Abstract
Hematopoietic stem cell/hematopoietic progenitor cell (HSC/HPC) homing to specific microenvironmental niches involves interactions between multiple receptor ligand pairs. Although CXCL12/CXCR4 plays a central role in these events, CXCR4 regulators that provide the specificity for such cells to lodge and be retained in particular niches are poorly defined. Here, we provide evidence that the sialomucin endolyn (CD164), an adhesion receptor that regulates the adhesion of CD34+ cells to bone marrow stroma and the recruitment of CD34+CD38lo/− cells into cycle, associates with CXCR4. The class II 103B2 monoclonal antibody, which binds the CD164 N-linked glycan-dependent epitope or CD164 knockdown by RNA interference, significantly inhibits the migration of CD133+ HPCs toward CXCL12 in vitro. On presentation of CXCL12 on fibronectin, CD164 associates with CXCR4, an interaction that temporally follows the association of CXCR4 with the integrins VLA-4 and VLA-5. This coincides with PKC-ζ and Akt signaling through the CXCR4 receptor, which was disrupted on the loss of CD164 though MAPK signaling was unaffected. We therefore demonstrate a novel association among 3 distinct families of cell-surface receptors that regulate cell migratory responses and identify a new role for CD164. We propose that this lends specificity to the homing and lodgment of these cells within the bone marrow niche.
Introduction
An important determinant of successful stem cell transplantation is the ability of transplanted cells to mobilize, home, migrate, and efficiently engraft and repair damaged tissues with functional cells. It is now standard practice to mobilize CD133+CD34+ cells from bone marrow into the circulation by administering G-CSF or to collect such cells from umbilical cord blood and to use these for transplantation.1–4 Once administered, these cells home to the bone marrow, where they engraft in specific stromal or vascular niches.5 Homing begins with the chemoattraction of CD133+ cells to the bone marrow and progresses to their extravasation across bone marrow sinusoidal endothelium and their transmigration through the basal lamina to specific hematopoietic stem cell/hematopoietic progenitor cell (HSC/HPC) niches.5–16
The chemokine CXCL12 plays a central role as a chemoattractant for CD133+ HSCs/HPCs, regulating their motility, homing to, and retention, survival, and proliferation in the bone marrow (for reviews, see Burger and Kipps,12 Broxmeyer et al,17 Kollet et al,18 and Kim et al19 ). CXCL12 is the ligand for CXCR4, a 7-transmembrane G-protein–coupled receptor (for reviews, see Burger and Kipps12 and Zou et al14 ). Although lethal in the perinatal period, identical and significant reductions in B-lymphopoiesis and myelopoiesis have been described during fetal development in mice deficient in CXCL12 and CXCR4.20,21 B-lymphopoiesis and myelopoiesis are both regulated by the interactions of their precursors, with specific microenvironmental niches within the bone marrow (for reviews, see Nilsson and Simmons,5 Ma et al,21 Tokoyoda et al,22 and Chan and Watt23 ), and studies indicate that CXCL12 is required for the retention of these precursors within, and the metastatic spread of both myeloid and lymphoid leukemic cells to, these specific niches.12,21,24 Antagonists of CXCR4, when administered alone or with G-CSF, have been used in clinical trials to mobilize HSCs/HPCs from the bone marrow into the circulation,25–27 and the presence of functional CXCR4 receptors on CD34+ cells has been shown to predict time to engraftment after HSC/HPC transplantation into patients with hematologic disorders. Cooperation and cross talk between CXCL12/CXCR4, other cell surface receptors, and signaling molecules are required not only to fine-tune HSC/HPC mobilization, migration, and homing but also to define the specificity of their retention in microenvironmental niches. For example, this chemokine/receptor axis promotes the intramedullary migration of HSCs/HPCs by activation of the integrins very late antigen-4 (VLA-4) and VLA-5.10,28,29
Here, we provide evidence that endolyn (CD164) is a key molecule regulating the CXCL12-mediated migration of human umbilical cord blood (UCB) CD133+ cells, and we demonstrate that CD164 acts as a component of a CXCR4 complex. CD164 is a type 1 integral transmembrane sialomucin (for reviews, see Watt and Chan,30 Chan et al,31 Zannettino et al,32 Watt et al,33 Watt et al,34 Jorgensen-Tye et al,35 Doyonnas et al,36 and Buhring et al37 ) containing 2 extracellular O-glycan–substituted mucin domains (I and II) linked by a cysteine-rich nonmucin domain. There are 3 classes of CD164 monoclonal antibodies (mAbs).34–36 Class I and II mAbs identify glycosylated variants/epitopes of CD164 that have restricted patterns of cell expression.34–36 Class III mAbs recognize a ubiquitously expressed epitope on the CD164 peptide backbone.34–36 Epitopes recognized by the CD164 mAbs are expressed by most CD133+ HSCs/HPCs, albeit at varying levels,34 with the class II epitope the most extensively studied. Interestingly, CD164 class II epitope distribution on HSCs/HPCs and pre-B precursors from human bone marrow is reminiscent of functionally absent cells in CXCR4- and CXCL12-deficient mice, suggesting a potential association between the 2 receptors and bone marrow homing or retention. The CD164 class II epitope is also highly expressed on the more primitive human Lin−CD133+CD34+CD38lo/− and Lin−CD133+CD34−CD38lo/− cells, which have the ability to repopulate long-term definitive hematopoiesis, on early erythromyeloid cells, and on associated endothelial and bone marrow stromal cells, monocytes, and osteoblasts but not on most other cell types.34,37 On binding to cells, the 103B2 mAb inhibits the adhesion of bone marrow–derived CD34+ cells to stroma and the cytokine-induced recruitment of primitive CD34+CD38lo/− precursors into cycle.32
In this study, we showed that the class II 103B2, but not the class III N6B6, CD164 mAb significantly inhibited the migration on fibronectin of UCB CD133+ cells toward CXCL12. This effect is recapitulated by the knockdown of CD164 levels with the use of RNAi technology. We further examined the temporal association of CD164, the integrins VLA-4 and VLA-5, and CXCR4 on these cells and the effects of reducing CD164 expression on CXCR4-mediated signaling through protein kinase C-ζ (PKC-ζ), Akt, and mitogen-activated protein kinase (MAPK) after CXCL12 stimulation. Our results indicate that CD164 cooperates with CXCR4 in promoting CXCL12-mediated cell migration.
Materials and methods
Cells
UCB units were collected with informed written consent in accordance with the Declaration of Helsinki and ethics permission from the Oxford Radcliffe Research Ethics Committee (Oxford, United Kingdom). CD133+ cells were purified with the MACS CD133+ cell isolation kit (Miltenyi Biotech, Bergisch Gladbach, Germany).38 Cells were cultured at 5 × 105/mL in StemBioA medium (SBA) (StemBio Research, Villejuif, France) supplemented with 100 ng/mL each Flt3-ligand, SCF, and IL-6 and 10 ng/mL TPO (R&D Systems, Minneapolis, MN).38 Jurkat (clone E6-1) and HS-5 cell lines (both purchased from LGC Promochem, Middlesex, United Kingdom) were maintained in RPMI-1640 and DMEM culture medium, respectively (Gibco-BRL, Life Technologies, Paisley, United Kingdom), supplemented with 10% (vol/vol) fetal calf serum (PAA Laboratories GmbH, Linz, Austria).
Antibodies
Monoclonal antibodies were unconjugated or conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), or allophycocyanin (APC). Class II CD164 103B2/9E10 (mIgG3; hereafter, termed 103B2) was produced as described.32 The anti-CXCR4 mAb 6H8 (mIgG1) was kindly provided by Dr C. Arenzana (Institut Pasteur, Paris, France). Antibodies to class III CD164 epitope (N6B6, mIgG2a), CXCR4 (12G5, mIgG2a), PSGL-1 (KPL-1, mIgG1), CD5 (6E3, mIgG3), CD38 (HIT2, mIgG2a), CD3 (UCHT-1, mIgG1), CD41b (HIP2, mIgG3), CD49d (9F10, IgG1), CD49e (VC5, IgG1), CD38-FITC (HIT2, mIgG1), CD34-APC (8G12, IgG1), and to isotype controls IgG1-FITC and IgG1-APC were supplied by BD Biosciences, San Jose, CA. The CD133 mAbs AC133/1 (AC133, mIgG1) and AC133/2-PE (293C3, mIgG2b) and the PE-conjugated isotype negative control were supplied by Miltenyi Biotec. CD49d (HP2/1, mIgG1) and CD49e (JBS5, mIgG1) mAbs were supplied by Serotec (Oxford, United Kingdom). ICAM-3 (ICAM3.3, mIgG1) was from R&D Systems; CD43 (MEM-59, mIgG1) was from Monosan, Uden, The Netherlands; and α-tubulin mAb (B-5-1-2, mIgG1) was from Sigma-Aldrich (St Louis, MO). The CXCR4 polyclonal (ab2074) and CD45 (T29/33, mIgG1) antibodies and the isotype-negative controls mIgG1 and mIgG2a were obtained from Abcam (Cambridge, United Kingdom) and DakoCytomation (Glostrup, Denmark), respectively; the mIgG3 and mIgM control mAbs and FITC- or PE-conjugated secondary antibodies were from Southern Biotechnology Associates (Birmingham, AL); and the Alexa-488 and Alexa-546 secondary antibodies were from Molecular Probes (Leiden, The Netherlands). Santa Cruz Biotechnology (Santa Cruz, CA) supplied the p-PKC-ζ (sc-12894-R) and PKC-ζ (sc-216) antibodies. Phospho-Akt (Ser 473) (no. 9271), Akt (no. 9272), phospho-p44/42 MAPK (Thr202/Tyr204) (no. 9101), and p44/42 MAPK (no. 9102) polyclonal antibodies were purchased from Cell Signaling Technology (Danvers, MA). ImmunoPure peroxidase-conjugated goat anti–rabbit and anti–mouse (IgG) antibodies were purchased from Pierce Biotechnology (Rockford, IL).
Flow cytometry
For single-color, dual-color, or multicolor flow cytometric analyses, cells were incubated with Fc receptor blocking agent (Miltenyi Biotec), followed by the application of relevant antibodies or similarly conjugated isotype-matched, negative-control mAbs and the viability dye 7-AAD (BD Biosciences) and were analyzed on a BD LSR II flow cytometer with FACSDiva software (BD Biosciences).34,38 Isolated CD133+ cells were assessed for purity (95.2% ± 0.5%; n = 26) with the PE-conjugated AC133/2-PE antibody and the isotype-negative control mIgG1-PE.
RNAi and nucleofection
Two small interfering RNA (siRNA) duplexes (CD164-1 5′-CCCGAACGUGACGACUUUA-3′ and CD164-2 5′-GGACUGGUGAUUCAUUUGU-3′) were designed to specifically knock down CD164 (Samchully Pharmaceuticals, Seoul, Korea). Two control duplexes were synthesized (control-1, 5′-AUGAACGUGAAUUGCUCAA-3′; control-2, 5′-UAGCGACUAAACACAUCAA-3′) based on siControl nontargeting siRNAs (Dharmacon, Lafayette, CO). The HS-5 cell line was transfected with or without siRNAs using Lipofectamine 2000 (Invitrogen, Paisley, United Kingdom) according to manufacturer's instructions and was cultured for 48 hours at 37°C in RPMI-1640 medium with 10% (vol/vol) FCS. With the use of nucleofection technology, CD133+ or Jurkat cells (4 × 105) were nucleofected with or without siRNAs (1.2 μM final concentration) with the nucleofector program U-01 and T-14, respectively (Amaxa Biosystems, Cologne, Germany) and were transferred to their respective serum-free SBA medium with cytokines or to RPMI-1640 medium containing 10% (vol/vol) FCS and incubated at 37°C for 48 hours. Immediately after nucleofection with or without siRNAs, cell recovery rates ranged from 44% to 54% for Jurkat cells and 71% to 80% for CD133+ cells, with no significant difference between groups. Viability of these recovered cells after another 48 hours of culture was assessed with the viability stain To-pro (Molecular Probes, Paisley, United Kingdom) and ranged from 96% to 99% for Jurkat cells and from 95% to 98% for CD133+ cells. Cell-surface phenotypes were analyzed before nucleofection and at the end of the culture after nucleofection by single-color or multicolor flow cytometry, as described. Cells were then used in the migration assay in vitro or for the assessment of signaling activity or they were immunoblotted. In the latter case, cells were resuspended in lysis buffer (75 mM Tris-HCl [pH 6.8], 3.8% [wt/vol] SDS, 4 M urea, and 20% [vol/vol]) glycerol), protein concentrations were normalized with the DC Protein Assay kit (Bio-Rad, Hemel Hempstead, United Kingdom), and aliquots were subjected to SDS-PAGE and immunoblotting.
Transwell migration assay
Purified CD133+ or Jurkat cells (1-5 × 105/100 μL) were cultured overnight and then used untreated, pretreated with 10 to 50 μg/mL relevant mAbs, or nucleofected with relevant control or CD164 siRNA duplexes. After nucleofection, cells were cultured in the same medium for another 48 hours and allowed to migrate across 5-μm pore inserts (Corning-Costar, Cambridge, MA), uncoated or coated with 20 μg/mL fibronectin (Sigma-Aldrich), toward optimized CXCL12, CCL1, CCL5, CCL17, CCL19, CCL20, CCL21, CCL22, and CXCL13 concentrations (200 ng/mL; R&D Systems). Transmigrated cells were harvested after 4 hours, and a known quantity of BCECF-AM–labeled (Molecular Probes) Jurkat cells was added and compared with unlabeled cells counted on the flow cytometer. Twenty thousand events were collected using the Epics V Coulter Elite flow cytometer and EXPO 32 MultiCOMP software (Beckmann-Coulter, Miami, FL). Aliquots of cells were taken before and after the migration assays and were assessed for surface expression of CD164 and, in CD133+ cells, of CD34 and CD38 markers by flow cytometry.
Coimmunoprecipitation and immunoblotting
Plates were uncoated or coated with 20 μg/mL fibronectin, and 100 ng/mL CXCL12 was added. Cells (1 × 106) were exposed to CXCL12 for indicated times and lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [vol/vol] IGEPAL-CA-630, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, containing protease inhibitors) before immunoprecipitation with 1 to 5 μg CXCR4 or control polyclonal antibodies (Abcam). Immune complexes were captured with protein A Sepharose (Amersham-Pharmacia, Uppsala, Sweden) and subjected to SDS-PAGE and immunoblotting. For coimmunoprecipitation and RNAi studies, immunoblots were probed with CD164 (N6B6), VLA-4 (9F10) VLA-5 (VC5), ICAM-3 (ICAM3.3; all at 0.5 μg/mL), CXCR4 (12G5; 2 μg/mL), or α-tubulin antibodies. The reaction was developed with peroxidase-conjugated secondary antibodies and the SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology). Blot Restore Membrane Rejuvenation Kit (Chemicon International, Temecula, CA) was used to strip immunoblots before reblotting. Immunoblots were quantified using Scion Image densitometry software (Scion, Frederick, MD). CD164 levels were normalized to α-tubulin levels and expressed as a percentage of control nucleofection or transfection.
Confocal microscopy
Slides were coated with fibronectin and then with CXCL12, as described, before the addition of 1 × 105 CD133+ cells. Cells were exposed to CXCL12 presented on fibronectin for 10 minutes before fixation in 4% (wt/vol) paraformaldehyde and immunolabeling with primary antibodies, CXCR4 (6H8), CD164 (N6B6), ICAM-3 (ICAM3.3), VLA-4 (9F10), VLA-5 (VC5) or isotype-matched controls (all at 5 μg/mL) followed by Alexa-488 and Alexa-546 subclass-specific goat anti–mouse IgG antibodies (5 μg/mL). Cells were imaged using an LSM510 confocal microscope (Carl Zeiss, Oberkochen, Germany) equipped with a planapochromat 63×/1.4 numerical aperture oil objective, with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Images were processed using LSM software version 3.2 (Carl Zeiss) and Adobe Photoshop 7 (Adobe Systems, San Jose, CA).
CXCR4 signaling analysis
Because of limited CD133+ cell numbers, CXCR4 signaling through Akt, PKC-ζ, and MAPK was assessed in Jurkat cells. Cells (4 × 105 per time point) were transfected with control siRNAs or CD164 siRNAs, as described, and were then stimulated with 200 ng/mL CXCL12 in serum-free RPMI-1640 medium for the indicated times at 37°C. Cells were lysed in 25 mM Tris, pH 7.5, 1% (vol/vol) Triton, 0.5 mM EDTA, 150 mM NaCl, 50 mM NaF, NaVO3, PMSF, and 1% (vol/vol) protease inhibitor cocktail (Sigma-Aldrich) and were subjected to SDS-PAGE and immunoblotting with p-PKC-ζ, PKC-ζ, P-Akt, Akt, phospho-p44/42 MAPK, p44/42 MAPK, and anti–α-tubulin antibodies (all at 0.2 μg/mL). For each signaling experiment, phosphorylated protein levels were normalized to the total signal and were expressed as a percentage of the time point with the highest protein level.
Statistical analysis
Data are presented as mean ± SEM from multiple independent experiments (3 or more) with statistical significance (P < .05; Student t test).
Results
103B2 CD164 mAb inhibits CXCL12-induced cell migration
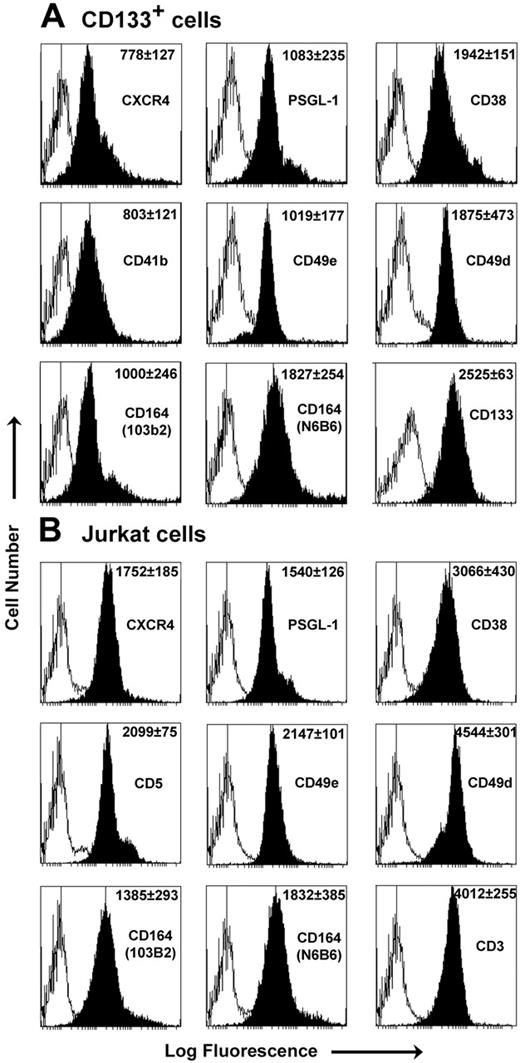
In determining the role of CD164 in CXCL12/CXCR4-mediated cell migration, we analyzed UCB CD133+ progenitor cells and the Jurkat cell line. Both cell types express the receptors of interest (Figure 1). Of note, and in agreement with other findings,11 was the up-regulation of CXCR4 surface expression on CD133+ cells after 24-hour incubation with cytokines compared with freshly isolated cells. Coexpression of the class II CD164 epitope on their surfaces was not significantly altered by cytokine stimulation (Figure 1).
Phenotype analysis of CD133+ and Jurkat cells. Representative FACS histograms of CD133+ cells after 24-hour cytokine stimulation (A) and Jurkat cells (B) with antibodies (black peak) and relevant isotype controls (white peak). MFI ± SEM (n = 3) is shown for each antibody in the top right corner of the plot. CXCR4 surface expression was up-regulated on CD133+ cells after incubation with cytokines (from 15.1% ± 2.1% to 77.0 ± 3.6% CXCR4+, with MFI increasing from 234 ± 10.6 to 778 ± 127; n = 3). No significant alterations in 103B2 CD164 staining were observed before or after 24-hour cytokine stimulation (staining, 67% ± 4.5% to 69.4% ± 9.1%; MFI values, 1003 ± 121 to 1000 ± 246 (n = 3) for freshly isolated and 24-hour cytokine-stimulated cells, respectively). In separate dual-labeling experiments, 73.1% ± 6.4% of CD133+ cells coexpressed CXCR4 and the class II CD164 epitope on the surface after 24-hour cytokine stimulation (P = .41 for CXCR4 expression alone vs CXCR4 coexpression with CD164).
Phenotype analysis of CD133+ and Jurkat cells. Representative FACS histograms of CD133+ cells after 24-hour cytokine stimulation (A) and Jurkat cells (B) with antibodies (black peak) and relevant isotype controls (white peak). MFI ± SEM (n = 3) is shown for each antibody in the top right corner of the plot. CXCR4 surface expression was up-regulated on CD133+ cells after incubation with cytokines (from 15.1% ± 2.1% to 77.0 ± 3.6% CXCR4+, with MFI increasing from 234 ± 10.6 to 778 ± 127; n = 3). No significant alterations in 103B2 CD164 staining were observed before or after 24-hour cytokine stimulation (staining, 67% ± 4.5% to 69.4% ± 9.1%; MFI values, 1003 ± 121 to 1000 ± 246 (n = 3) for freshly isolated and 24-hour cytokine-stimulated cells, respectively). In separate dual-labeling experiments, 73.1% ± 6.4% of CD133+ cells coexpressed CXCR4 and the class II CD164 epitope on the surface after 24-hour cytokine stimulation (P = .41 for CXCR4 expression alone vs CXCR4 coexpression with CD164).
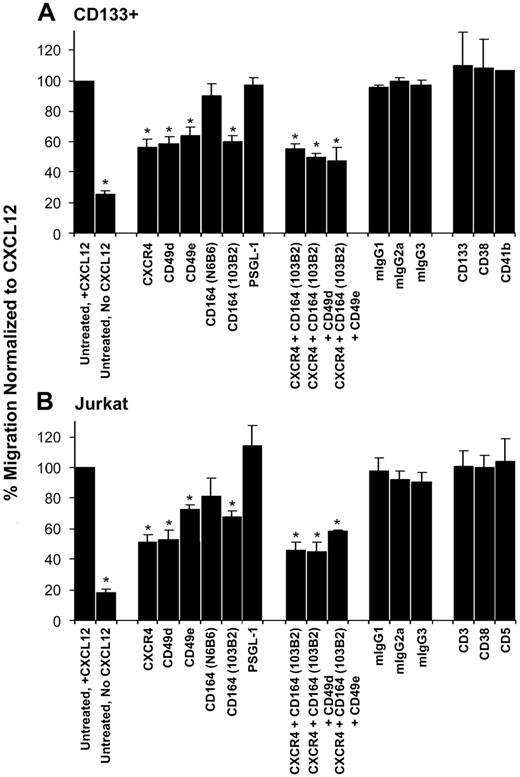
With the use of transwell migration assays, we observed 54.3% ± 5.1% of the total untreated CD133+ cells migrated on fibronectin toward CXCL12 compared with 13.5% ± 1.9% of cells in the absence of CXCL12 over a 4-hour period. Similarly, 46.9% ± 6.8% of untreated Jurkat cells migrated in the presence of CXCL12 compared with 10.4% ± 2.2% in its absence. The migration of these untreated cells to CXCL12 was normalized to 100%, and all other migration was determined as a ratio of this normalized value (Figure 2). Our results confirmed previous observations10,11 that the incubation of CD133+ and Jurkat cells with the anti-CXCR4 mAb to inhibit CXCL12 binding and with CD49d (anti–VLA-4α) and CD49e (anti–VLA-5α) mAbs to reduce binding to fibronectin significantly inhibited migration. For CD133+ cells, this migration was inhibited by 45.9% ± 6.7%, 38.1% ± 2.1%, and 33.9% ± 5.8% for the respective antibodies and was reduced by 48.5% ± 4.5%, 46.9% ± 5.9%, and 27.9% ± 3.2%, respectively, for Jurkat cells compared with untreated control cells (Figure 2A-B; P < .05 for all). Strikingly, binding of the class II CD164 mAb 103B2 to CD133+ and Jurkat cells reduced their migration toward CXCL12, decreasing by 41.1% ± 4.2% and 32.5% ± 4.6%, respectively, compared with the normalized untreated control (Figure 2A-B; P ≤ .001 for both). Binding of the class III CD164 mAb N6B6 did not affect CD133+ or Jurkat cell migration to CXCL12 (Figure 2A-B). When compared with anti-CXCR4 alone, there was no significant additive effect on the inhibition of cell migration with a combination of mAbs against CXCR4, CD164, and VLA-4 or VLA-5 (Figure 2A-B; P > .05 for all), suggesting that these antibodies interfered with components of a common migration pathway. Notably, there was no significant effect on cell migration using the isotype-matched nonbinding and binding mAbs, including that to another sialomucin, PSGL-1 (Figure 2A-B; P > .05 for all antibodies tested on CD133+ and Jurkat cells). In the absence of CXCL12 and in the presence of fibronectin, the 103B2 mAb had no effect on the spontaneous migration of CD133+ cells on fibronectin (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). Conversely, in the presence of CXCL12 and the absence of fibronectin, the migration of CD133+ and Jurkat cells to CXCL12 was reduced by 62.5% ± 5.2% and 72.8% ± 3.4%, respectively, compared with migration on fibronectin, but neither 103B2 nor relevant control mAbs further inhibited this cell migration (Figure S2A-B). To investigate the specificity of the effect of CD164 on CXCL12-mediated migration, we analyzed the migration of Jurkat cells to 8 additional chemokines, CCL1, CCL5, CCL17, CCL19, CCL20, CCL21, CCL22, and CXCL13, (Figure S3A). Although levels were lower than for CXCL12, cell migration was significant only for CCL5, but this was not reduced by addition of the 103B2 CD164 mAb (Figure S3B).
CD164 binding inhibits CD133+ and Jurkat cell migration. Transwell migration assays using CD133+ cells (A) and Jurkat cells (B). Cells were incubated with the antibodies indicated and stimulated to migrate to CXCL12 presented on fibronectin. The value for untreated migration to CXCL12 was normalized to 100%, and all other migration was compared with this normalized value. Data shown represent the mean ± SEM of 3 to 7 independent experiments. *Antibody treatment caused statistically significant reductions in the number of cells migrating to CXCL12 compared with untreated control (P < .05; 1-paired Student t test). No significant effect occurred on cell migration using a variety of isotype-matched binding and nonbinding mAbs, including that to the sialomucin PSGL-1, in either cell type.
CD164 binding inhibits CD133+ and Jurkat cell migration. Transwell migration assays using CD133+ cells (A) and Jurkat cells (B). Cells were incubated with the antibodies indicated and stimulated to migrate to CXCL12 presented on fibronectin. The value for untreated migration to CXCL12 was normalized to 100%, and all other migration was compared with this normalized value. Data shown represent the mean ± SEM of 3 to 7 independent experiments. *Antibody treatment caused statistically significant reductions in the number of cells migrating to CXCL12 compared with untreated control (P < .05; 1-paired Student t test). No significant effect occurred on cell migration using a variety of isotype-matched binding and nonbinding mAbs, including that to the sialomucin PSGL-1, in either cell type.
Knockdown of CD164 levels inhibits CXCL12-mediated cell migration
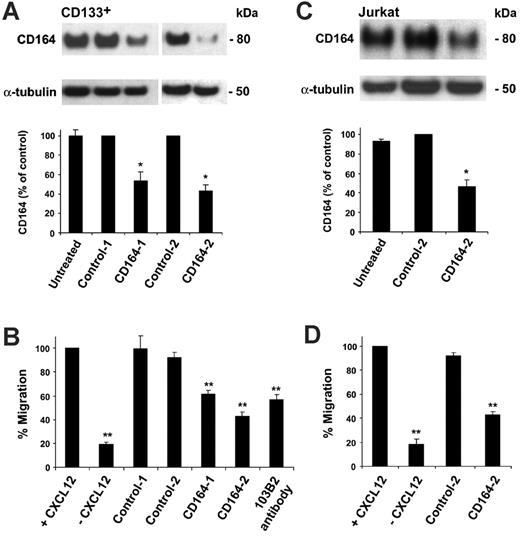
To confirm that the inhibitory effect on cell migration was CD164 specific, 2 siRNA duplexes to the cDNA coding region (CD164-1) and the 3′-untranslated region (CD164-2) were designed to knock down expression of the CD164 protein (Figure S4A). The efficacy of these siRNAs in reducing CD164 levels was first assessed by transfection into the human stromal cell line HS-5, which expresses high CD164 levels, and immunoblot of whole cell lysates. CD164-1 and CD164-2 siRNAs reduced CD164 levels in HS-5 cells by 73.9% and 93.5% compared with control-1 siRNA levels (Figure S4B), with CD164-2 siRNA more effective. CD164 siRNAs were introduced into CD133+ and Jurkat cells by nucleofection. For these studies, cells were either untreated (nucleofected without siRNA) or nucleofected with control, CD164-1, or CD164-2 siRNA duplexes. After nucleofection of CD133+ cells with CD164-1 and CD164-2 siRNA, CD164 protein levels were reduced by 46.5% ± 9.2% and 56.6% ± 6.2%, respectively, compared with siRNA controls (Figure 3A; P < .05 for both). Cell migration toward CXCL12 after nucleofection with control siRNAs was not significantly different from that for untreated cells (Figure 3B). In contrast, cells treated with the CD164-1 and CD164-2 siRNAs showed significantly reduced ability to migrate to CXCL12, with migration inhibited by 38.6% ± 2.9% and 57.2% ± 3.7%, respectively, compared with the normalized control (Figure 3B; P < .001). This level of inhibition by the CD164 siRNAs was comparable to, or slightly higher than, the level seen with the 103B2 CD164 mAbs (Figure 3B). In Jurkat cells transfected with CD164-2 siRNA, CD164 protein levels were reduced by 53.6% ± 6.8% compared with control levels (Figure 3C). Cell migration toward CXCL12 after nucleofection with or without control siRNA was similar to that for untreated cells (Figure 3D). However, migration of CD164-2 siRNA nucleofected cells to CXCL12 was reduced by 57.1% ± 2.5% (P < .001 compared with normalized control). In the absence of fibronectin, though cell migration was decreased, the migration of siRNA control–treated cells to CXCL12 was not significantly different from that of untreated cells (P = .08). This was not altered further when CD164 was knocked down in CD133+ or Jurkat cells (P = .2 and P = .1, respectively, compared with untreated nucleofected cells; see Figure S5), indicating that CD164 exerts its action in the presence of CXCL12 and fibronectin.
Knockdown of CD164 modulates CD133+ and Jurkat cell migration to CXCL12. (A) CD164 knockdown using RNAi in CD133+ cells. (Top) Western blot of CD164 protein levels in CD133+ cells that were untreated (nucleofected without siRNA) or nucleofected with control or CD164 siRNA duplexes, using α-tubulin as a loading control. (Bottom) Densitometry analysis of CD164 protein levels in RNAi-treated CD133+ cells (*P < .05; n = 2). (B) Transwell migration assay of CD133+ cells that were untreated or nucleofected with control or CD164 siRNA duplexes and induced to migrate to CXCL12 on fibronectin (**P < .001). (C) CD164 knockdown using RNAi in Jurkat cells. (Top) Western blot of CD164 protein in Jurkat cells that were untreated or nucleofected with control-2 or CD164-2 siRNA duplexes, using α-tubulin as a loading control. (Bottom) Densitometry analysis of CD164 protein levels in RNAi-treated Jurkat cells (*P < .05; n = 2). (D) Transwell migration assay of Jurkat cells that were untreated or nucleofected with control-2 or CD164-2 siRNA duplexes and induced to migrate to CXCL12 (**P < .001).
Knockdown of CD164 modulates CD133+ and Jurkat cell migration to CXCL12. (A) CD164 knockdown using RNAi in CD133+ cells. (Top) Western blot of CD164 protein levels in CD133+ cells that were untreated (nucleofected without siRNA) or nucleofected with control or CD164 siRNA duplexes, using α-tubulin as a loading control. (Bottom) Densitometry analysis of CD164 protein levels in RNAi-treated CD133+ cells (*P < .05; n = 2). (B) Transwell migration assay of CD133+ cells that were untreated or nucleofected with control or CD164 siRNA duplexes and induced to migrate to CXCL12 on fibronectin (**P < .001). (C) CD164 knockdown using RNAi in Jurkat cells. (Top) Western blot of CD164 protein in Jurkat cells that were untreated or nucleofected with control-2 or CD164-2 siRNA duplexes, using α-tubulin as a loading control. (Bottom) Densitometry analysis of CD164 protein levels in RNAi-treated Jurkat cells (*P < .05; n = 2). (D) Transwell migration assay of Jurkat cells that were untreated or nucleofected with control-2 or CD164-2 siRNA duplexes and induced to migrate to CXCL12 (**P < .001).
Effects of CD164 knockdown on migration of CD133+ subsets
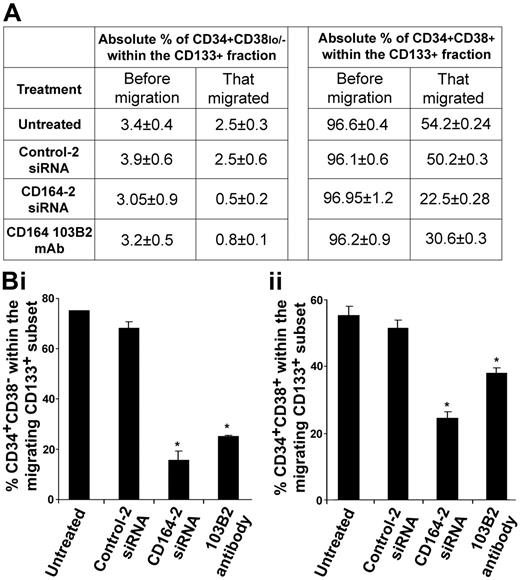
To explore whether knocking down the CD164 protein affected the migration of specific CD133+ cell subsets expressing CD34 or CD38, we examined the distribution of CD34+CD38lo/− and CD34+CD38+ cell populations in the migrated and nonmigrated cell fractions. Immediately before migration assay, we examined the percentages of these subsets within the untreated and siRNA-treated CD133+ cell samples and showed no significant differences in the percentages of cells that were CD34+CD38+ or CD34+CD38lo/− after each specified treatment (Figure 4A). In migration experiments using CD133+ cells, we observed a significant reduction of CD34+CD38lo/− and CD34+CD38+ cells migrating after treatment with CD164-2 siRNA or the 103B2 mAb compared with relevant controls (Figure 4A-Bi, ii).
CD164 knockdown modulates CXCL12-mediated migration of CD133+ subsets on fibronectin. (A) Absolute percentages of CD34+CD38lo/− and CD34+CD38+ cells before migration in the original CD133+ cell population and in the migrated fraction. The absolute percentage of each cell subset in the overall population was not significantly different before migration, irrespective of treatment (P > .05). (B) Percentage of each CD34+CD38lo/− and CD34+CD38+ cell subset within the CD133+ population that migrated. In untreated cells (nucleofected without siRNA) or cells nucleofected with control siRNAs, 73.4% ± 0.2% and 66.8% ± 2.6%, respectively, of the total CD133+CD34+CD38lo/− cell subset migrated to CXCL12 (Bi). This decreased to 15.2% ± 3.6% and 24.8% ± 0.1% for cells nucleofected with CD164-2 siRNAs or treated with the 103B2 mAb, respectively (P < .05 for both). In the CD34+CD38+ subset, 56.1% ± 2.7% of untreated and 52.2% ± 2.5% of cells nucleofected with control siRNA migrated to CXCL12 (Bii). This was reduced to 23.2% ± 1.7% and 31.8% ± 1.7% of cells treated with CD164-2 siRNAs or the 103B2 mAb, respectively (P < .05 for both).
CD164 knockdown modulates CXCL12-mediated migration of CD133+ subsets on fibronectin. (A) Absolute percentages of CD34+CD38lo/− and CD34+CD38+ cells before migration in the original CD133+ cell population and in the migrated fraction. The absolute percentage of each cell subset in the overall population was not significantly different before migration, irrespective of treatment (P > .05). (B) Percentage of each CD34+CD38lo/− and CD34+CD38+ cell subset within the CD133+ population that migrated. In untreated cells (nucleofected without siRNA) or cells nucleofected with control siRNAs, 73.4% ± 0.2% and 66.8% ± 2.6%, respectively, of the total CD133+CD34+CD38lo/− cell subset migrated to CXCL12 (Bi). This decreased to 15.2% ± 3.6% and 24.8% ± 0.1% for cells nucleofected with CD164-2 siRNAs or treated with the 103B2 mAb, respectively (P < .05 for both). In the CD34+CD38+ subset, 56.1% ± 2.7% of untreated and 52.2% ± 2.5% of cells nucleofected with control siRNA migrated to CXCL12 (Bii). This was reduced to 23.2% ± 1.7% and 31.8% ± 1.7% of cells treated with CD164-2 siRNAs or the 103B2 mAb, respectively (P < .05 for both).
Defining the mechanism: CD164 associates with CXCR4 in response to CXCL12 stimulation
There are several potential mechanisms by which interfering with CD164 expression and function could affect cell migration. These include (1) induction of apoptosis and, hence, a reduction in the number of viable cells able to migrate; (2) alteration of CXCR4 surface expression and, hence, a reduction in the ability of cells to sense CXCL12; and (3) direct influence on the function of the CXCR4 receptor. To address the first possibility, we investigated the effects of CD164 binding by the 103B2 mAb or CD164 knockdown by siRNAs in CD133+ and Jurkat cells on inducing apoptosis. None of the treatments induced apoptosis in these cells (Figure S6; data not shown). The second possible mechanism was addressed by examining CXCR4 surface expression levels after incubation with the 103B2 and N6B6 CD164 mAbs or after nucleofection with CD164 siRNA duplexes. CXCR4 expression was unaffected by these treatments. As demonstrated in Figure S6, no significant change in the percentage of cells expressing CXCR4 or in the mean fluorescence intensity (MFI) of CXCR4 staining by flow cytometry was observed when CD164 siRNAs were compared with siRNA or nucleofected controls (with an average change in cells expressing CXCR4 of only 3.5% ± 0.8% for CD133+ cells and 0.5% ± 0.1% for Jurkat cells, respectively).
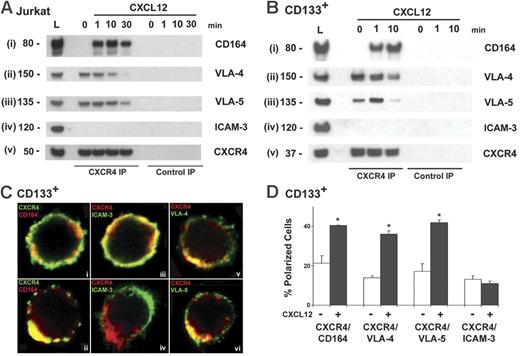
We investigated whether CD164 might be a functional component of a CXCR4 protein complex. Our results demonstrate that in the presence of fibronectin but in the absence of CXCL12, CD164 did not appreciably coimmunoprecipitate with CXCR4 in Jurkat cells (Figure 5Ai, lane 0). However, when CXCL12 was presented on fibronectin, both receptors coimmunoprecipitated (Figure 5Ai, lanes 1, 10, 30). This was observed as a rapid recruitment of CD164 to the CXCR4 complex, seen maximally 10 minutes after CXCL12 stimulation. This result contrasts with the integrins VLA-4 and VLA-5, which coimmunoprecipitated with CXCR4 in the absence of CXCL12. The association of VLA-4 and VLA-5 with CXCR4 increased to a maximal level 1 minute after CXCL12 on fibronectin was presented to the cells (Figure 5Aii-iii). As this recruitment increased, the integrins appeared to dissociate from the complex before CD164. However, in the absence of fibronectin, the recruitment of CD164, VLA-4, and VLA-5 to the CXCR4 complex was significantly attenuated (Figure S7B). The recruitment of CD164 to the CXCR4 complex after the integrins was confirmed by their coimmunoprecipitation from CXCL12-stimulated CD133+ cells (Figure 5B), suggesting that CD164 is recruited to a CXCR4 complex after the integrins and in response to CXCL12 stimulation, where fibronectin is required for the association of these receptors.
CD164 associates with CXCR4 and the integrins VLA-4 and VLA-5. (A) Coimmunoprecipitation of CXCR4 with CD164 and the integrins VLA-4 and VLA-5 in Jurkat cells in response to CXCL12 stimulation presented on fibronectin. Cells were stimulated with CXCL12 on fibronectin for 0, 1, 10, and 30 minutes. Immunoprecipitated CXCR4 was immunoblotted with mAbs to (i) CD164, (ii) VLA-4, (iii) VLA-5, (iv) ICAM-3, and (v) CXCR4. L = 50 μg total cell lysate. (B) Coimmunoprecipitation of CXCR4 and CD164 in CD133+ cells. Cells were stimulated with CXCL12 for 0, 1, and 10 minutes, and lysates were immunoprecipitated with anti-CXCR4 and immunoblotted with mAbs to (i) CD164, (ii) VLA-4, (iii) VLA-5, (iv) ICAM-3, and (v) CXCR4. (C) Single Z-stack confocal images of CD133+ cells in the absence (i, iii) and presence (ii, iv, v, vi) of CXCL12 for 10 minutes and coimmunolabeled with indicated antibodies. In the absence of CXCL12, CXCR4 and CD164 were equally distributed on the cell membrane. After CXCL12 stimulation, CD164, VLA-4, and VLA-5 redistribute to the leading edge of the cell with CXCR4, whereas ICAM-3 clusters to the rear and opposite pole. (D) Quantification of receptor redistribution in the absence (white bars) or presence (black bars) of CXCL12 stimulation. The number of cells that showed a polarized and colocalized distribution of CXCR4 with CD164, VLA-4, or VLA-5 was counted and expressed as a percentage of the total number of polarized cells observed, as defined by CXCR4 staining. Significant redistribution of CXCR4 with CD164, VLA-4, and VLA-5 was observed after CXCL12 presentation for 10 minutes (P < .05). ICAM-3 was counted as a control (P > .05).
CD164 associates with CXCR4 and the integrins VLA-4 and VLA-5. (A) Coimmunoprecipitation of CXCR4 with CD164 and the integrins VLA-4 and VLA-5 in Jurkat cells in response to CXCL12 stimulation presented on fibronectin. Cells were stimulated with CXCL12 on fibronectin for 0, 1, 10, and 30 minutes. Immunoprecipitated CXCR4 was immunoblotted with mAbs to (i) CD164, (ii) VLA-4, (iii) VLA-5, (iv) ICAM-3, and (v) CXCR4. L = 50 μg total cell lysate. (B) Coimmunoprecipitation of CXCR4 and CD164 in CD133+ cells. Cells were stimulated with CXCL12 for 0, 1, and 10 minutes, and lysates were immunoprecipitated with anti-CXCR4 and immunoblotted with mAbs to (i) CD164, (ii) VLA-4, (iii) VLA-5, (iv) ICAM-3, and (v) CXCR4. (C) Single Z-stack confocal images of CD133+ cells in the absence (i, iii) and presence (ii, iv, v, vi) of CXCL12 for 10 minutes and coimmunolabeled with indicated antibodies. In the absence of CXCL12, CXCR4 and CD164 were equally distributed on the cell membrane. After CXCL12 stimulation, CD164, VLA-4, and VLA-5 redistribute to the leading edge of the cell with CXCR4, whereas ICAM-3 clusters to the rear and opposite pole. (D) Quantification of receptor redistribution in the absence (white bars) or presence (black bars) of CXCL12 stimulation. The number of cells that showed a polarized and colocalized distribution of CXCR4 with CD164, VLA-4, or VLA-5 was counted and expressed as a percentage of the total number of polarized cells observed, as defined by CXCR4 staining. Significant redistribution of CXCR4 with CD164, VLA-4, and VLA-5 was observed after CXCL12 presentation for 10 minutes (P < .05). ICAM-3 was counted as a control (P > .05).
CD164 and CXCR4 cluster at the leading edge of cells when presented with CXCL12
To explore whether a redistribution of CXCR4 and CD164 occurred on the cell surface after CXCL12 sensing, we performed confocal microscopy on CD133+ cells. In the absence of CXCL12, CXCR4 and CD164 were equally distributed on the cell membrane (Figure 5Ci). With CXCL12 presentation on fibronectin, both receptors redistributed, colocalizing at the leading edge of migrating cells (Figure 5Cii). This was in contrast to ICAM-3, which concentrates at the rear pole of migrating cells, opposite the site of accumulation of CXCR4 (Figure 5Civ), as previously described.39 Ten-minute stimulation with CXCL12 resulted in a significant increase in the percentage of cells demonstrating a polarized, colocalized distribution of CXCR4 and CD164 at the CXCL12–fibronectin interface compared with unstimulated cells (P < .05; Figure 5D). VLA-4 and VLA-5 also demonstrated a polarized, clustered phenotype with CXCR4 in response to CXCL12 presentation on fibronectin at 10 minutes (Figure 5Cv-vi, D). Taken together, these data suggest that CD164 and the integrins VLA-4 and VLA-5 are part of a functional CXCR4 complex involved in CXCL12-mediated migration.
CD164 modulates the CXCR4 downstream signaling pathway
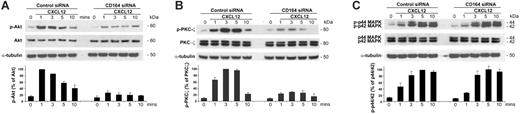
Given that CD164 associates with CXCR4 on CXCL12 stimulation, we hypothesized that the loss of CD164 expression may alter CXCR4 cell signaling responses, resulting in decreased cell migration. It has been demonstrated that CXCL12 stimulates the activation of PI3K, PKC—particularly the atypical PKC-ζ—and p42/44 MAPK (ERK1/2).40–42 We explored the effects of CD164 knockdown in CXCL12/CXCR4-mediated phosphorylation of these pathways in Jurkat cells. These cells were nucleofected, with control-2 or CD164-2 siRNA duplexes as described, and 48 hours later they were stimulated with 200 ng/mL CXCL12 presented on fibronectin for 0, 1, 3, 5, and 10 minutes. Basal levels of phosphorylated Akt (p-Akt) were detectable in unstimulated cells nucleofected with control-2 siRNA, but, on stimulation with CXCL12, a sharp increase occurred in p-Akt levels that decreased over 10 minutes (Figure 6A). In cells treated with CD164-2 siRNA, basal levels were also detected, but the phosphorylated response was significantly muted (Figure 6A). After 1 minute of stimulation, p-Akt levels were reduced to 23.7% ± 2.9% of control levels. Similarly, basal levels of p-PKC-ζ were detectable in unstimulated cells nucleofected with control-2 siRNA, but p-PKC-ζ levels increased dramatically to a maximum after 3 minutes of CXCL12 exposure, after which phosphorylation levels decreased (Figure 6B). In cells treated with CD164-2, basal p-PKC-ζ levels were unchanged, but a significant attenuation of PKC-ζ phosphorylation occurred on CXCL12 stimulation (Figure 6B). In cells treated with control-2 siRNA, p-p42/44 MAPK signal was induced by CXCL12, with maximal induction at 5 minutes (Figure 6C). A similar pattern and level of induction were observed in cells treated with CD164-2 siRNA, suggesting that knockdown of CD164 does not alter the p-p42/44 signal (Figure 6C). This contrasts to the significant attenuation observed in the PI3K and PKC-ζ pathways, suggesting that CD164 is involved in the activation of these pathways but not in the p-42/44 MAPK pathway.
CD164 modulates CXCL12-mediated migration by modulating CXCR4 downstream signaling pathways. p-Akt (A), p-PKC-ζ (B), and phospho-p44/42 MAPK (C) signaling in Jurkat cells nucleofected with control and CD164 siRNA duplexes. Cells were nucleofected, cultured for 48 hours, and stimulated with CXCL12 in the presence of fibronectin for 0, 1, 3, 5, and 10 minutes. (A) In control siRNA-treated cells, p-Akt signaling was induced and found to be maximal at 1 minute. This induction was significantly reduced in cells treated with CD164 siRNA. Total Akt and α-tubulin were used as loading controls with p-Akt protein levels compared with total Akt signal and were normalized to the Akt signal at 1 minute in cells treated with control siRNA (n = 2). (B) In control siRNA-treated cells, p-PKC-ζ signaling was induced at 1 minute and was maximal at 3 minutes after stimulation. This pattern was evident in cells treated with CD164 siRNA, but the activation of phosphorylated PKC-ζ was significantly attenuated. For example, after 3-minute CXCL12 stimulation, p-PKC-ζ levels were reduced to 28.5% ± 1.7% of the control levels. Total PKC-ζ (t-PKC-ζ) and α-tubulin were used as loading controls. Densitometry analysis of p-PKC-ζ protein levels compared with t-PKC-ζ in RNAi-treated Jurkat cells was normalized to the strongest p-PKC-ζ signal (3 minutes in control siRNA-treated cells; n = 3). (C) In control siRNA-treated cells, sp-p44/42 MAPK signaling was induced and found to be maximal at 5 minutes. The pattern and level of signaling induction were similar in CD164 siRNA–treated cells, indicating that reducing the level of CD164 did not significantly affect p-p-44/42 MAPK signaling. p44/42 MAPK and α-tubulin were used as loading controls with p-p44/42 MAPK protein levels compared with total p44/42 MAPK signal and were normalized to p44/42 MAPK protein levels at 5 minutes in cells treated with control siRNA (n = 2).
CD164 modulates CXCL12-mediated migration by modulating CXCR4 downstream signaling pathways. p-Akt (A), p-PKC-ζ (B), and phospho-p44/42 MAPK (C) signaling in Jurkat cells nucleofected with control and CD164 siRNA duplexes. Cells were nucleofected, cultured for 48 hours, and stimulated with CXCL12 in the presence of fibronectin for 0, 1, 3, 5, and 10 minutes. (A) In control siRNA-treated cells, p-Akt signaling was induced and found to be maximal at 1 minute. This induction was significantly reduced in cells treated with CD164 siRNA. Total Akt and α-tubulin were used as loading controls with p-Akt protein levels compared with total Akt signal and were normalized to the Akt signal at 1 minute in cells treated with control siRNA (n = 2). (B) In control siRNA-treated cells, p-PKC-ζ signaling was induced at 1 minute and was maximal at 3 minutes after stimulation. This pattern was evident in cells treated with CD164 siRNA, but the activation of phosphorylated PKC-ζ was significantly attenuated. For example, after 3-minute CXCL12 stimulation, p-PKC-ζ levels were reduced to 28.5% ± 1.7% of the control levels. Total PKC-ζ (t-PKC-ζ) and α-tubulin were used as loading controls. Densitometry analysis of p-PKC-ζ protein levels compared with t-PKC-ζ in RNAi-treated Jurkat cells was normalized to the strongest p-PKC-ζ signal (3 minutes in control siRNA-treated cells; n = 3). (C) In control siRNA-treated cells, sp-p44/42 MAPK signaling was induced and found to be maximal at 5 minutes. The pattern and level of signaling induction were similar in CD164 siRNA–treated cells, indicating that reducing the level of CD164 did not significantly affect p-p-44/42 MAPK signaling. p44/42 MAPK and α-tubulin were used as loading controls with p-p44/42 MAPK protein levels compared with total p44/42 MAPK signal and were normalized to p44/42 MAPK protein levels at 5 minutes in cells treated with control siRNA (n = 2).
Discussion
We have previously shown that binding of the 103B2 class II mAb to cells significantly inhibits the adhesion of CD34+ cells to stroma and the recruitment of CD34+CD38lo/− cells into cycle.32 We now provide compelling evidence that CD164 also modulates the migration of UCB CD133+ precursors and that this effect is mediated through the CXCL12/CXCR4 axis. Our results indicated that CD164 associated specifically with CXCR4 because it colocalized with CXCR4 at the leading edge of the cell on the presentation of CXCL12 on fibronectin; CXCR4 and CD164 coimmunoprecipitated after CXCL12 stimulation in the presence of fibronectin; siRNA silencing or class II CD164 mAb binding of the N-terminal N-linked glycan epitope of CD164 significantly inhibited cell migration; it did not modulate CCL5-mediated migration; and treatment with CD164 siRNA attenuated p-Akt and p-PKC-ζ, but not p-p42/44 MAPK, signaling through CXCR4. We also observed that a higher proportion of the CD133+CD34+CD38lo/− cells were inhibited after CD164 knockdown or 103B2 mAb binding compared with the CD133+CD34+CD38+ subset, suggesting a more stringent effect on more primitive hematopoietic precursors. Therefore, altering the expression or binding of CD164 modulates the CXCL12-migratory function for CD38lo/− and CD38+ CD133+ progenitor cells.
We and others have demonstrated that bone marrow stromal, osteoblast, and vascular niche cells express receptors or ligands for the specific homing and retention of HSCs/HPCs (reviewed in Nilsson and Simmons,5 Chan and Watt,23 and Taichman43 ). Because the chemokine CXCL12 and its receptor, CXCR4, play central roles in orchestrating such events (reviewed in Burger and Kipps12 ), our hypothesis is that CXCR4 must cooperate with other receptors, on the same or opposing cells, to lend specificity to their homing and retention in particular niches. Indeed, if we examine the available evidence, we can observe an emerging trend with significant cross talk between the CXCR4/CXCL12 axis and other receptors and ligands. For example, CXCL12 can activate the VLA-4 and VLA-5 integrins, resulting in cell arrest under conditions of shear flow in vitro.10 These activated integrins are also essential for engraftment of human CD34+ cells into NOD/SCID murine bone marrow.10 Complement C3a enhances homing to CXCL12 by stimulation of the VLA-4 receptor and the matrix metalloproteinase-9 (MMP-9) in human CD34+ cells.44 Of the other molecules interacting with CXCR4/CXCL12, the peptidase CD26, the phosphatase CD45, and syndecan-4 are of particular note.45–47 Inhibition or deficiency of CD26 in murine HSCs/HPCs increases homing by the removal of dipeptides at the CXCL12 amino terminus,45 whereas CD45 dephosphorylates kinases that may be downstream components of the PKC-ζ signaling pathway.46 PKC-ζ, a member of the superfamily of PKC serine-threonine kinase enzymes, has been shown to have a direct involvement in CXCL12-induced downstream signaling.40 Finally, syndecan-4 acts as a receptor for CXCL12 on HeLa cells and modulates CXCR4 signaling through the p44/p42 MAPK and Jun N-terminal/stress-activated protein kinase (JNK/SAP) pathways.47 This suggests that specific molecules, such as syndecan-4 and endolyn, could modulate CXCR4 signaling pathways differentially in a tissue-dependent manner. Binding of CXCL12 to CXCR4 activates PI3K, which in turn mediates the activation of PKC-ζ and subsequently the downstream effectors Pyk2 and MAPK. Our data are in agreement with the activation of PI3K and PKC-ζ by CXCL12 and suggest that CD164 is required for the activation of these pathways through CXCR4 in hematopoietic cells. In contrast, though p42/p44 MAPK is activated by CXCL12 through CXCR4, knockdown of CD164 does not seem to affect this pathway, suggesting that the role of CD164 may be more specific than that of CXCR4. This signaling cascade results in cell adhesion and chemotaxis through cell polarization and MMP-9 secretion, resulting in enhanced migration to appropriate microenvironmental niches.40 Although 103B2 mAb binding prevents adhesion and cell-cycle recruitment in CD133+ cell subsets, the interaction of CD164 with other specific molecules has not been defined until now; here we show that CD164, on CD133+ cells and the leukemic Jurkat cell line, interacts in cis with CXCR4. This interaction in cis and modulation of CXCR4 function in CXCL12-mediated migration is reminiscent of the role of another sialomucin, MUC IV, which interacts in cis with the ErbB2 receptor for EGF, allowing full expression of its function on epithelial cells.48,49
Our current finding that the knockdown of CD164 or the blockade of the 103B2 CD164 class II epitope on CD133+ cells inhibits their migration and our previous demonstration that antibody binding to the 103B2 class II epitope of CD164 on CD133+ HSC/HPC blocks their adhesion to bone marrow stroma32 together raise several interesting possibilities. The VLA-4/VLA-5 integrins may rapidly associate with CXCR4 when CXCL12 is presented on fibronectin, leading to CD164 recruitment to the migratory complex. CD164 may then either modulate the conformation of CXCR4 or stabilize or interact with CXCL12, allowing CXCR4 to effectively signal through PI3K and PKC-ζ. Alternatively, it may act to modulate the adhesion of the integrins to fibronectin by controlling the dynamic severing and reestablishment of integrin-dependent focal adhesions. In the bone marrow, we could also envisage that CD164 strengthens the effects we observed in vitro by interaction with homophilic or heterophilic ligands expressed by cells within the bone marrow microenvironmental niche. Precedents exist for other sialomucins functioning in multiple interactions, though these have not been described for the same cell types. For instance, we have previously shown that the sialomucin CD45, a molecule present on HSCs/HPCs50 but not on stromal osteoblast or vascular endothelial niche cells, interacts in trans with heparan sulfate, an extracellular matrix molecule known to bind and concentrate growth factors or chemokines such as GM-CSF and CXCL12 within specific niches.51–54 Others46 have demonstrated the potential of CD45 in Jurkat cells to interact in cis with CXCR4, where it functions as a tyrosine phosphatase, deactivating components of the CXCR4 signaling pathway.
In summary, our studies indicate that CD164, or endolyn, on CD133+ cells and leukemic Jurkat cells, regulates their CXCL12-mediated homing. These studies provide the first demonstration of the effects of CD164 on CXCR4 function in vitro and pave the way for more detailed analysis of the effects of CD164 knockdown on hematopoietic stem cell homing and engraftment in vivo and the further identification of additional cognate ligands for CD164.
Authorship
Contribution: S.M.W., S.F., B.J.T., and S.E.N. contributed to research funding and design, data analysis, and manuscript writing; S.F., B.J.T., S.E.N., and M.R. performed the experiments; S.M.W., S.F., B.J.T., M.R., S.E.N., J.S., C.P.M., and R.P. designed the study protocols; and S.M.W. and J.S. obtained ethics approval and cord blood provision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
S.F. and B.J.T. contributed equally to this study.
Correspondence: Suzanne Watt, Stem Cell Laboratory, National Blood Service, National Blood and Transplant Authority, The John Radcliffe Hospital, Headington, Oxford; OX3 9BQ, United Kingdom; e-mail: suzanne.watt@nbs.nhs.uk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work is supported by grants from the Leukaemia Research Fund (S.M.W., B.J.T., M.R.), the Wellcome Trust (S.E.N., S.M.W.), and the National Blood and Transplant Authority (S.M.W., S.F., B.J.T., S.E.N., M.R., J.S.). The research was carried out at the laboratories of the National Health Service (NHS) Blood and Transplant Authority in Oxford and the Nuffield Department of Clinical Laboratory Sciences, University of Oxford, and benefits from research and development funding received from the NHS Research and Development (R&D) Directorate. S.E.N. holds a Wellcome Trust Travelling Fellowship.
We thank Professor M. Contreras for her support, Dr Kingsley Michlem for assistance with the confocal microscope, and Mr Robin Roberts-Gant for help with the images.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal