Abstract
Innate recognition of bacteria is a key step in the activation of inflammation and coagulation, and it is dependent on pathogen-associated molecular pattern (PAMP) ligation to Toll-like receptors (TLRs) and CD14. The dominant receptors activated when cells encounter a whole bacterium, which express several PAMPs, are poorly defined. Herein, we have stimulated various human cells with prototypic Gram-negative and Gram-positive bacteria. Receptor-dependent responses to whole bacteria were assessed using both TLR-transfected cells and specific monoclonal antibodies against TLRs, MD-2, and CD14. Enterobacteria-activated leukocytes and endothelial cells in a TLR4/MD-2–dependent manner, most likely via lipopolysaccharide (LPS). TLR2 activation was observed with a high bacterial inoculum, and in epithelial cells expressing TLR2 but not TLR4. Pseudomonas aeruginosa stimulated cells by both TLR2 and TLR4/MD-2. Gram-positive bacteria activated cells only at high concentrations, in a partially TLR2-dependent but TLR4/MD-2–independent manner. Either TLR or CD14 neutralization blocked activation to all bacterial strains tested with the exception of some Gram-positive strains in whole blood in which partial inhibition was noted. This study identifies dominant TLRs involved in responses to whole bacteria. It also validates the concept that host cell activation by bacterial pathogens can be therapeutically reduced by anti-TLR4, -TLR2, and -CD14 mAbs.
Introduction
The recognition of bacteria as nonself agents by mammalian cells is key in mounting an innate response to control infection. Several bacterial antigens, known as pathogen-associated molecular patterns (PAMPs), are sensed as “nonself” molecules by host immune cells, using receptors of the innate immune system. PAMPs are, for the majority, cell-wall molecules. Some PAMPs are found in both Gram-negative and Gram-positive bacteria, (lipopeptides, peptidoglycan, flagellin, and bacterial DNA). Others are specific either for Gram-negative bacteria (LPS), Gram-positive bacteria (lipoteichoic acid), or mycobacteria (lipoarabinomannan).
Toll-like receptors (TLRs) belong to a family of leucine-rich repeat (LRR) proteins that are either expressed at the cell surface or in intracellular compartments.1 They recognize and bind a wide variety of bacterial PAMPs, including LPS (typically recognized by TLR4, although some LPS species can be recognized by TLR2), lipopeptides (TLR1, TLR2, TLR6), lipoarabinomannan and lipoteichoic acid (TLR2 and other TLRs), flagellin (TLR5), and bacterial DNA (TLR9).2-4 The ligation of PAMPs to TLRs induces a profound reprogramming of immune cells and activates several innate pathways such as inflammation, coagulation, and cell death. Some of the nucleotide-binding oligomerization domain (NOD) LRR proteins, such as Nod1 and Nod2, are also intracellular sensors for bacterial molecules and mediate inflammation on recognition of specific motifs in bacterial peptidoglycan.5 Other receptors also recognize PAMPs and are essential for the phagocytosis of bacteria, as well as the clearance of bacterial molecules.6
CD14, a GPI-anchored glycoprotein, binds PAMPs, associates with TLRs, and markedly enhances cell responsiveness to several bacterial molecules.7,8 It is, for example, an essential component of the LPS receptor along with TLR4 and MD-2.9-11 Plasma proteins such as LPS-binding protein (LBP) and the soluble forms of CD14 and MD-2 also play an important role in the recognition of PAMPs and present them efficiently to receptors of the innate immune system.8,12-14
Most studies addressing TLR specificity have been performed using purified bacterial ligands and transfected cells. Only a few studies have investigated PAMPs and their receptors in mediating human cell activation using whole bacteria.15-21 Bacteria are essentially sensed by human immune cells as whole microorganisms. PAMPs can also stimulate immune cells in the form of microparticules and soluble mediators released from bacteria. The predominant PAMPs stimulating TLRs are largely unknown in the context of a whole bacterium. The present work defines the receptors predominantly mediating cell activation to bacterial PAMPs when presented to cells as “whole bacteria.” Using either TLR-transfected cells or human leukocytes, endothelial cells, or epithelial cells treated with specific anti-TLRs, –MD-2, and -CD14 monoclonal antibodies (mAbs), we have determined the contribution of cell-surface TLRs and their accessory proteins toward cellular responses to prototypic Gram-negative and Gram-positive bacterial strains responsible for human septic shock.
Materials and methods
Reagents
α–Human CD14 MAb 28C5 (mouse IgG1 κ) and α–human TLR2 mAb T2.5 (mouse IgG1 κ) were previously described.22,23 T2.5 was purchased from eBioscience (San Diego, CA). α-Human TLR4 mAb 15C1 (mouse IgG1 κ) and the α–human MD-2 MAb 18H10 (mouse IgG2b κ) were generated as previously described.24 Isotype controls were purchased from Becton Dickinson ([BD], Franklin Lakes, NJ). All mAbs were tested and found to have no effect on cell viability using TruCOUNT tubes (BD; data not shown). Ultrapure Escherichia coli K12LCD25 LPS and Pseudomonas aeruginosa 10 LPS were purchased from InvivoGen (San Diego, CA) and Sigma Aldrich (St Louis, MO), respectively. P aeruginosa 10 LPS was subsequently phenol extracted to remove contaminating lipoproteins as previously described.25 Flagellin and PAM3CSK4 were purchased from InvivoGen. E coli (wild type), P aeruginosa, Klebsiella pneumoniae, Listeria monocytogenes, Bacillus subtilis, and Staphylococcus aureus were isolates from patients who developed clinically relevant infection (a gift from Peter Rohner, University Hospital of Geneva). Gram-negative bacteria were flagellated. E coli strain K12W3110 was from Pierre Genevaux, University of Geneva. Bacteria were grown at 37°C overnight, centrifuged, resuspended to obtain an OD660 of 1 (corresponding to 5 × 108 cfu/mL) or higher, and killed either by heating (30 minutes at 100°C) or by gentamicin (50 μg/mL) treatment. Inactivation was verified by plating on LB agar to check for the absence of colonies. Human umbilical vein endothelial cells (HUVECs) were from Dr F. Mach, Centre Médicale Universitaire (CMU), Geneva. Human bronchial BEAS-2B and embryonic kidney HEK 293 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Stably transfected cell lines expressing either human TLR4/MD-2 or TLR2 were generated as described previously,24 with expression levels similar in both cell lines.
Flow cytometry
For HUVECs and BEAS 2B cells, 106 cells/mL was incubated in PBS, 1% BSA, and either 10 μg/mL of the appropriate antibody or an isotope control. Cells were washed and incubated in the same buffer with APC-conjugated goat anti–mouse IgG antibody (1:250 dilution; Molecular Probes, Leiden, The Netherlands). Cells were analyzed using a fluorescence-activated cell sorting (FACS)Calibur flow cytometer (BD) in the FL-4 channel. For circulating leukocytes, the appropriate antibodies (10 μg/mL) were added to human whole blood. Following 2 washes, cells were incubated with secondary antibody (APC-conjugated anti–mouse IgG diluted 1:250) containing 100 μg/mL human IgG (Sigma) to prevent Fc-mediated interactions. Red blood cells were lysed, and the remaining cells were washed twice and were analyzed. Different leukocyte populations were distinguished on the basis of forward and side scatter.
HEK 293, HUVEC, and BEAS 2B assays
Cells were plated in 96-well plates at 6 × 104 cells/well. HUVECs were cultured on gelatin-coated plates (Sigma Aldrich). The medium was removed on the day of the experiment, and 30 μL medium containing 6% heat-inactivated human plasma was added. mAbs were diluted in 30 μL basal medium to the appropriate concentration and added to the cells for 1 hour at 37°C. PAMPs or heat-inactivated bacteria were diluted in 30 μL medium, added to the cells, and left to incubate for 21 hours at 37°C. IL-6 (HUVEC and BEAS 2B) or IL-8 (HEK 293) secretion in the culture supernatant was monitored by enzyme-linked immunoabsorbent assay (ELISA; Endogen, Boston, MA).
Whole-blood assays
Fresh heparinated blood from healthy volunteers was obtained by venipuncture and diluted 1:2 with RPMI 1640. Blood was plated at 60 μL/well in a 96-well plate and incubated for 15 minutes at 37°C. mAbs were diluted in RPMI 1640 (30 μL final volume) and added to the blood. One hour later, 30 μL of either heat-inactivated bacteria, E coli K12 LPS (4 ng/mL final), P aeruginosa LPS (100 ng/mL), PAM3CSK4 (100 ng/mL final), or flagellin (300 ng/mL final) was added to the blood and incubated for 6 hours. These concentrations corresponded to the maximal response observed with these agonists. IL-6, IL-8, or TNF-α content was measured by ELISA (Endogen). In experiments with heat-killed bacteria, mAbs were added to whole blood, diluted 1:4 with RPMI to a final concentration of 5 μg/mL 1 hour prior to the addition of bacteria. Stimulation time was 6 hours for Gram-negative bacteria and 24 hours for Gram-positive bacteria. IL-6 levels were measured by ELISA. In some experiments, TNF-α, IL-8, and IP-10 were measured in parallel by ELISA (R&D Systems, Minneapolis, MN) or multiple cytokine concentrations (TNF-α, IL-1β, IL-6, IL-8, IL-10, IL-12p70) were measured by cytometric bead array (CBA; BD). All blood samples used in this study were obtained anonymously, and there is no reference to the donors within. IRB approval/informed consent was therefore not required by our institution.
Results
TLR2, TLR4, MD-2, and CD14 expression profiles
To predict cellular responsiveness to different TLR ligands, we examined cell-surface expression of TLR2, TLR4, and the TLR accessory molecules MD-2 and CD14 using specific mAbs.
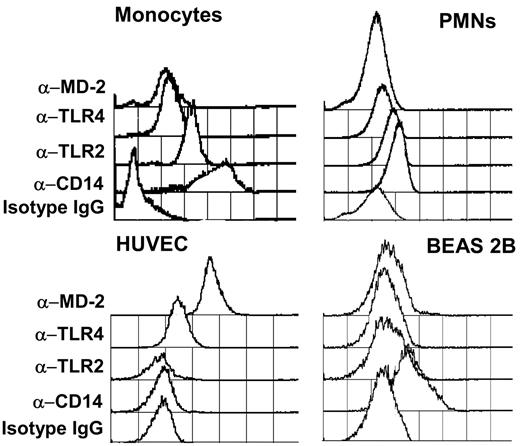
Human blood cell populations were distinguished by size and granularity. Monocytes expressed significant surface levels of TLR4, MD-2, and TLR2 and a high surface level of CD14. Polymorphonuclear neutrophils (PMNs) were positive for CD14 and weak for TLR2, TLR4, and MD-2 (Figure 1, top panel). Lymphocytes did not express detectable levels of any of the proteins tested (data not shown).
Flow cytometry quantification of the expression of CD14, TLR2, TLR4, and MD-2 on human monocytes, polymorphonuclear neutrophils, human umbilical vein endothelial cells (HUVECs), and human bronchial BEAS 2B cells. Antibodies (5 μg/mL) were incubated with cells, and binding was revealed with an APC-conjugated goat anti–mouse IgG secondary antibody.
Flow cytometry quantification of the expression of CD14, TLR2, TLR4, and MD-2 on human monocytes, polymorphonuclear neutrophils, human umbilical vein endothelial cells (HUVECs), and human bronchial BEAS 2B cells. Antibodies (5 μg/mL) were incubated with cells, and binding was revealed with an APC-conjugated goat anti–mouse IgG secondary antibody.
HUVECs were positive for TLR4 and MD-2 expression but negative for TLR2 and CD14, whereas BEAS 2B cells expressed weak but detectable levels of TLR2 and a significant level of CD14 (Figure 1, bottom panel). Higher expression levels of TLR2 were observed in permeabilized BEAS 2B cells, suggesting a partially intracellular expression of TLR2 in these cells (data not shown).
Induction of proinflammatory cytokine production in transfected HEK 293 cells with bacterial PAMPs
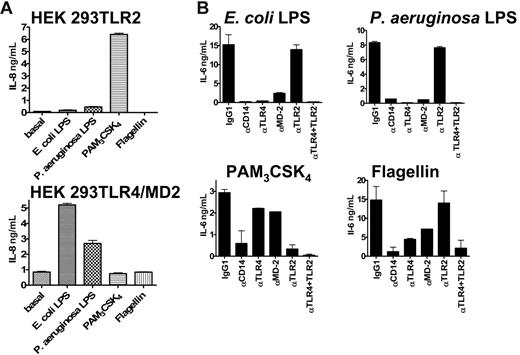
To determine the activity and specificity of a selection of PAMPs relevant to this study on TLR2 and TLR4/MD-2, we used HEK 293 cells transfected with either TLR2 or TLR4/MD-2 (Figure 2A). Cells transfected with TLR2 were strongly stimulated to produce IL-8 in the presence of PAM3CSK4 and weakly with P aeruginosa LPS as previously shown.26 No stimulation was detected in the presence of E coli LPS and flagellin. Cells transfected with TLR4/MD-2 were stimulated by both E coli and P aeruginosa LPS. No response was detected with PAM3CSK4 and flagellin.
PAMP activity on various human cell types. (A) Activity of PAMPs on HEK 293 cells transfected with either TLR2 (top) or TLR4/MD-2 (bottom). Concentration of PAMPs were as follows: E coli LPS (100 ng/mL) and P aeruginosa LPS (1 μg/mL), PAM3CSK4 (200 ng/mL), and flagellin (1 μg/mL). (B) Inhibitory effects of mAbs specific to CD14, TLR4, MD-2, and TLR2 (10 μg/mL) on the activation of human whole blood (IL-6 secretion) by PAMPs. E coli LPS (4 ng/mL) and P aeruginosa LPS (100 ng/mL), the synthetic PAM3CSK4 lipopeptide (100 ng/mL) and flagellin (300 ng/mL). Duplicates were performed for all conditions tested. This represents 1 representative experiment of 3. Error bars show ± standard error of the mean (SEM).
PAMP activity on various human cell types. (A) Activity of PAMPs on HEK 293 cells transfected with either TLR2 (top) or TLR4/MD-2 (bottom). Concentration of PAMPs were as follows: E coli LPS (100 ng/mL) and P aeruginosa LPS (1 μg/mL), PAM3CSK4 (200 ng/mL), and flagellin (1 μg/mL). (B) Inhibitory effects of mAbs specific to CD14, TLR4, MD-2, and TLR2 (10 μg/mL) on the activation of human whole blood (IL-6 secretion) by PAMPs. E coli LPS (4 ng/mL) and P aeruginosa LPS (100 ng/mL), the synthetic PAM3CSK4 lipopeptide (100 ng/mL) and flagellin (300 ng/mL). Duplicates were performed for all conditions tested. This represents 1 representative experiment of 3. Error bars show ± standard error of the mean (SEM).
Inhibition of proinflammatory cytokine production in human whole blood with blocking TLR2, TLR4, MD-2, and CD14 mAbs
We investigated the specificity and potency of neutralizing TLR2, TLR4, MD-2, and CD14 mAbs on human whole blood stimulated with E coli LPS,27 Pseudomonas LPS,26 PAM3CSK4,28 and flagellin.29 Results are shown in Figure 2B.
E coli LPS–induced IL-6 production was inhibited by anti-TLR4, –MD-2, and -CD14 mAbs. The anti-TLR2 MAb showed no significant neutralizing capacity. These results are in accordance with a role for TLR4, MD-2, and CD14 in the recognition of E coli LPS. Pseudomonas LPS–induced IL-6 production was also completely inhibited by anti-TLR4, –MD-2, and -CD14 mAbs.
PAM3CSK4 is a synthetic tripalmitoylated lipopeptide that mimics the acylated amino terminus of bacterial lipoproteins and signals through TLR2. As expected, PAM3CSK4-dependent induction of IL-6 in human whole blood was greatly diminished with TLR2 blocking mAbs. In addition, anti-CD14 treatment also strongly inhibited this effect, suggesting a role for this adaptor protein in the recognition of the molecule by TLR2. PAM3CSK4 also stimulated BEAS 2B cells in a TLR2-dependent manner, but it had no effect on HUVECs (data not shown).
Flagellin is the major component of the bacterial flagella, which confers motility on a wide range of bacterial species. It has been shown to signal via TLR5, possibly as a heterodimer with TLR4.30 In human whole blood, flagellin strongly induced the production of IL-6. Anti-TLR2 MAb treatment had no effect on this induction. In contrast, anti-TLR4, MD-2, and CD14 mAbs could significantly inhibit this response, suggesting a role for these proteins in the innate immune response to flagellin, as previously reported.30 Flagellin also activated HUVECs but not BEAS 2B cells (data not shown).
Together, these results confirm the specificity of the anti-TLR2, TLR4, MD-2, and anti-CD14 blocking mAbs used in this study and demonstrate their ability to neutralize the activity of their respective targets.
Induction of proinflammatory cytokine production in transfected HEK 293 cells
To demonstrate the ability of different bacterial strains to stimulate proinflammatory cytokine production, we treated cells types with increasing doses of heat-inactivated E coli (wild-type and K12 strains), P aeruginosa, K pneumoniae, L monocytogenes, B subtilis, and S aureus.
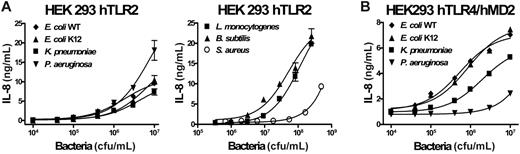
We examined the stimulation of either TLR2 or TLR4/MD-2–transfected HEK 293 cells (Figure 3). In TLR2-transfected HEK 293 cells, all Gram-negative strains tested induced a detectable response from 5 × 105 cfu/mL upward with a maximum IL-8 response at the highest dose tested (107 cfu/mL). P aeruginosa produced the highest response of all strains tested. These results confirm, as previously reported, the presence of PAMPs such as lipoproteins capable of stimulating TLR2 within the Gram-negative bacterial strains tested in this study. Gram-positive bacteria (> 106 cfu/mL) induced TLR2-dependent cell activation, except for S aureus which required higher concentrations. HEK 293 cells transfected with TLR4 and MD-2 responded robustly to both E coli strains and to K pneumoniae, starting at 5 × 104 cfu/mL. The cells responded to P aeruginosa with a weak but significant response observed above 5 × 106 cfu/mL. The cells were unresponsive to Gram-positive bacteria (highest concentration tested, 5 × 108 bacteria/mL, data not shown). Untransfected HEK 293 cells were not activated by any of the bacterial strains tested (data not shown).
Dose-dependent induction of IL-8 secretion by heat-inactivated bacteria in HEK 293 cells stably transfected with human TLR2 or human TLR4/MD-2. TLR2 is shown in panel A or human TLR4/MD-2 in panel B. Duplicates were performed for all conditions tested. This represents 1 of 2 representative experiments. Error bars show ± SEM.
Dose-dependent induction of IL-8 secretion by heat-inactivated bacteria in HEK 293 cells stably transfected with human TLR2 or human TLR4/MD-2. TLR2 is shown in panel A or human TLR4/MD-2 in panel B. Duplicates were performed for all conditions tested. This represents 1 of 2 representative experiments. Error bars show ± SEM.
Induction of proinflammatory cytokine production in HUVECs and BEAS 2B cells
To understand the contribution made by different TLRs to the cellular response to whole bacteria, we exposed different and relevant human cell types to heat-inactivated bacteria (clinical isolates) following a preincubation with blocking TLR2, TLR4, MD-2, and CD14 mAbs.
We first investigated the responsiveness of HUVECs to LPS and PAM3CSK4 (Figure 4A). As expected from the expression profile of TLR2 and TLR4/MD-2 seen in Figure 1, LPS stimulated HUVECs, whereas PAM3CSK4 had no effect. We next investigated HUVEC stimulation with whole bacteria. IL-6 production was induced by doses above 104 cfu/mL for E coli and K pneumoniae (Figure 4B). P aeruginosa was a poor inducer of IL-6 production, with a very weak response seen only at the higher doses tested (> 106cfu/mL). S aureus failed to activate HUVECs at any of the doses tested. We next tested the effect of blocking TLR2 and TLR4 signaling on IL-6 production in HUVECs treated with either E coli WT or K pneumoniae at 107 cfu/mL. As expected (HUVECs are CD14-, TLR2-, TLR4/MD-2+), activation was inhibited by anti-TLR4 and anti–MD-2 blocking mAbs, whereas TLR2 blockade had no effect (Figure 4C). The inhibition seen with the anti-CD14 MAb is due to the blockade of soluble CD14 present in human plasma, necessary for the activation of endothelial cells in response to LPS.23 IL-8 production by HUVECs also mirrored IL-6 levels on bacterial stimulation (data not shown).
TLR-specific activation of HUVECs. (A) Induction of IL-6 production in HUVECs with either E coli LPS (100 ng/mL) or PAM3CSK4 (200 ng/mL). (B) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (107/mL E coli and K pneumoniae). Error bars show ± SEM. (C) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by E coli and K pneumoniae (5 different experiments). Because the magnitude of IL-6 production was variable among different HUVEC donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
TLR-specific activation of HUVECs. (A) Induction of IL-6 production in HUVECs with either E coli LPS (100 ng/mL) or PAM3CSK4 (200 ng/mL). (B) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (107/mL E coli and K pneumoniae). Error bars show ± SEM. (C) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by E coli and K pneumoniae (5 different experiments). Because the magnitude of IL-6 production was variable among different HUVEC donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
BEAS 2B cells responded to PAM3CSK4 but not E coli LPS (Figure 5A), in accordance with a detectable level of TLR2 expression on the cell surface and an absence of TLR4/MD-2 as seen in Figure 1. We next investigated BEAS 2B stimulation with whole bacteria. All Gram-negative bacteria tested above 106 cfu/mL. The maximal response for each strain was observed at 107 cfu/mL, the highest dose tested (Figure 5B). Despite low expression levels of TLRs (Figure 1D), BEAS 2B cells responded in a relatively robust fashion. Neither TLR4 nor MD-2 blockade affected IL-6 production. In contrast, TLR2 blockade strongly diminished IL-6 production, suggesting that TLR2 is functionally responsible for bacterial stimulation of this epithelial cell line. The contribution of CD14 to the recognition of enterobacteria and the subsequent BEAS 2B cell activation was also demonstrated using the anti-CD14 blocking mAb (Figure 5C). Slightly higher Gram-positive bacterial concentrations were required to activate BEAS 2B cells (Figure 6A). Again, cell activation was essentially TLR2 and CD14 dependent for all strains tested (Figure 6B). These results suggest that human airway epithelial cells sense and are activated by both Gram-negative and Gram-positive bacteria via ligands activating TLR2 but not TLR4/MD-2 and that CD14 also plays a role in the recognition and activation process of these cells.
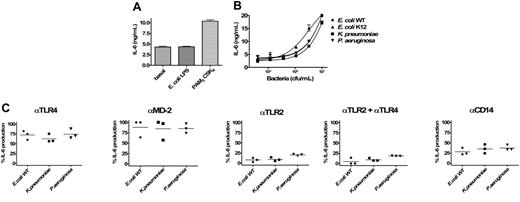
TLR-specific activation of BEAS 2B cells by Gram-negative bacteria. (A) Induction of IL-6 production in BEAS 2B with either E coli LPS (100 ng/mL) or PAM3CSK4 (200 ng/mL). (B) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa) in BEAS 2B cells. Error bars show ± SEM. (C) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by 107/mL Gram-negative bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa; 3 different experiments). Because the magnitude of IL-6 production was variable between different experiments, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
TLR-specific activation of BEAS 2B cells by Gram-negative bacteria. (A) Induction of IL-6 production in BEAS 2B with either E coli LPS (100 ng/mL) or PAM3CSK4 (200 ng/mL). (B) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa) in BEAS 2B cells. Error bars show ± SEM. (C) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by 107/mL Gram-negative bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa; 3 different experiments). Because the magnitude of IL-6 production was variable between different experiments, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
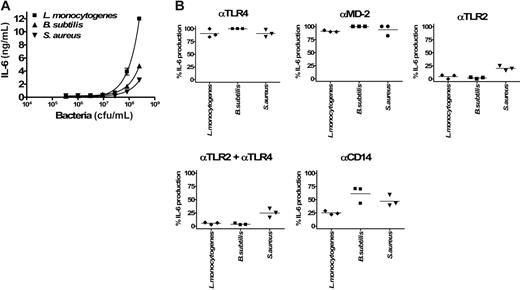
TLR-specific activation of BEAS-2B cells by Gram-positive bacteria. (A) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (L monocytogenes, B subtilis, and S aureus) in BEAS 2B cells. Error bars show ± SEM. (B) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by 2 × 108/mL Gram-positive bacteria (L monocytogenes, B subtilis, and S aureus; 3 different experiments). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
TLR-specific activation of BEAS-2B cells by Gram-positive bacteria. (A) Dose-dependent induction of IL-6 secretion by heat-inactivated bacteria (L monocytogenes, B subtilis, and S aureus) in BEAS 2B cells. Error bars show ± SEM. (B) Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs in the activation of cells by 2 × 108/mL Gram-positive bacteria (L monocytogenes, B subtilis, and S aureus; 3 different experiments). Duplicates were performed for all conditions tested. Horizontal lines represent the mean.
Induction of proinflammatory cytokine production in whole blood
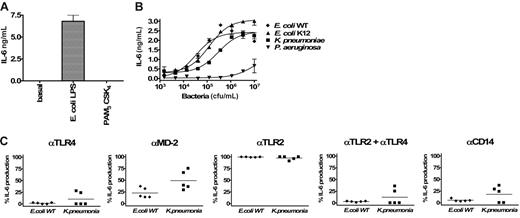
In human whole blood, a robust response to all Gram-negative bacterial strains was observed (Figure 7A). IL-6 induction was observed with as little as 2 × 103 cfu/mL. At 104 cfu/mL (Figure 7B), TLR4 blockade strongly reduced the IL-6 production induced by E coli and K pneumoniae and partially reduced the levels seen with P aeruginosa. MD-2 blockade largely mirrored TLR4 blockade, with the exception that P aeruginosa–induced IL-6 production was more potently inhibited. TLR2 blockade had no significant effect on E coli– and K pneumoniae–induced IL-6 production and partially inhibited P aeruginosa. Combined TLR2/TLR4 blockade strongly inhibited all Gram-negative responses. Anti-CD14 treatment largely resembled the anti-TLR2/TLR4 cotreatment. Together, these results suggest that, at this chosen dose of bacteria, PAMPs signaling via TLR4/MD-2 are largely dominant in inducing an immune response to E coli and K pneumoniae. P aeruginosa appears to signal via TLR4/MD-2 and TLR2-containing receptor complexes. The inhibition pattern seen with the anti-CD14 MAb implies a role for this protein in the recognition of ligands signaling via both TLR2 and TLR4.
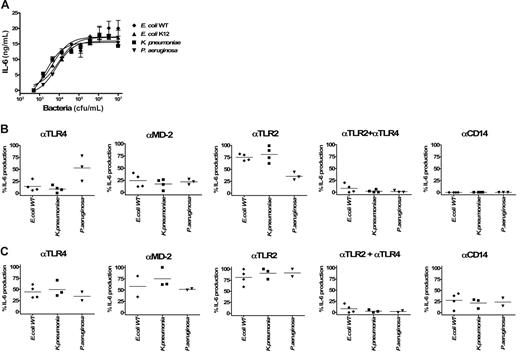
TLR-specific activation of whole blood by Gram-negative bacteria. Dose-dependent induction of IL-6 secretion by heat-inactivated Gram-negative bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa) in human whole blood (A). Error bars show ± SEM. Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs on the activation of cells by E coli WT, E coli K12, K pneumoniae, and P aeruginosa. Human whole blood was stimulated with 104 (B) or 106 bacteria (C). The different points per condition represent experiments performed with different healthy blood donors. Because the magnitude of IL-6 production was variable among different donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Horizontal bars represent the mean.
TLR-specific activation of whole blood by Gram-negative bacteria. Dose-dependent induction of IL-6 secretion by heat-inactivated Gram-negative bacteria (E coli WT, E coli K12, K pneumoniae, and P aeruginosa) in human whole blood (A). Error bars show ± SEM. Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs on the activation of cells by E coli WT, E coli K12, K pneumoniae, and P aeruginosa. Human whole blood was stimulated with 104 (B) or 106 bacteria (C). The different points per condition represent experiments performed with different healthy blood donors. Because the magnitude of IL-6 production was variable among different donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Horizontal bars represent the mean.
At 106 cfu/mL in human whole blood (Figure 7C), E coli–, K pneumoniae–, and P aeruginosa–induced IL-6 production could be partially inhibited by TLR4 and MD-2 blockade, whereas TLR2 blockade was ineffective. Interestingly, anti-TLR2 and anti-TLR4 combination treatment resulted in a complete inhibition of E coli– and K pneumoniae–induced IL-6 production. This suggests that at higher doses of bacteria, TLR2 ligands can stimulate IL-6 production in the absence of TLR4 signaling. Induction of IL-6 by 106 cfu/mL P aeruginosa was partially inhibited by TLR4/MD-2 blockade. No effect was seen by TLR2 blockade, but a strong inhibition was seen following cotreatment with anti-TLR2 and TLR4 mAbs, reflecting what was observed at 104 cfu/mL. The pattern of CD14 blockade at 106 cfu/mL with E coli, K pneumoniae, or P aeruginosa also mirrored that seen at 104 cfu/mL, although inhibition was generally slightly less potent. The inhibitory effect of antibodies was similar when IL-8 and TNF-α were measured by ELISA (data not shown). Compared with IL-6 levels shown in Figure 7, identical inhibition profiles were observed when multiple cytokines were measured using a cytometric beads assay (TNF-α, IL-1β, IL-6, IL-8, and IL-10 levels in conditioned supernatants from diluted whole blood stimulated with K pneumoniae, data shown in Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). IL-12 was absent in most samples following incubation with Gram-negative bacteria. Levels of IP-10 remained below the level of detection in whole blood stimulated 6 and 24 hours with Gram-negative bacteria.
In general, Gram-positive bacteria required a longer incubation time and higher concentration to stimulated blood leukocytes. Although some activation was observed with Gram-positive bacteria after 6 hours of incubation, a maximal level of activation was noted after 18 to 24 hours. Results of a 24-hour exposure of whole blood to 3 strains of Gram-positive bacteria are shown in Figure 8A. Leukocyte stimulation was detected at 105 cfu/mL for L monocytogenes and B subtilis and at 106 cfu/mL for S aureus, with a response plateau around 107 cfu/mL. Gram-positive bacteria stimulated leukocytes in a TLR2-dependent manner (Figure 8B). The involvement of CD14 was variable, depending both on the bacterial strain and the blood donor (Figure 8B). Compared with IL-6 levels shown in Figure 8, identical inhibition profiles were observed when multiple cytokines were measured using a cytometric beads assay (TNF-α, IL-1β, IL-6, IL-8, IL-10 in conditioned supernatants from diluted whole blood stimulated with L monocytogenes; data shown in Figure S1). Neither IL-12 nor IP-10 was detected in plasma from whole blood incubated 24 hours with Gram-positive bacteria.
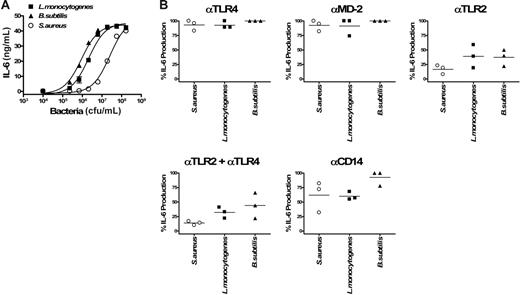
TLR-specific activation of whole blood by Gram-positive bacteria. Dose-dependent induction of IL-6 secretion by heat-inactivated Gram-positive bacteria (L monocytogenes, B subtilis, S aureus) in human whole blood (A). Error bars show ± SEM. Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs on the activation of cells by 106/mL L monocytogenes and B subtilis, and 107/mL S aureus (B). The different points per condition represent experiments performed with 3 different healthy blood donors. Because the magnitude of IL-6 production was variable among different donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Horizontal bars represent the mean.
TLR-specific activation of whole blood by Gram-positive bacteria. Dose-dependent induction of IL-6 secretion by heat-inactivated Gram-positive bacteria (L monocytogenes, B subtilis, S aureus) in human whole blood (A). Error bars show ± SEM. Inhibitory effects of anti-TLR4, anti–MD-2, anti-TLR2, and anti-CD14 mAbs on the activation of cells by 106/mL L monocytogenes and B subtilis, and 107/mL S aureus (B). The different points per condition represent experiments performed with 3 different healthy blood donors. Because the magnitude of IL-6 production was variable among different donors, results are given as the percentage of inhibition of IL-6 release compared with the condition with the isotype control antibody (corresponding to 100% activation). Horizontal bars represent the mean.
Evaluation of heat inactivation versus antibiotic inactivation
We studied the effect of heat inactivation on the integrity of PAMPs and their ability to stimulate innate immune responses by comparing IL-6 production induced by heat-inactivated versus antibiotic-inactivated bacteria. We chose gentamicin because it acts at the level of ribosomal RNA; therefore, structural PAMPs remain intact with this method of inactivation. As shown in Figure S2, bacteria inactivated by both methods retained an equivalent capacity, at a dose of approximately 106 cfu/mL, to stimulate leukocytes. Inhibition of cytokine production using anti-TLR2, TLR4, MD-2, and CD14 was also comparable, suggesting that the relevant TLR ligands retained their potency no matter what the method of inactivation used. Similar observations were made with HUVECs and BEAS 2B cells (data not shown), suggesting that heat-inactivated and antibiotic-inactivated bacteria produced equivalent levels of cellular activation. These results suggest that heat inactivation of bacteria is a valid method to study the stimulation of the innate immune system by TLR agonists derived from bacteria.
Discussion
Understanding the contribution of receptors and accessory proteins involved in the innate recognition of bacteria is essential in the context of potential therapeutic intervention in severe bacterial infections.31,32 The identification of TLRs as pattern recognition receptors (PRRs) was a major step forward in our understanding of how microorganisms, and particularly bacteria, are sensed by the immune system.2 TLRs are differentially expressed depending on cell types. The established function of TLRs is in the recognition of exogenous PAMPs, although recognition of endogenous ligands has also been speculated.33 In addition, intracellular proteins of the NOD family have also been identified as PRR for peptidoglycan degradation products.5
In less than 5 years, microbial ligands for virtually all TLRs, as well as Nod1 and Nod2, have been identified. Biochemical, cellular, and animal studies proving this have principally been carried out using purified or synthetic PAMPs. Although it has been established that cells of the innate immune system can be activated by whole bacteria, little is known about which TLRs are essential or dominant to activate these cells. Indeed, several PAMPs recognized by a variety of TLRs are typically expressed by a given bacterial strain. For example, Gram-negative bacteria are sources of ligands such as LPS, flagellin, lipopeptides, peptidoglycan, and bacterial DNA, all of which can bind receptors of the TLR or NOD family and activate cells. Gram-positive bacteria can at the same time display peptidoglycan, lipoteichoic acid, lipopeptides, bacterial DNA, and flagellin. To add to the complexity of the system, several TLR and NOD proteins are expressed by cells of the innate immune system.
Using a novel panel of blocking monoclonal antibodies, we have identified herein the relevant receptors of the innate immune system that are engaged after encounter with bacterial pathogens. We have studied cell types relevant to the human innate immune system (blood leukocytes, endothelial cells, and epithelial cells) and have exposed them to prototypic Gram-negative and Gram-positive bacterial strains cultured from clinical samples.
Heat-inactivated enterobacteria efficiently stimulated HEK 293 cells expressing the TLR4/MD-2 complex, a finding consistent with the activation of these cells by enterobacterial LPS.27 Roughly 100 times more enterobacteria were necessary to induce a similar level of cell activation in TLR2 transfectants. This suggested that for bacteria such as E coli and K pneumoniae, TLR4/MD-2 ligands were dominant. These bacteria strongly stimulated blood leukocytes and endothelial cells expressing TLR4/MD-2 but poorly stimulated TLR4-negative BEAS 2B cells. Experiments with specific blocking mAbs confirmed unequivocally the dominance of the TLR4/MD-2 complex in leukocytes and endothelial cells stimulated with enterobacteria, being suggestive of a dominant role for LPS in the activation of these cells during infection with such pathogens.34 Note that, despite the presence of lipopeptides in enterobacteria, these ligands, and therefore TLR2, seem to be irrelevant in comparison with TLR4 ligands. Enterobacteria also display flagellin. It has been shown previously that flagellin can stimulate cells through a TLR4/TLR5 pathway, although formal proof for the existence of a TLR4/TLR5 heterodimer is still lacking.30 Using an anti-TLR4 MAb, we confirmed the involvement of TLR4 in cell activation in response to this particular PAMP. Because flagellin did not activate HEK 293 cells transfected with TLR4/MD-2 and was insensitive to polymyxin treatment, a potential LPS contamination is unlikely (G.E., I.D.S., and J.P.; unpublished results, January 2005).
A robust activation of whole blood was noted in response to heat-killed P aeruginosa, which was dependent on both TLR2 and TLR4. It has been reported that P aeruginosa can vary LPS acylation during its biosynthesis. TLR4 classically recognizes hexa-acylated LPS, whereas TLR2 binds tetra- and penta-acylated pseudomonal LPS.35-38 The dependence on both TLR2 and TLR4 of the Pseudomonas tested suggests that this particular clinical strain is capable of making LPS of variable levels of acylation. It is also possible that TLR2 ligands are more potent in Pseudomonas than in other Gram-negative bacteria. The Pseudomonas LPS that we have used in our study after repurification according to the method of Hirschfeld et al25 stimulated cells via a TLR4-dependent pathway, indicating that this LPS was essentially hexa-acylated.
TLR2-dependent stimulation of BEAS 2B epithelial cells was relatively poor when incubated with enterobacteria and P aeruginosa. This is likely to be due to the lack of a significant expression of the TLR4–MD-2 complex in these cells under normal conditions, which physiologically prevents overwhelming inflammation to Gram-negative commensals.39 It has however been shown that under pathologic conditions, lung and gut epithelial cells can express TLR4, which renders them more sensitive to Gram-negative bacteria and LPS.40
It has now been well established that TLR4 ligands activate 2 different pathways. The classical MyD88-dependent pathway, shared by TLR2 and TLR4, results in the production of cytokines such as TNF-α, IL-6, and IL-8. Ligands for TLR4 and TLR3 also activate a distinct TIR domain-containing adaptor-inducing interferon β (IFN-β) (TRIF)–dependent pathway necessary for the production of IP-10, IL-12, and IFN-β. Interestingly, we were unable to generate IP-10 or IL-12 in human leukocytes stimulated with whole bacteria, despite the presence of classical TLR4 ligands such as LPS. An explanation for this observation is that the concomitant presence of TLR-2 ligands in whole bacteria specifically blocks the TRIF pathway. The existence of such a negative cross talk of TLR2 on TLR4's TRIF pathway has recently been unraveled by Re and Strominger41 in dendritic cells, implicating IL-10 as a critical down-regulating factor. We postulate that when circulating leukocytes sense whole bacteria displaying both TLR2 and TLR4 ligands, a prominent MyD-88–dependent response takes place as a result of an IL-10–dependent blockade of the TRIF pathway secondary to TLR2 stimulation. Supporting this, IL-12 could be detected in human leucocytes pretreated with anti-TLR2 neutralizing mAb on stimulation with Gram-negative bacteria (G.E. and I.D.S.; unpublished data, August 2006).
A consistent finding was that higher concentrations of heat-killed Gram-positive bacteria (> 106 cfu/mL) were necessary to detect cell activation in whole blood and BEAS 2B cells, compared with Gram-negative bacteria. In addition, a stimulation time of 24 hours was required to give a signal comparable to that of Gram-negative bacteria, even though a detectable level of stimulation could be seen at 6 hours (data not shown). Our results strongly suggest that the Gram-positive bacteria used in this study stimulate cells mainly via a TLR2-dependent pathway and do not display TLR4 ligands. Of course, other Gram-positive bacteria such as Streptococcus pneumoniae may produce ligands (eg, pneumolysin) capable of inducing a TLR4-dependent response.42 The incomplete inhibition obtained with an anti-TLR2 MAb in whole blood stimulated with L monocytogenes and B subtilis suggests that other PAMPs such as flagellin may participate in leukocyte activation by whole Gram-positive bacteria. Other receptors such as complement receptors may play a role in the innate recognition of S aureus.43 Gram-positive cell wall components are known stimulants for TLR2-expressing cells.44 It was initially believed that S aureus peptidoglycan was the major TLR2 ligand.44 Although this issue is not completely settled,45 it is probable that contaminants, such as lipoteichoic acid, accounted for cell activation through TLR2, rather than peptidoglycan itself.46,47 It is also possible that the peptidoglycan-derived muramyl dipeptide activates cells in a TLR-independent manner, via Nod2, after phagocytosis of S aureus.48 Other staphylococcal products, such as modulin, proteins A and L, as well as exotoxins, are also known to activate cells via mechanisms that do not involve TLRs.49
Although CD14 was the first receptor identified as a PRR more than a decade ago, it remains unclear how it recognizes structurally different bacterial PAMPs.7 Although the anti-CD14 mAb had only a partial inhibitory effect on Gram-positive bacteria-induced cell activation, this antibody invariably abrogated cell activation to all Gram-negative bacterial strains in all cell types tested. It is possible that a different CD14 epitope than that blocked by our mAb is used by Gram-positive cell wall components, as previously suggested.45 Interestingly, CD14 also seems to play a significant role in the innate recognition of Gram-negative bacteria in bronchial BEAS 2B epithelial cells. We also report for the first time that cell stimulation via flagellin is CD14 dependent. We postulate that the flagellin receptor is a heterocomplex comprising CD14, TLR4, and TLR5. The binding of palmitoylated lipopeptides to CD14, mediated by LBP, and the involvement of CD14 in cell activation to these ligands have recently been described.50,51 Together, our results demonstrate that CD14 blockade is efficient and shows broad specificity across various bacteria and cell lines tested.
It is possible that some important bacterial PAMPs may have been denatured during heat inactivation of the bacteria. To address this, experiments were repeated with bacteria killed with gentamicin, an antibiotic that does not influence cell wall structure and integrity. The levels of cell activation, as well as the dependence on TLRs, MD-2, and CD14, were similar when heat-killed or antibiotic-killed bacteria were assayed. We did not report the effect of live bacteria in our assays, but only heat-killed or antibiotic-killed bacteria. In studies not presented here, we found inconsistent results when using live bacteria, resulting from bacterial growth during the experiment and toxic effects on cells. We can therefore not exclude the participation of secreted bacterial molecules in the TLR-dependent and -independent activation of cells. It remained also unclear whether cells were being activated with bacterial surface molecules directly associated with the microorganism, by soluble molecules or by both. Experiments in which we exposed cells to bacteria washed from the growth culture supernatant demonstrated that cells could be activated by whole bacteria and by soluble bacterial molecules (Figure S3). Interestingly, TLR2-expressing BEAS-2B cells were much more sensitive to soluble molecules than were whole bacteria. The opposite was found with Gram-negative bacteria in whole blood. At any rate, the TLR dependence was very similar whether cells saw whole bacteria in the presence or absence of growth culture medium. This suggested that the stimulating soluble molecules are probably the same as those displayed at the surface of bacteria.
Our study has some limitations. First, because of a lack of reagents, we have not investigated the precise contribution of heterodimer-forming TLRs that might be important for cell responses to flagellin, such as TLR5 within a TLR4/TLR5 heterodimer,26 and Gram-positive bacteria such as TLR1 and TLR6 within TLR1/TLR2 and TLR2/TLR6 heterodimers.52 The potential involvement of TLR9 and the role of bacterial CpG DNA were only indirectly appreciated, because of the lack of available blocking reagents. On one hand, complete inhibition of cell activation by Gram-negative bacteria was obtained with anti-TLR4, anti–MD-2, and anti-CD14 mAbs. It is therefore unlikely that TLR9 plays a significant role in the activation induced by these bacteria. On the other hand, TLR2 and CD14 blockade only partially inhibited cell activation to whole Gram-positive bacteria and leaves a doubt as to whether Gram-positive bacterial CpG may be partially responsible for cell activation. In a recent study, Mogensen et al20 showed a partial TLR9 dependence of human leukocyte activation by live streptococci, an effect that was basically lost in heat-killed bacteria. Similarly, a primary role of Nod1 and Nod2 proteins and peptidoglycan-derived molecules in cell activation is certainly unlikely in Gram-negative bacteria-induced cell activation. An enhancing effect can, however, not be excluded. Again, the fact that TLR2 MAb does not completely inhibit cell activation by Gram-positive bacteria makes the involvement of NODs (and their peptidoglycan-derived ligands) possible as a cofactor.53 Second, we have tested a restricted panel of bacterial strains, although enterobacteria, Pseudomonas, and staphylococci are together causative microorganisms in greater than 70% of severe bacterial infections. Finally, subsequent studies investigating a wider range of primary cells and cell lines which could be deemed relevant to the overall innate immune response could give information complimentary to the current study.
In conclusion, we have shown that whole Gram-negative and Gram-positive bacteria activate human leukocytes, endothelial, and epithelial cells through TLRs and CD14. The blockade of these receptors with specific mAbs efficiently abrogates cell activation in response to their exposure to whole bacteria. TLR4 is the main TLR involved in cell activation by Gram-negative bacteria. In the absence of TLR4 or at high concentration of Gram-negative bacteria, TLR2 is also activated. Higher concentrations of Gram-positive bacteria are needed to activate cells in a TLR2-dependent fashion. This activation appears to be completely TLR4/MD-2 independent. These results largely confirm studies performed in genetically modified mice54 and demonstrate that antibody blockade of CD14 and TLRs can potentially decrease excessive and detrimental inflammation in response to bacteria, making it an attractive therapeutic avenue to explore in patients with severe sepsis and septic shock. Indeed, during the initial phase of shock, the blockade of TLR-dependent hyperactivation of cells without interfering with CD14-dependent internalization of bacteria and bacterial products34 might be beneficial.55 Interfering with the initial inflammatory response with TLR antagonists carries also the theoretical risk of bacterial overgrowth, a problem that should be minimized by an appropriate antibiotic therapy.
Authorship
Contribution: G.E. and I.D.-S. designed and performed the research and wrote the paper; B.D. contributed vital new reagents; and J.P. designed the research and wrote the paper.
Conflict-of-interest disclosure: Two of the authors (G.E. and B.D.) are employed by NovImmune SA, Geneva, Switzerland, whose potential products, anti–MD-2 (18H10) and -TLR4 (15C1) mAbs, were studied in this work. I.D.-S. and J.P. declare no competing financial interests.
G.E. and I.D.-S. contributed equally to the work.
Correspondence: Greg Elson, NovImmune S.A., 64, av. de la Roseraie, 1211 Geneva 5, Switzerland; e-mail: gelson@novimmune.com.
The online version of this article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We would like to thank Dr T. Martin and Dr C. Frevert, Seattle, WA, for helpful and informative discussions. We also thank Dr P. Tissières for his help in collecting blood and for fruitful discussions, Valery Moine for assistance with the CBA assay, and Dr Y. Dean for assistance with the TruCOUNT assay.
This work was supported by research funding from NovImmune SA and the Swiss National Foundation for Scientific Research (grant 3200B0-105770) (J.P.).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal