4A). Wild-type FADD expression increased the apoptotic response of U937 cells to CH11 anti-Fas antibody, whereas the dominant-negative construct protected the cells from Fas-mediated cell death (Figure 4A). Wild-type FADD overexpression enhanced the kinetics of TPA-induced macrophagic differentiation in U937 cells, as indicated by studying the expression of CD11b at the cell surface (Figure 4B) and the cell adhesion to plastic flasks (not shown). FADD wild-type overexpression in peripheral-blood monocytes also accelerated their differentiation into macrophages on M-CSF exposure, without affecting their differentiation into dendritic cells on exposure to GM-CSF and IL-4 (Figure 4C). On the other hand, expression of the FADD-mutated construct did not affect TPA-induced differentiation in U937 cells (Figure 4B). Altogether, these observations suggested that the role of FADD in macrophagic differentiation was independent of its interaction with death receptors.
Influence of FADD constructs on the differentiation pathway. (A-B) U937 cells were infected with lentiviral constructs encoding either EGFP alone (T or ○) or wild-type FADD (WT or ▪) or mutated FADD in which the death effector domain has been partly deleted (DN or •), then selected on EGFP expression by cell sorting. (A) Expression of FADD (24 kDa) and FADD-DN (17 kDa) was analyzed by immunoblotting. HSC70 indicates loading control. Molecular weights are in kilodaltons. Cells were either left untreated or treated with 100 ng/mL CH11 Fas antibody plus 0.8 μg/mL CHX for 6 hours before measuring the percentage of apoptotic cells after Hoechst 33352 staining of the nuclear chromatin. (B) Cells were exposed to 20 nM TPA for indicated times before measuring CD11b expression by flow cytometry. (C) Monocytes were infected with lentiviral constructs encoding either EGFP alone (▪) or wild-type FADD (□) and treated for 4 days as in Figure 1 to induce their differentiation into macrophages (Mac) or dendritic cells (DC) before flow cytometry analysis of the cell-surface expression of CD71 or CD1a. Results are normalized to EGFP-infected monocytes. Results are the mean ± SD of at least 3 independent experiments. **P < .01; ***P < .005; NS indicates not significant.
Influence of FADD constructs on the differentiation pathway. (A-B) U937 cells were infected with lentiviral constructs encoding either EGFP alone (T or ○) or wild-type FADD (WT or ▪) or mutated FADD in which the death effector domain has been partly deleted (DN or •), then selected on EGFP expression by cell sorting. (A) Expression of FADD (24 kDa) and FADD-DN (17 kDa) was analyzed by immunoblotting. HSC70 indicates loading control. Molecular weights are in kilodaltons. Cells were either left untreated or treated with 100 ng/mL CH11 Fas antibody plus 0.8 μg/mL CHX for 6 hours before measuring the percentage of apoptotic cells after Hoechst 33352 staining of the nuclear chromatin. (B) Cells were exposed to 20 nM TPA for indicated times before measuring CD11b expression by flow cytometry. (C) Monocytes were infected with lentiviral constructs encoding either EGFP alone (▪) or wild-type FADD (□) and treated for 4 days as in Figure 1 to induce their differentiation into macrophages (Mac) or dendritic cells (DC) before flow cytometry analysis of the cell-surface expression of CD71 or CD1a. Results are normalized to EGFP-infected monocytes. Results are the mean ± SD of at least 3 independent experiments. **P < .01; ***P < .005; NS indicates not significant.
A role for caspase-8–induced RIP1 cleavage in macrophagic differentiation
We then analyzed whether RIP1 cleavage played a role in macrophagic differentiation. This cleavage as well as the proteolytic cleavage of FLIPL were specifically observed in monocytes induced to differentiate into macrophages under M-CSF exposure (Figure 5A). Cleavage of RIP1 and c-FLIPL requires caspase-8 activation because these cleavages are strongly delayed in U937 cells stably transfected with a caspase-8 dominant-negative mutant exposed to TPA (Figure 5B). Overexpression of the baculovirus p35 or the cowpox virus CrmA caspase inhibitory proteins also prevented RIP1 and c-FLIPL cleavage in TPA-treated U937 cells (data not shown). In these cells, lentivirus-mediated expression of a mutated RIP1 construct, in which the caspase-mediated cleavage site had been modified, decreased the cleavage of RIP1 (Figure 5C) and negatively interfered with the differentiation process (Figure 5D).
Caspase-mediated RIP1 and FLIP cleavage in cells undergoing macrophagic differentiation. (A) Peripheral-blood monocytes (Mo) were treated as in Figure 1 for indicated times (days) to trigger their differentiation into macrophages (Mac) or dendritic cells (DC) before analyzing the expression of indicated proteins by immunoblotting. (B) U937 cells transfected with either an empty vector (Co) or a mutated caspase-8–expressing vector (C8-DN) were treated with 20 nM TPA for indicated times (hours) before immunoblot analysis of indicated proteins. (C-D) U937 cells transduced with a lentivirus encoding an empty vector (ΔMCS or ▪) or RIP1 mutated on the caspase cleavage site (RIPm or □) were exposed to TPA as in panel B before (C) immunoblot analysis of indicated proteins. RIP1 cleavage fragment was normalized to FADD expression (fold increase). *Cleavage fragment. (D) CD11b expression measured by FACS analysis. One representative of at least 3 independent experiments is shown, or mean ± SD of 3 independent experiments. **P < .01.
Caspase-mediated RIP1 and FLIP cleavage in cells undergoing macrophagic differentiation. (A) Peripheral-blood monocytes (Mo) were treated as in Figure 1 for indicated times (days) to trigger their differentiation into macrophages (Mac) or dendritic cells (DC) before analyzing the expression of indicated proteins by immunoblotting. (B) U937 cells transfected with either an empty vector (Co) or a mutated caspase-8–expressing vector (C8-DN) were treated with 20 nM TPA for indicated times (hours) before immunoblot analysis of indicated proteins. (C-D) U937 cells transduced with a lentivirus encoding an empty vector (ΔMCS or ▪) or RIP1 mutated on the caspase cleavage site (RIPm or □) were exposed to TPA as in panel B before (C) immunoblot analysis of indicated proteins. RIP1 cleavage fragment was normalized to FADD expression (fold increase). *Cleavage fragment. (D) CD11b expression measured by FACS analysis. One representative of at least 3 independent experiments is shown, or mean ± SD of 3 independent experiments. **P < .01.
Caspase-mediated RIP1 cleavage is required for NF-κB activity modulation
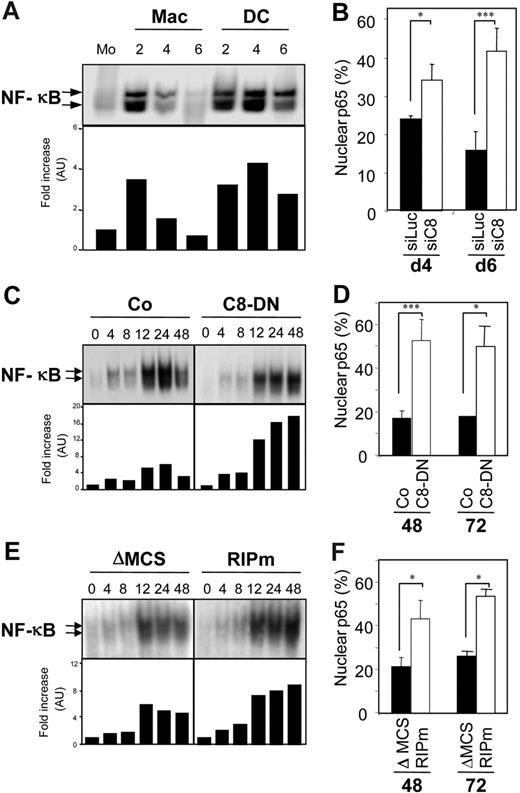
The family of Rel/NF-κB transcription factors plays an essential role in macrophagic and myeloid dendritic differentiation,22,23 and caspase-mediated cleavage of RIP1 negatively regulated NF-κB activation.19 By using an EMSA, we observed, in accordance with previously published observations,22 that NF-κB DNA-binding activity was transiently increased in monocytes undergoing macrophagic differentiation, whereas this increase was sustained in cells undergoing dendritic differentiation (Figure 6A). These results were further confirmed by showing that nuclear expression of p65 was higher at day 4 and day 6 of M-CSF treatment when caspase-8 was down-regulated by specific siRNA (Figure 6B). A transient activation of NF-κB activity was also observed in U937 cells treated with TPA, reaching a maximum 24 hours after the beginning of TPA treatment, then decreasing at 48 hours (Figure 6C,E). The supershift obtained with antibodies against p50 and p65/RelA NF-κB subunits confirmed the previously reported activation of a p50/p65 NF-κB complex (data not shown). In U937 cells stably expressing the caspase-8 dominant-negative mutant, NF-κB activation did not decrease after 24 hours of TPA treatment (Figure 6C). Similar results were obtained in cells expressing CrmA and p35 (not shown), and in those expressing a noncleavable RIP1 (Figure 6E). These results were also confirmed by studying p65 nuclear expression. Expression of caspase-8 dominant-negative (Figure 6D) or noncleavable RIP1 (Figure 6F) mutants in U937 cells induced a higher nuclear expression of p65 along the macrophagic differentiation as compared to corresponding empty vectors. Altogether, these results suggested that caspase-8–mediated RIP1 cleavage was required to down-regulate NF-κB activation during the macrophagic differentiation pathway.
Caspase-8 activity is required for modulation of NF-κB activation. (A) Primary monocytes (Mo) were exposed for indicated times (days) to 100 ng/mL M-CSF to trigger their macrophagic differentiation (Mac) or 100 ng/mL GM-CSF plus 10 ng/mL IL-4 for inducing their differentiation into dendritic cells (DC). NF-κB DNA-binding activity was assessed by EMSA (top panel) and the results were analyzed using a PhosphorImager and expressed relative to untreated cells (bottom panel). (B) Monocytes transfected with either luciferase (siLuc) or caspase-8 siRNA (siC8) were treated for 4 days (d4) or 6 days (d6) with M-CSF before quantifying nuclear p65+ cells by immunofluorescence. (C-D) U937 cells stably transfected with a caspase-8 dominant-negative mutant (C8-DN) or the corresponding empty vector (Co) were exposed to 20 nM TPA for indicated times (hours). (C) NF-κB DNA-binding activity was assessed by EMSA as in panel A. (D) The percentage of cells with nuclear p65 was determined by immunofluorescence as in panel B. (E-F) U937 cells stably transfected with a lentivirus encoding RIP1 mutated on its caspase cleavage site (RIPm) or the corresponding empty vector (ΔMCS) were exposed to 20 nM TPA for the indicated times (hours). (E) NF-κB DNA-binding activity was assessed by EMSA as in panel A. (F) The percentage of cells with nuclear p65 was determined by immunofluorescence as in panel B; one representative of at least 3 independent experiments, or mean ± SD of 3 independent experiments. * P < .05; ***P < .005.
Caspase-8 activity is required for modulation of NF-κB activation. (A) Primary monocytes (Mo) were exposed for indicated times (days) to 100 ng/mL M-CSF to trigger their macrophagic differentiation (Mac) or 100 ng/mL GM-CSF plus 10 ng/mL IL-4 for inducing their differentiation into dendritic cells (DC). NF-κB DNA-binding activity was assessed by EMSA (top panel) and the results were analyzed using a PhosphorImager and expressed relative to untreated cells (bottom panel). (B) Monocytes transfected with either luciferase (siLuc) or caspase-8 siRNA (siC8) were treated for 4 days (d4) or 6 days (d6) with M-CSF before quantifying nuclear p65+ cells by immunofluorescence. (C-D) U937 cells stably transfected with a caspase-8 dominant-negative mutant (C8-DN) or the corresponding empty vector (Co) were exposed to 20 nM TPA for indicated times (hours). (C) NF-κB DNA-binding activity was assessed by EMSA as in panel A. (D) The percentage of cells with nuclear p65 was determined by immunofluorescence as in panel B. (E-F) U937 cells stably transfected with a lentivirus encoding RIP1 mutated on its caspase cleavage site (RIPm) or the corresponding empty vector (ΔMCS) were exposed to 20 nM TPA for the indicated times (hours). (E) NF-κB DNA-binding activity was assessed by EMSA as in panel A. (F) The percentage of cells with nuclear p65 was determined by immunofluorescence as in panel B; one representative of at least 3 independent experiments, or mean ± SD of 3 independent experiments. * P < .05; ***P < .005.
Discussion
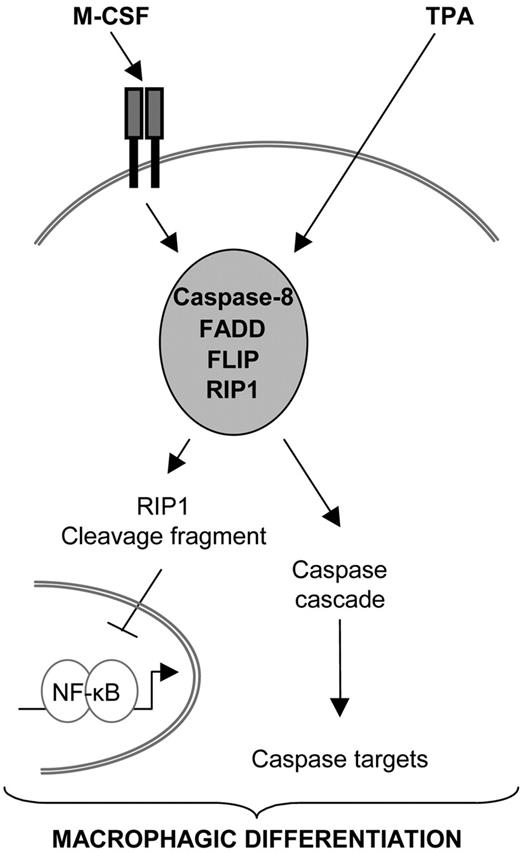
Previous studies had suggested a role for caspase-8 in cytokine-induced proliferation of hematopoietic progenitors8 and in M-CSF–induced differentiation of myeloid precursors.16 However, the molecular pathway leading to caspase-8 activation in myeloid precursors on cytokine exposure remained unidentified. The present study suggests that, in the proteolytic caspase cascade that mediates monocyte differentiation into macrophages, caspase-8 occupies an apical position. Its activation involves a multiprotein complex that includes the adaptor molecule FADD, the serine/threonine kinase RIP1, and the 2 FLIP isoforms. Overexpression of FADD accelerates the differentiation process, whereas a FADD mutant that prevents Fas-mediated apoptosis does not interfere with macrophagic differentiation. Together with the lack of detection of any interaction of caspase-8 with a death receptor, these observations suggest that death receptors, which in other circumstances are required for inducing FADD interaction with caspase-8,11 may not play a role in differentiation-associated caspase-8 activation. Active caspase-8 appears to cleave RIP1 that, in turn, down-regulates NF-κB, which may favor the macrophagic differentiation process. A schematic model of this pathway is proposed in Figure 7.
Proposed role for caspase-8 in monocytes undergoing macrophagic differentiation. Exposure of primary monocytes to M-CSF or U937 cells to TPA triggers caspase-8 interaction with FADD, FLIP, and RIP1. Caspase-8 is activated in this multimolecular platform, which induces the cleavage of RIP1. In turn, RIP1 cleavage fragment could down-regulate NF-κB activity. Caspase-8 may also activate downstream caspases that could further contribute to the differentiation pathway by cleaving other cellular targets.
Proposed role for caspase-8 in monocytes undergoing macrophagic differentiation. Exposure of primary monocytes to M-CSF or U937 cells to TPA triggers caspase-8 interaction with FADD, FLIP, and RIP1. Caspase-8 is activated in this multimolecular platform, which induces the cleavage of RIP1. In turn, RIP1 cleavage fragment could down-regulate NF-κB activity. Caspase-8 may also activate downstream caspases that could further contribute to the differentiation pathway by cleaving other cellular targets.
How caspases are activated to fulfill nonapoptotic functions remains poorly known.14 Here, we show that caspase-8, which, in the setting of apoptosis, behaves as either an upstream initiator24 or a downstream, effector25 enzyme, is activated upstream of caspase-9 and caspase-3 in monocytes undergoing macrophagic differentiation. The observation that caspase-8 behaves as an upstream enzyme in this setting is in accordance with the specific inhibition of macrophagic differentiation observed in a mouse model of conditional caspase-8 gene knockout in bone marrow cells.16 In mice, deletion of Bid gene, which encodes a BH3-only protein connecting death receptor-mediated activation of caspase-8 to the mitochondrial pathway of cell death,15 provokes accumulation of monocytes in the peripheral-blood and spleen, thus mimicking a human disease known as chronic myelomonocytic leukemia.26 Whereas this observation was related to a default in monocyte death, another interpretation is a decreased differentiation leading to cell accumulation. Interestingly, all the differentiation-associated caspase cascades may not be initiated in the same way, as caspase-8 is not activated in erythroid cells whose differentiation involves caspase-3 activation.13,27
Caspase-8 recruitment by adaptor molecules induces its oligomerization and its subsequent activation, either through its full processing or not.28 On death receptor engagement, caspase-8 is recruited by FADD through interaction between the death effector domain of each protein. Here, we show that, in monocytic cells undergoing macrophagic differentiation, caspase-8 interacts with FADD. Whereas a strong overexpression of FADD induces cell death29 (and S.C., E.S., unpublished data, June 2006), a limited increase in wild-type FADD expression, as obtained by lentivirus-mediated transfer,17 accelerates the macrophagic differentiation of monocytic cells. Interestingly, a FADD-DN construct lacking most of the death effector domain competitively inhibits Fas-mediated caspase-8 activation and cell death17 without affecting the macrophagic differentiation. In this latter setting, the FADD mutant may not compete with the formation of the caspase-8–activating platform, suggesting that FADD-mediated recruitment of caspase-8 is independent of its death domain–mediated interaction with a death receptor. Accordingly we did not identify any interaction of caspase-8 with Fas, DR4, and DR5, nor with TRADD and TRAF2, 2 molecules that are involved in the formation of the soluble complex II that recruits caspase-8 on TNF-mediated death signaling.18
A critical question raised by the participation of active caspases in nonapoptotic functions is how cells survive after activating these enzymes.14 Various explanations have been proposed such as activation of a protective signaling pathway involving NF-κB.30,31 Activation of this transcription factor is transient when monocytes differentiate into macrophages and sustained when they differentiate into dendritic cells.22,23 Because siRNA-mediated caspase-8 down-regulation into macrophages or expression of a dominant-negative mutant of caspase-8, or CrmA, or p35 in U937 cells maintains a high level of nuclear p65 and NF-κB activity, caspase-8 activation may be necessary to regulate NF-κB activity along the macrophagic differentiation pathway.
Our data suggest that NF-κB regulation involves caspase-8–mediated RIP1 cleavage. A dual role was assigned to RIP1 in cell death, that is, survival through NF-κB activation and apoptosis induction. The kinase can be cleaved by caspase-8 at Asp324 to generate a C-terminal cleavage product that blocks NF-κB activation, thus promoting cell death,19,32,33 its subcellular localization can change, depending on the molecular complex it is associated with18 and the protein can be degraded by the proteasome machinery through ubiquitination by the A20 NF-κB inhibitory molecule.34,35 The present study shows that caspase-8–mediated cleavage of RIP1 modulates NF-κB activity in cells undergoing macrophagic differentiation. Interestingly, another RIP kinase, RIP4, was recently shown to be involved in keratinocyte differentiation as RIP-4 null mice demonstrate a severely affected keratinocyte differentiation associated with a complete absence of cornified layer.36 Similar to RIP1, the pro-NF-κB activity of RIP4 is inhibited by caspase-mediated cleavage, most probably generating a dominant-negative C-terminal, ankyrin-containing fragment.37 Because caspases have been reported to play a role in keratinocyte differentiation, one could speculate that caspase-mediated cleavage of RIP4 may be one of the consequences of caspase activation that is required for appropriate differentiation in these cells.
c-FLIPL, which acts either as a competitive inhibitor for caspase-8 recruitment38 or associates with and activates caspase-839 in death receptor–mediated signaling, was shown to stimulate T-cell proliferation by associating with RIP1 and caspase-8 when overexpressed in T cells. In turn, caspase-8 cleaves c-FLIPL to a p43 form that recruits more efficiently RIP1 than full-length c-FLIPL20,40 and activates rather than inhibits NF-κB.41 Thus, caspase-8–mediated FLIPL cleavage identified in monocytes undergoing macrophagic differentiation could potentially contribute to NF-κB activity regulation together with RIP1 fragment. Caspase-8 also activates downstream caspases such as caspase-9 and caspase-3 whose targets may also play a role in the macrophagic differentiation pathway.42 Further studies will determine the connection between caspase-8 and other caspases and the deregulation of these pathways in human monocytic diseases such as chronic myelomonocytic leukemia. If a default in caspase activation is observed in M-CSF–treated monocytes from patients with this later disease, then a limited activation of these enzymes could possibly restore differentiation, thus preventing monocyte accumulation.
Authorship
Conflict-of-interest disclosure: the authors declare no competing financial interests.
Correspondence: Eric Solary, INSERM U517, IFR 100, Faculty of Medicine, 7 boulevard Jeanne d'Arc, 21079 Dijon cedex, France; e-mail: esolary@u-bourgogne.fr.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This study was supported by a special grant from the Ligue Nationale Contre le Cancer (E.S.) (équipe labélisée) and a grant from the Direction Régionale de la Recherche Clinique de l'Assistance Publique des Hôpitaux de Paris (M.F.). S.L., S.C., and R.F. were supported by the Ligue Nationale Contre le Cancer, C.R. by the Association pour la Recherche contre le Cancer, and E.G. by the INSERM.
We thank Arlette Hammann, Anabelle Legrand, André Bouchot. and Jean Christophe Deschemin for technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal