Abstract
Gfi1b and Gfi1 are 37- and 55-kDa transcriptional repressors that share common features such as a 20-amino acid (aa) N-terminal SNAG domain, a nonconserved intermediary domain, and 6 highly conserved C-terminal zinc fingers. Both gene loci are under autoregulatory and cross-regulatory feedback control. We have generated a reporter mouse strain by inserting the cDNA for green fluorescent protein (GFP) into the Gfi1b gene locus which allowed us to follow Gfi1b expression during hematopoiesis and lymphopoiesis by measuring green fluorescence. We found highly dynamic expression patterns of Gfi1b in erythroid cells, megakaryocytes, and their progenitor cells (MEPS) where Gfi1 is not detected. Vice versa, Gfi1b could not be found in granulocytes, activated macrophages, or their granulomonocytic precursors (GMPs) or in mature naive or activated lymphocytes where Gfi1 is expressed, suggesting a complementary regulation of both loci during hematopoiesis. However, Gfi1b was found to be up-regulated in early stages of B-cell and in a subset of early T-cell development, where Gfi1 is also present, suggesting that cross-regulation of both loci exists but is cell-type specific.
Introduction
Normal hematopoiesis is dependent on the proper regulation of self-renewal, proliferation, and differentiation of stem cells and multipotent and lineage-committed progenitors, from which all hematopoietic lineages originate. The generation and maintenance of a steady state level of multiple types of mature blood cells is a complex process, which requires several levels of control. This is provided by the orchestrated activity of many regulatory proteins, including cytokines such as interleukins or erythropoietin, which are important for the communication among different cells of the hematopoietic system or between supporting stoma cells and hematopoietic cells. The information transmitted by these factors is received by specific cell-surface receptors and is translated into an adequate response, frequently by activating the JAK/STAT (Janus kinase/signal transducer and activator of transcription) and the MAPK (mitogen activated protein kinase) signal transduction cascades but also other signaling pathways. Feedback inhibitors such as the SOCS (suppressor of cytokine signaling) and PIAS (protein inhibitor of activated STAT) family of proteins are involved in the fine-tuning of these signaling processes, which finally terminate in the nucleus by stimulating the synthesis of activating or repressing numerous transcription factors, which, in turn, can regulate the expression or the activity of these modulators.
The transcriptional repressors Gfi1 and Gfi1b are paradigmatic examples of transcription factors with regulatory function in hematopoiesis. It has been shown that Gfi1b can repress SOCS-1 and SOCS-3 expression,1 whereas Gfi1 modulates the activity of PIAS-3 and the STAT3 pathway by directly interacting with PIAS-3.2 Gfi1b and Gfi1 encode nuclear proteins with 3 identifiable domains, an N-terminal 20-amino acid (aa) repressory snail-Gfi1 (SNAG) domain,3 a nonconserved intermediate domain of 144 aa or 237 aa, respectively, of unknown function, and 6 highly conserved C-terminal zinc-fingers (ZFs). ZFs 3 to 5 of Gfi1 and Gfi1b have been shown to be necessary for binding to their common consensus DNA recognition sequence taAATCac(t/a)gca.4,5 The function of the intermediate domain and the first 3 ZFs remains to be precisely defined, but it is conceivable that they provide protein-protein interaction interfaces.
Gfi1 is expressed mainly in thymic T cells, granulocytes, and activated macrophages but also in the lung, sensory epithelia, neuronal cells, hematopoietic precursors, and stem cells, and the expression dynamics during the development of hematopoietic cells has been well elucidated, also with the help of a GFP reporter mouse.3,6–9 In contrast, much less is known about the expression pattern of Gfi1b during development and hematopoietic differentiation, although expression has been detected in hematopoietic stem cells, erythroid cells, and megakaryocytes.10 Overexpression of Gfi1b can induce peripheral T lymphopenia and inhibits myeloid differentiation of cultured M1 cells.4 Deletion of Gfi1b in mice leads to an arrest of embryonic development at day E15, probably as a result of delayed maturation of primitive erythrocytes and failure to produce definitive enucleated erythrocytes.11 The deletion of one Gfi1b allele has no apparent consequences, and heterozygous animals develop normally.
Very little is known about the expression of Gfi1b in other cell types outside the hematopoietic system, and the expression dynamics during development or differentiation of hematopoietic cells is virtually unknown. To obtain more information and a better insight in which cells of the fetal and adult hematopoietic system and at what stages of differentiation Gfi1b is expressed, we decided to generate Gfi1b:GFP knock-in mutant mice by targeting the EGFP cDNA into the coding region of the Gfi1b locus by homologous recombination. Heterozygous Gfi1b+/GFP mice express Gfi1b from one allele and GFP from the other and proved to be a fully functional reporter mouse that allowed us to follow Gfi1b expression by GFP fluorescence. We present here an extensive analysis of relative levels of Gfi1b expression by monitoring GFP fluorescence, from hematopoietic stems cells (HSCs) and early progenitors to fully differentiated mature cells of all hematopoietic lineages. We find a surprisingly dynamic expression pattern of Gfi1b that is largely complementary to Gfi1 with the exception of few cell types that coexpress both proteins. Our findings underline the importance of Gfi1 family proteins during commitment and maturation of many hematopoietic-cell populations.
Materials and methods
Generation of Gfi1b:GFP knock-in mice
The pBluescript-based targeting vector contained a 5.4-kilobase (kb) upstream fragment from the Gfi1b locus from an 5′-XmnI site to the first ATG of Gfi1b and a polymerase chain reaction (PCR)–generated 1.7-kb downstream fragment starting from the STOP codon in exon 7 preserving the complete 3′-untranslated region of the cDNA of Gfi1b. SalI sites were introduced at both ends of the 3′ fragment, and the fragment was cloned in a pBluescript-based vector where the cDNA for the diphtheria-toxin A protein was introduced as a selection marker to increase the number of embryonic stem-cell (ES) clones carrying an integration of the construct at the right locus. The EGFP open reading frame was fused to the 5′ fragment by overlap PCR from a NdeI site 570 base pair (bp) 5′ of the ATG to the STOP codon of GFP generating an XbaI site at the 3′ end. A thymidine kinase-neomycin resistance fusion cassette, flanked by loxP sites, was cloned between the SalI site at the 3′ end of the GFP cassette and the XbaI site of the 3′-untranslated region as a selection marker. Gfi1bGFP+/neo mice were generated by homologous recombination into R1 ES cells using previously described procedures. Transfection of the ES cells and selection of clones were done as described before.12 Successful homologous recombination was analyzed by Southern blot on EcoRI/SalI-digested genomic DNA using an external 3′ probe. Excision of the floxed TK-neo cassette was achieved by crossing of Gfi1b:GFP+/GFP-neo heterozygotes with cytomegalovirus (CMV)–]RE recombinase transgenic animals. Successful excision of the TK-neo cassette was verified by Southern blot using HindIII/SalI-digested mouse tail genomic DNA and a 5′-external probe.
Genotyping of mice and mouse embryos by PCR
The genotype of mice and wt, heterozygous, and homozygous knock-in embryos was determined by multiplex PCR covering either sequence from intron 1 to intron 2 of the Gfi1b allele or from intron 1 into GFP from the targeted allele. The 6- to 10-week-old mice analyzed in this study were maintained under specific pathogen-free conditions and backcrossed onto the C57Bl/6 background for more than 6 generations.
Antibodies and flow cytometry
For flow cytometry, single-cell suspensions were prepared from all tissues analyzed in staining solution (PBS supplemented with 1% fetal calf serum [FCS]). Erythrocytes were lysed in Red Blood Cell Lysing Buffer (Sigma-Aldrich Chemie, Taufkirchen, Germany) when necessary. Cells were washed in staining buffer and incubated for 20 minutes with antibodies against surface antigens as mentioned in figure legends, which were either biotinylated or directly fluorochrome coupled. All data were collected using a fluorescence-activated cell sorting (FACS)Calibur (BD Biosciences, Heidelberg, Germany) and were analyzed with FlowJo software (TreeStar, San Carlos, CA). For selection of lineage-negative (Lin−) cells, biotinylated antibodies were used against CD3, CD4, CD8, Mac-1, GR-1, Ter119, DX, and IL7 receptor (for CLP staining the IL7R AB was not included; for CMP, GMP, MEP staining Sca1 AB was added) labeled with streptavidin-APC. For the analysis of MEPs, CMP, GMPs, and CLPs, Lin− cells were presorted using a FACSDiVa machine (BD Biosciences) and then stained with antibodies against c-Kit, CD34, Fcγ-receptor, Sca1, or IL7-receptor coupled to PE, PerCP-Cy5.5, or APC as needed. For analysis of the thymic DN population, cells were stained with biotinylated lineage marker followed by streptavidin-PerCP-Cy5.5 and APC- or PE-labeled anti-CD44 and anti-CD25 antibodies. For the analysis of B cells and B-cell development, APC-, PE-, or PerCP-Cy5.5–coupled antibodies against B220, IgM, CD25, or CD43 were used. LPS- or CD40- or CD40 + IL4-induced activation of B cells was verified using an antibody against CD86. Splenic memory T cells were identified using a combination of antibodies against CD44 and CD62L plus antibodies against CD4 or CD8. For flow cytometric analysis of megakaryocyte progenitor cells, bone marrow cells were isolated from Gfi1bwt/wt and Gfi1bwt/GFP mice, and the expression of green fluorescence among highly pure megakaryocyte progenitor cells were determined. Bone marrow cells were incubated with the following lineage-marker antibodies: CD3 (17A2), CD4 (GK1.5), CD8 (53.6.7), B220 (RA3-6B2), Gr-1 (8C5), Mac-1 (M1/70), TER119, Thy1.1 (HIS51), IL-7Rα (A7R34), and Sca-1 (E13-161-7). Then the cells were incubated with anti–rat IgG micro beads (Miltenyi Biotec, Bergisch Gladbach, Germany), and Lin+IL7Rα+Sca-1+ cells were removed. Resulting cells were stained with APC-conjugated anti–rat IgG polyclonal antibodies (Pharmingen, Heidelberg, Germany) and then washed twice with PBS containing 2% FCS to remove remaining antibodies. Then the cells were stained with PE-Cy7–conjugated anti-FcγRII/III (93; eBioscience, San Diego, CA), PE-conjugated anti-CD41 (MWReg30; Pharmingen), PE-Cy5–conjugated anti–c-Kit (2B8; eBioscience), and biotin-conjugated anti-CD9 (KMC8; Pharmingen). The cells were washed once and further incubated with PE-Texas Red–conjugated streptavidin. The cells were analyzed using a Moflo flow cytometric analyzer (Dako, Mississauga, ON, Canada). All antibodies used in FACS analysis were purchased from BD Biosciences. Antibodies against Gfi1b or GFP were from Santa Cruz Biotechnology (Heidelberg, Germany) (sc-8559, sc-9996).
Cell culture
Megakaryocytes were generated from bone marrow from freshly harvested femurs and tibias of Gfi1b+/GFP mice. Femurs and tibias were flushed with phosphate-buffered salt solution (PBS) containing 1% BSA. Mature red blood cells were removed by a 5-minute room temperature incubation in 3 mL Red Blood Cell Lysing Buffer (Sigma), and cells were resuspended in growth medium. Megakaryocytes were enriched by cultivation in Dulbecco Modified Eagles Medium (DMEM) supplemented with 10% fetal calf serum (Gibco-BRL, Eggenstein, Germany), 100 U penicillin G sodium/mL, 0.1 mg streptomycin sulfate, 10 ng/mL thrombopoietin (TPO; R&D Systems, Wiesbaden, Germany) and 10 ng/mL IL3 (PeproTech, Rocky Hill, NJ) for 5 days. Cytospins were prepared from about 5 × 104 nonadherent cells, and GFP expression was detected by fluorescence microscopy. Wright-Giemsa staining was performed following a standard procedure as proposed by the supplier (Sigma-Aldrich, Deisenhofen, Germany). B cells or T cells were either harvested from bone marrow (as described under “Antibodies and flow cytometry”), or as single-cell suspensions from other freshly prepared tissues. Red blood cells were removed as described above under “Antibodies and flow cytometry.” B cells and T cells were cultured in RPMI supplemented with 10% fetal calf serum, 100 U penicillin G sodium/mL, 0.1 mg streptomycin sulfate, 10 μm HEPES-KOH, 50 nM β-mercaptho ethanol, and 1 × MEM (all from Gibco-BRL).
Quantitative real-time PCR
For analysis of Gfi1b mRNA during erythroid development, bone marrow cells prepared from heterozygous Gfi1b+/GFP knock-in mice were subjected to mature red blood cell lysis and stained with an APC-labeled anti-Ter119 antibody. Ter119+ cells were sorted using a FACSDiVa machine (BD Biosciences) for low, medium, or high GFP expression. For analysis of Gfi1b or Gfi1 mRNA during early T-cell development or early B-cell development, cells were stained as described in “Antibodies and flow cytometry” and purified using FACS. RNA was prepared from 5 × 105 cells using peqGOLD RNAPure reagent as proposed by the supplier (peQLab, Erlangen, Germany), and 1 μg total RNA was DnaseI digested and used for reverse transcriptase (RT) reactions (20 μL). For quantitative analysis of Gfi1b or GFP expression, real-time PCR was performed in a 20-μL reaction volume containing 900 nM of each primer, 250 nM TaqMan probe, and TaqMan Universal PCR Master Mix (ABI Instruments, Freiberg, Germany) according to the manufacturer's instructions in an ABI PRISM 7700 Sequence detection system (Applied Biosystems, Weiterstadt, Germany). Assays on demand were used for Gfi1b (Mm 00492319_m1) Gfi1 (Mm 00515853_m1), and glyceraldehyde phosphate dehydrogenase (GAPDH) (Mm 99999915_g1). A specific probe was generated for the amplification of EGFP (forward, ggacgacggcaactacaaga; reverse, tcagctcgatgcggttcac; probe, aagttcgagggcgacacc). PCR was performed in triplicates. The expression of Gfi1b or GFP was calculated as difference of PCR cycles (ct probe minus ct GADPH) at middle of logarythmic phase as calculated by the acquisition software (SDS 2.2; Applied Biosystems, Foster City, CA). Animal experiments were carried out at the Institute für Zellbiologie in Essen, Germany, and were performed under a license granted by the Bezirksregierung Düsseldorf (NRW), Germany (Nr. G022/02Z).
Results
Generation of Gfi1b:GFP knock-in mice
Gfi1b:GFP reporter mice were generated by replacing the entire coding sequence of Gfi1b by the GFP-cDNA to avoid expression of residual Gfi1b coding sequences (Figure 1A-C). The TK-neo selection cassette was removed by crossing chimeric mice into a transgenic CMV-CRE recombinase expressing deleter strain. After CRE recombinase-mediated excision of the selection cassette (Figure 1B), the mutated allele carried the GFP gene inserted in frame with the Gfi1b ATG translation initiation (exon 2) and the TGA termination codon (exon 7) and both the 5′-transcription regulatory elements and the 3′-untranslated region, which may regulate RNA stability, were preserved. As expected from the published Gfi1b knock-out,11 intercrossing of Gfi1b+/GFP mice never gave rise to adult homozygous mutant mice, whereas heterozygous (wt/KI) mice were born and were indistinguishable from wt mice (data not shown). Gfi1bGFP/GFP knock-out embryos (termed KI/KI) developed to the E15 stage but were pale and showed hemorrhages (Figure 1D, top). Fluorescence microscopy on freshly prepared E13.5 Gfi1b+/GFP embryos revealed that expression of Gfi1b was largely restricted to the fetal liver, where hematopoiesis takes place (Figure 1D, middle). A whole-mount staining for the presence of CD31 revealed that the vascularization process of the Gfi1bGFP/GFP embryos was indistinguishable from that in wt or Gfi1b+/GFP embryos (Figure 1D, bottom). To verify the absence of Gfi1b expression in Gfi1bGFP/GFP embryos and the proper induction of GFP expression, protein extracts from E14.5 fetal liver cells were analyzed with antibodies against Gfi1b or GFP. Gfi1b was readily detected in wt cells but was absent in Gfi1bGFP/GFP cells, whereas GFP was strongly expressed in Gfi1bGFP/GFP cells and absent in wt cells (Figure 1E).
Generation of Gfi1b:GFP knock-in mice. (A) Schematic representation of the targeting construct design and simplified restriction maps of the wt and targeted alleles after CRE recombinase excision of the thymidine kinase-neomycin cassette (TK-neo) by crossing of heterozygous mice with CRE-transgenic mice. All coding sequences of Gfi1b are replaced by the GFP open reading frame. lxP indicates lox-P site; H, HindIII; E, EcoRI; N, NotI; Sl, SalI; B, BamHI; DTA, diphtheria toxin; 1-7, exon 1 to 7; GFP, green fluorescent protein. (B) Southern blotting to detect excision of the thymidine kinase-neomycin resistance gene fusion protein (TK-neo) cassette in mice after gene targeting and CRE-recombination showed the expected 8-kb fragment using a 5′-external probe. (C) PCR strategy and results to detect the insertion of the GFP cDNA into the Gfi1b locus in mouse embryo tail DNA representing the embryos shown in panel D. Because all 5′ sequences are retained, but intron 2 to intron 6 were deleted in the targeted allele, the wt, wt/KI (Gfi1b+/GFP), and KI/KI (Gfi1bGFP/GFP) mice were analyzed by multiplex PCR covering either intron 1 to 2 or intron 1 to GFP (Figure 1C) to demonstrate proper targeting. (D) Appearance of (top panel) and detection of GFP fluorescence in day 13.5 postconception (pc) embryos which were wt, heterozygous, or homozygous for the GFI1b:GFP allele (wt, wt/KI [Gfi1b+/GFP] and KI/KI [Gfi1bGFP/GFP], bottom panel). In heterozygous embryos, GFP expression is restricted to the fetal liver, where erythropoiesis takes place. (Bottom panel) CD31 whole-mount staining showing normal vascularization in wt and mutant embryos. Embryos were analyzed with the Leica MZ/FLIII stereomicroscope (Leica, Wetzlar, Germany) and photographed with the KAPPA CF 15/4 MC camera (KAPPA Opto-Electronics, Gleichen, Germany) using 0.5× magnification (0.125 NA). (E) Western blot analysis of whole-cell extracts from fetal liver cells of wt or homozygous GFP knock-in embryos at day 13.5 pc showing the loss of Gfi1b expression (top) and gain of GFP expression (bottom) in Gfi1b:GFP knock-in mice.
Generation of Gfi1b:GFP knock-in mice. (A) Schematic representation of the targeting construct design and simplified restriction maps of the wt and targeted alleles after CRE recombinase excision of the thymidine kinase-neomycin cassette (TK-neo) by crossing of heterozygous mice with CRE-transgenic mice. All coding sequences of Gfi1b are replaced by the GFP open reading frame. lxP indicates lox-P site; H, HindIII; E, EcoRI; N, NotI; Sl, SalI; B, BamHI; DTA, diphtheria toxin; 1-7, exon 1 to 7; GFP, green fluorescent protein. (B) Southern blotting to detect excision of the thymidine kinase-neomycin resistance gene fusion protein (TK-neo) cassette in mice after gene targeting and CRE-recombination showed the expected 8-kb fragment using a 5′-external probe. (C) PCR strategy and results to detect the insertion of the GFP cDNA into the Gfi1b locus in mouse embryo tail DNA representing the embryos shown in panel D. Because all 5′ sequences are retained, but intron 2 to intron 6 were deleted in the targeted allele, the wt, wt/KI (Gfi1b+/GFP), and KI/KI (Gfi1bGFP/GFP) mice were analyzed by multiplex PCR covering either intron 1 to 2 or intron 1 to GFP (Figure 1C) to demonstrate proper targeting. (D) Appearance of (top panel) and detection of GFP fluorescence in day 13.5 postconception (pc) embryos which were wt, heterozygous, or homozygous for the GFI1b:GFP allele (wt, wt/KI [Gfi1b+/GFP] and KI/KI [Gfi1bGFP/GFP], bottom panel). In heterozygous embryos, GFP expression is restricted to the fetal liver, where erythropoiesis takes place. (Bottom panel) CD31 whole-mount staining showing normal vascularization in wt and mutant embryos. Embryos were analyzed with the Leica MZ/FLIII stereomicroscope (Leica, Wetzlar, Germany) and photographed with the KAPPA CF 15/4 MC camera (KAPPA Opto-Electronics, Gleichen, Germany) using 0.5× magnification (0.125 NA). (E) Western blot analysis of whole-cell extracts from fetal liver cells of wt or homozygous GFP knock-in embryos at day 13.5 pc showing the loss of Gfi1b expression (top) and gain of GFP expression (bottom) in Gfi1b:GFP knock-in mice.
Dynamic expression of Gfi1b during fetal erythroid development
Next, we analyzed the relative expression levels of Gfi1b during fetal erythroid maturation in Gfi1b+/GFP embryos at E14.5 by following GFP fluorescence and the expression of the erythroid markers c-Kit and Ter11913 or CD71 and Ter119.14 We electronically gated different maturation stages (Figure 2A, left) and plotted their median GFP fluorescence against the gate numbers (Figure 2A, right). GFP fluorescence rose sharply with c-Kit expression, remained high at intermediate stages expressing Ter119lo/int levels and decreased with increasing Ter119 levels, suggesting a peak of Gfi1b expression in early cKithi/Ter119int erythroblasts (Figure 2A). Similarly, GFP expression rose with CD71 up-regulation and decreased when Ter119 levels increased (Figure 2B, left). In the absence of Gfi1b, the expression of GFP increased significantly compared with cells from Gfi1+/GFP mice (Figure 2B, right). In Gfi1bGFP/GFP embryos, only a small fraction of the cells fully differentiated, and an accumulation of a Ter119int/CD71int or a Ter119int/cKitint immature population was observed (Figure 2B, right).
Gfi1b is a developmental marker during erythroid maturation in fetal liver. (A) Analysis of GFP expression (right) of Gfi1b+/GFP fetal liver cells stained with cKit-PE and Ter119-APC and gated for erythroid maturation stages as indicated (left). Given are the MFIs (median fluorescence intensities) of the indicated populations. (B) FACS analysis of GFP:Gfi1b expression of fetal liver cells from Gfi1b+/GFP or Gfi1bGFP/GFP embryos at 14.5 days pc stained with CD71-PE and Ter119-APC and gated as indicated. Given are the MFIs of the indicated populations. FACS analyses are representative for at least 3 independent experiments with individual sets of mice.
Gfi1b is a developmental marker during erythroid maturation in fetal liver. (A) Analysis of GFP expression (right) of Gfi1b+/GFP fetal liver cells stained with cKit-PE and Ter119-APC and gated for erythroid maturation stages as indicated (left). Given are the MFIs (median fluorescence intensities) of the indicated populations. (B) FACS analysis of GFP:Gfi1b expression of fetal liver cells from Gfi1b+/GFP or Gfi1bGFP/GFP embryos at 14.5 days pc stained with CD71-PE and Ter119-APC and gated as indicated. Given are the MFIs of the indicated populations. FACS analyses are representative for at least 3 independent experiments with individual sets of mice.
Dynamic expression of Gfi1b in the erythroid and megakaryocytic lineage of adult mice
As in the fetal liver, Gfi1b expression was found to be highly regulated in adult bone marrow judged from the analysis of GFP fluorescence in cells from Gfi1b+/GFP mice using Ter119 and CD71 as developmental markers (Figure 3A). GFP was almost undetectable in Ter119−, CD71− cells, low in Ter119−, CD71+ cells which comprise the CFU-E stage, and highest in Ter119high, CD71high late erythroblasts (Figure 3A). In terminally differentiated Ter119high, CD71lo/int cells GFP expression decreased sharply (Figure 3A). Similar high levels of GFP expression were also observed in cultured bone marrow cells from Gfi1b+/GFP mice induced by erythropoietin (EPO) (Figure S2A, lower panel, available on the Blood website; see the Supplemental Figures link at the top of the online article).
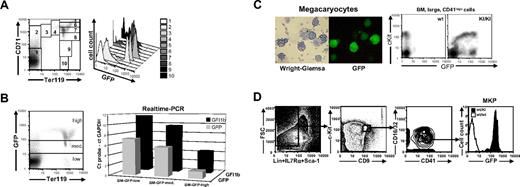
Gfi1b:GFP expression during erythroid development in bone marrow and cultured megakaryocytes. (A) Living cells from bone marrow of Gfi1b+/GFP mice were stained with CD71-PE and Ter119-APC and subjected to FACS analysis (left). Cells were gated for different developmental stages, and GFP expression of these cells is shown in a histogram (right). (B) Living cells from bone marrow of Gfi1b+/GFP mice were stained with Ter119-APC and sorted for low, medium, or high GFP expression (left). Transcripts from the Gfi1b allele (▪) and the GFP allele (⊡) were quantatized by real-time PCR. Shown are the Δct values where the Gfi1b or GFP transcripts become detectable relative to the transcripts of the glyceraldehyde-3-phosphate dehydrogenase gene on the same threshold line. (C) Megakaryocytes were generated from bone marrow of Gfi1b+/GFP mice by cultivation in DMEM in the presence of 10 ng/mL thrombopoietin (TPO) and 10 ng/mL IL3 for 5 days. Cytospins were prepared from nonadherent cells, and GFP expression was detected by fluorescence microscopy (right). Afterward, cells were stained according to the Wright-Giemsa protocol and photographed using visible light (left). (Rightmost) Flow cytometric analysis of c-Kit expression and GFP fluorescence in bone marrow–derived, large CD41-expressing cells. Cells were analyzed using a Leica DM IRB fluorescence microscope with an L 20×/0.4 NA Corr PH1 objective lens and photographed using a Nikon Coolpix 990 digital camera (Nikon, Düsseldorf, Germany) and LM-scope Adapter (Micro Tech Lab Rudnicki KEG, Graz, Austria). (D) Flow cytometric analysis of bone marrow cells after depletion of Lin+IL7Rα+Sca-1+ cells. Gatings for highly pure megakaryocyte progenitor population and expression of green fluorescense among these cells are shown. The data shown is representative of 3 similar data obtained from individual experiments.
Gfi1b:GFP expression during erythroid development in bone marrow and cultured megakaryocytes. (A) Living cells from bone marrow of Gfi1b+/GFP mice were stained with CD71-PE and Ter119-APC and subjected to FACS analysis (left). Cells were gated for different developmental stages, and GFP expression of these cells is shown in a histogram (right). (B) Living cells from bone marrow of Gfi1b+/GFP mice were stained with Ter119-APC and sorted for low, medium, or high GFP expression (left). Transcripts from the Gfi1b allele (▪) and the GFP allele (⊡) were quantatized by real-time PCR. Shown are the Δct values where the Gfi1b or GFP transcripts become detectable relative to the transcripts of the glyceraldehyde-3-phosphate dehydrogenase gene on the same threshold line. (C) Megakaryocytes were generated from bone marrow of Gfi1b+/GFP mice by cultivation in DMEM in the presence of 10 ng/mL thrombopoietin (TPO) and 10 ng/mL IL3 for 5 days. Cytospins were prepared from nonadherent cells, and GFP expression was detected by fluorescence microscopy (right). Afterward, cells were stained according to the Wright-Giemsa protocol and photographed using visible light (left). (Rightmost) Flow cytometric analysis of c-Kit expression and GFP fluorescence in bone marrow–derived, large CD41-expressing cells. Cells were analyzed using a Leica DM IRB fluorescence microscope with an L 20×/0.4 NA Corr PH1 objective lens and photographed using a Nikon Coolpix 990 digital camera (Nikon, Düsseldorf, Germany) and LM-scope Adapter (Micro Tech Lab Rudnicki KEG, Graz, Austria). (D) Flow cytometric analysis of bone marrow cells after depletion of Lin+IL7Rα+Sca-1+ cells. Gatings for highly pure megakaryocyte progenitor population and expression of green fluorescense among these cells are shown. The data shown is representative of 3 similar data obtained from individual experiments.
To test whether GFP expression faithfully reflects the expression of Gfi1b we stained bone marrow cells of Gfi1b+/GFP mice for Ter119 and analyzed GFP fluorescence (Figure 3B, left). Different cell populations were sorted by flow cytometry according to their GFP fluorescence intensities (GFP-high, -medium, and -low), and both GFP and Gfi1b mRNA expressions were analyzed by quantitative real-time RT-PCR. Although the GFP probe was more sensitive, a clear correlation was observed between GFP and Gfi1b expression levels, indicating that GFP expression directly correlates with Gfi1b mRNA expression.
According to a previous study, Gfi1b is also required for the maturation of megakaryocytes but not for their commitment.11 Murine bone marrow was enriched in culture for megakaryocytes by TPO and IL3 and analyzed for green fluorescence. Megakaryocytes from Gfi1b+/GFP mice were found to be morphologically normal but displayed large variations in GFP expression (Figure 3C). As an alternative source, CD41+ cells, which mainly contain megakaryocytes, were freshly prepared from wt and Gfi1b+/GFP bone marrow and analyzed for c-Kit and GFP expression. All c-Kit–positive cells also expressed GFP, whereas only a small fraction of c-Kit–negative cells were positive for GFP fluorescence (Figure 3C, rightmost panel), suggesting a regulation of Gfi1b expression during megakaryocyte maturation. To address this in more detail, highly purified megakaryocyte precursor cells from Gfi1b+/GFP bone marrow were analyzed for GFP expression (Figure 3D). Nearly all cells (> 97%), which resemble about 0.01% of total nucleated bone marrow cells show a high expression of GFP, corresponding well to the GFP level of the CD41+, cKithigh cells described in Figure 3C.
Gfi1b is differentially expressed in stem cells, early progenitors, and myeloid compartment
Next, we analyzed the expression of Gfi1b by use of GFP as a marker in HSCs, common lymphoid (CLP), common myeloid (CMP), granulomonocytic (GMP), and megakaryocytes/erythroid (MEP) progenitor cells. CLPs showed a heterogeneous GFP expression, ranging from high to near negative values (Figure 4C), but HSCs, CMPs, and MEPs expressed very distinct, readily detectable levels of GFP in single populations, whereas greater than 90% of GMPs were negative for GFP (Figure 4A-B). The GMP fraction contains some highly GFP-positive cells (about 9%), which were repeatedly detected in different experiments (data not shown) but whose nature is unknown. Because their GFP expression level does not correspond to CMPs or MEPs, a cross-contamination of these populations is unlikely. To test whether Gfi1b expression is maintained in the myeloid lineage, we measured GFP expression in Mac-1–positive cells of different tissues from Gfi1b+/GFP (wt/KI) mice and Gfi1bGFP/GFP (KI/KI) embryos (Figure 5A). Surprisingly, GFP was detected in a major proportion of Mac-1+ cells derived from the peripheral blood but was absent in the same cell type when derived from spleen, lymph nodes, bone marrow, or fetal liver, where also Gfi1bGFP/GFP cells remained GFP negative (Figure 5A). Also, when B220, Mac-1, and Gr1 were used as markers to further discriminate between granulocytes and monocytes, we found that both granulocytes (B220neg/Gr1high/Mac1high) and monocytes (Gr1int/Mac1high) were GFP negative when derived from bone marrow (Figure 5B, BM). However, both cell types showed a high, virtually equivalent GFP fluorescence, when derived from peripheral blood (Figure 5B, PB).
Gfi1b expression in early precursor and hematopoietic stem cells. (A-C) Bone marrow cells from wt or wt/KI (Gfi1b+/GFP) mice, respectively, were depleted for lineage-positive cells (Lin: CD3, CD4, CD8, Mac-1, B220, Ter119, Ly6G, Pan natural killer [NK] plus Sca1, IL7Rα for CMP-GMP-MEP analysis), and negative cells were restained for surface markers as indicated and analyzed by flow cytometry (A-C, left panels). HSCs (A), MEPs, CMPs, GMPs (B), or CLPs (C) were electronically gated as previously described (see “Material and methods”) and analyzed for GFP expression (right panels).
Gfi1b expression in early precursor and hematopoietic stem cells. (A-C) Bone marrow cells from wt or wt/KI (Gfi1b+/GFP) mice, respectively, were depleted for lineage-positive cells (Lin: CD3, CD4, CD8, Mac-1, B220, Ter119, Ly6G, Pan natural killer [NK] plus Sca1, IL7Rα for CMP-GMP-MEP analysis), and negative cells were restained for surface markers as indicated and analyzed by flow cytometry (A-C, left panels). HSCs (A), MEPs, CMPs, GMPs (B), or CLPs (C) were electronically gated as previously described (see “Material and methods”) and analyzed for GFP expression (right panels).
Expression of Gfi1b in granulocytes and monocytes of the adult mouse. (A) Cells from different tissues of the hematopoietic system of adult wt or Gfi1b+/GFP mice or from fetal livers of wt, wt/KI (Gfi1b+/GFP), or KI/KI (Gfi1bGFP/GFP) embryos, respectively, were stained with antibodies against the surface markers B220 and Mac-1 and subjected to FACS analysis. B220-negative but Mac-1–positive living cells were electronically gated and analyzed for GFP expression. (B) Cells from bone marrow (BM) or peripheral blood (PB) from adult mice of the indicated genotypes were additionally stained for GR1 and analyzed by FACS. GR1high granulocytes and GR1low monocytes were electronically gated (left) and analyzed for GFP expression (right).
Expression of Gfi1b in granulocytes and monocytes of the adult mouse. (A) Cells from different tissues of the hematopoietic system of adult wt or Gfi1b+/GFP mice or from fetal livers of wt, wt/KI (Gfi1b+/GFP), or KI/KI (Gfi1bGFP/GFP) embryos, respectively, were stained with antibodies against the surface markers B220 and Mac-1 and subjected to FACS analysis. B220-negative but Mac-1–positive living cells were electronically gated and analyzed for GFP expression. (B) Cells from bone marrow (BM) or peripheral blood (PB) from adult mice of the indicated genotypes were additionally stained for GR1 and analyzed by FACS. GR1high granulocytes and GR1low monocytes were electronically gated (left) and analyzed for GFP expression (right).
Gfi1b is switched on in early and off during late B-cell development
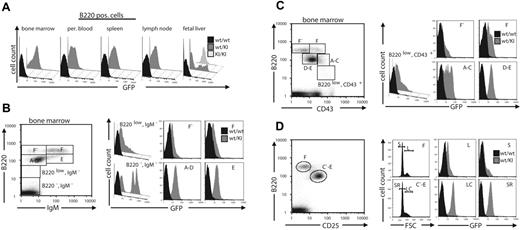
We knew from previous experiments that Gfi1b is expressed at least in a subset of B cells.15,16 Thus, we analyzed GFP fluorescence during B-cell development. Because only B220+ cells from bone marrow showed any GFP fluorescence (Figure 6A), we analyzed GFP levels in bone marrow–derived B-cell subpopulations characterized by the surface markers B220, IgM, CD43, and CD25 (Figure 6B-D). GFP was clearly present in a distinct population representing the pro–B-cell to immature B-cell stages (Hardy fractions A-E; Figure 6 B-C). B220low, IgM− cells included only few, low-level GFP–expressing cells (Figure 6B, right panel, upper left), whereas B220high, IgM− cells, which contain the pre-pro-B to pre-B stages all showed clearly detectable, homogeneous GFP fluorescence which was sustained in immature B cells (Hardy fraction E) and declined to near undetectable levels when B cells further maturate (Hardy fractions F-F′).
Dynamic expression of Gfi1b during B-cell development. (A) Cells from different tissues of the hematopoietic system of adult wt or Gfi1b+/GFP mice or from fetal livers of wt, wt/KI (Gfi1b+/GFP or KI/KI (Gfi1bGFP/GFP) embryos, respectively, were stained with antibodies against the surface markers B220 and Mac-1 and subjected to FACS analysis. Mac-1–negative, B220-positive living cells were electronically gated and analyzed for GFP expression. (B-D) Bone marrow cells from adult wt or wt/KI (Gfi1b+/GFP) mice were stained with antibodies against B220 and IgM (B), CD43 (C), or CD25 (D) and analyzed by FACS (left panels). Fractions of cells representing the different maturation stages of B cells (A indicates pre-pro-B; B/C, pro-B; C, large pre-B; D, small pre-B; E, immature B; F-F, mature B cell)17,18 were electronically gated as indicated (left panel) and analyzed for GFP expression (histograms in right panels). (D) B220-positive cells were gated for high and low CD25 expression, and cell size distribution was separated by forward angle light scatter analysis (right panel, left histograms). GFP fluorescence histograms for large and small cells (L indicates large B-cell; S, small B-cell; LC, large cycling B cell; SR, small resting B cell) were generated (right panel, right histograms) as indicated.
Dynamic expression of Gfi1b during B-cell development. (A) Cells from different tissues of the hematopoietic system of adult wt or Gfi1b+/GFP mice or from fetal livers of wt, wt/KI (Gfi1b+/GFP or KI/KI (Gfi1bGFP/GFP) embryos, respectively, were stained with antibodies against the surface markers B220 and Mac-1 and subjected to FACS analysis. Mac-1–negative, B220-positive living cells were electronically gated and analyzed for GFP expression. (B-D) Bone marrow cells from adult wt or wt/KI (Gfi1b+/GFP) mice were stained with antibodies against B220 and IgM (B), CD43 (C), or CD25 (D) and analyzed by FACS (left panels). Fractions of cells representing the different maturation stages of B cells (A indicates pre-pro-B; B/C, pro-B; C, large pre-B; D, small pre-B; E, immature B; F-F, mature B cell)17,18 were electronically gated as indicated (left panel) and analyzed for GFP expression (histograms in right panels). (D) B220-positive cells were gated for high and low CD25 expression, and cell size distribution was separated by forward angle light scatter analysis (right panel, left histograms). GFP fluorescence histograms for large and small cells (L indicates large B-cell; S, small B-cell; LC, large cycling B cell; SR, small resting B cell) were generated (right panel, right histograms) as indicated.
CD43high cells expressing B220 on a very low level (B220vlow) contained many GFP-negative and only very few GFP-positive cells, whereas the number of GFP-positive cells increased in pre-pro-B-cells (Hardy fraction A-C) and in pro-B to immature-B cells (Hardy fraction D-E) (Figure 6C, right panel). Pre-B cells defined by the markers B220 and CD25 can be subdivided in small resting (SR) and large cycling (LC), both of which were found to express identical elevated levels of GFP (Figure 6D). However, neither larger (L) nor smaller (S) mature B cells (Hardy fraction F) showed any noticeable GFP expression (Figure 6D, right panel). Even after stimulation with LPS, or on treatment with anti-CD40 or anti-CD40 and IL4, GFP expression could not be induced in mature B cells (Figure S2A, upper panel).
Gfi1b is absent from T cells with the exception of a very small early thymic pre–T-cell subset
GFP could not be detected in naive or memory CD4+ or CD8+ peripheral T cells from Gfi1b+/GFP mice and could not be induced on antigenic activation with anti-CD3 antibodies or on stimulation with concanavalin A (Figures S1, S2C). Similarly, thymic T cells from Gfi1b+/GFP mice stained for CD4 and CD8 expression did not reveal any noticeable GFP signal with the exception of very few cells in the CD4−CD8− (DN) population (Figure 7A). More detailed analysis of the lineage-negative (Lin−) subset of thymic T cells using the markers CD44 and CD25 to discriminate between subsets DN1 to DN4, revealed that only a fraction between DN1 and DN2 (called here “pre-DN2”) was clearly GFP positive, whereas the DN2 fraction still contained some positive cells, and the DN3 population was very weakly positive (Figure 7B, C, left). To test, whether GFP fluorescence in knock-in mice corresponds to Gfi1b expression in wt mice, quantitative RT-PCR (q-PCR) for Gfi1 and Gfi1b was performed on mRNA isolated from purified thymic T-cell precursor cells from wt mice (Figure 7C, right). Because the GFP-positive pre-DN2 population is too small for separate analysis, DN1 and pre-DN2 cells were coisolated, and their Gfi1 and Gfi1b mRNA levels were set as a reference. Although the Gfi1b mRNA level declines rapidly in DN2 to DN4, Gfi1 expression transiently increases in DN2 and decreases again. Because the number of Gfi1b-positive T-cell precursors is very low, it turned out to be below Western blot detection level (data not shown). Next, we analyzed Gfi1b and Gfi1 expression level in bone marrow–derived B-cell precursors from wt mice, which are a second cell lineage positive for both Gfi1 and Gfi1b (Figure 7D). Gfi1b protein decreases during B-cell maturation from B220+/Igm− to B220++/Igm− status (Figure 7D, left), nicely resembling GFP fluorescence in Gfi1b+/GFP cells shown before (Figure 6B). The same is shown for Gfi1b-mRNA level, which decreases more rapidly in comparison to Gfi1 during B-cell maturation (Figure 7D, right). For comparison, we included the analysis of erythroid precursors, which express Gfi1b but not Gfi1. Although Gfi1 message is at the detection limit, Gfi1b mRNA is 845-fold more frequent in erythroid precursors than in B220++/Igm− B cells and about 180-fold higher than in less mature B220+/Igm− precursor cells.
Gfi1b is expressed during a small window of early T-cell development. (A) Thymocytes from adult wt or wt/KI (Gfi1b+/GFP) mice were stained for the surface markers CD4 and CD8 and gated for different stages of T-cell development as described previously.9 GFP fluorescence of the respective subpopulations is shown in representative histograms (right). (B) Because only in the double-negative (DN) population GFP-positive cells were present, thymocytes from adult wt (▪) or wt/KI (Gfi1b+/GFP) mice (⊡) were stained for CD25 and CD44 expression and lineage-negative (Lin−) cells were electronically gated out as shown (left). The GFP expression of the respective gated cell population is shown in representative histograms (right). (C) Representative diagram showing the expression of GFP during early T-cell development, shown as the difference of the geometric mean fluorescence intensities between wt and wt/KI (Gfi1b+/GFP) cells (left). Quantitative RT-PCR (real-time analysis) showing mRNA levels for Gfi1 and Gfi1b during early T-cell development normalized to the respective expression levels in DN1-preDN2 cells (right). (D) Western blot analysis of Gfi1b protein showing the decrease of Gfi1b during early B-cell development (left) and quantitative mRNA analysis for Gfi1 and Gfi1b during early B-cell development and in early erythroid cells (right). Error bars represent the standard deviation of triplicate measurements.
Gfi1b is expressed during a small window of early T-cell development. (A) Thymocytes from adult wt or wt/KI (Gfi1b+/GFP) mice were stained for the surface markers CD4 and CD8 and gated for different stages of T-cell development as described previously.9 GFP fluorescence of the respective subpopulations is shown in representative histograms (right). (B) Because only in the double-negative (DN) population GFP-positive cells were present, thymocytes from adult wt (▪) or wt/KI (Gfi1b+/GFP) mice (⊡) were stained for CD25 and CD44 expression and lineage-negative (Lin−) cells were electronically gated out as shown (left). The GFP expression of the respective gated cell population is shown in representative histograms (right). (C) Representative diagram showing the expression of GFP during early T-cell development, shown as the difference of the geometric mean fluorescence intensities between wt and wt/KI (Gfi1b+/GFP) cells (left). Quantitative RT-PCR (real-time analysis) showing mRNA levels for Gfi1 and Gfi1b during early T-cell development normalized to the respective expression levels in DN1-preDN2 cells (right). (D) Western blot analysis of Gfi1b protein showing the decrease of Gfi1b during early B-cell development (left) and quantitative mRNA analysis for Gfi1 and Gfi1b during early B-cell development and in early erythroid cells (right). Error bars represent the standard deviation of triplicate measurements.
Discussion
The transcription factor Gfi1b is required for the proper development of the erythroid- and megakaryocytic-cell lineages in the hematopoietic system, but the embryonic lethality of targeted deletion of Gfi1b has restricted the analysis of its function and expression in adult hematopoiesis.19,20,21 Although Gfi1b is known to be expressed in hematopoietic stem cells and evidence exists for a rather restricted expression during early T-cell development, nothing or very little is known about its expression during development and maturation of all other cell types of the hematopoietic system. To determine the precise expression pattern of Gfi1b especially during development of the different cell lineages of the hematopoietic system we generated a Gfi1b:GFP knock-in mouse mutant. The GFP cDNA was inserted precisely at the translation initiation codon of the Gfi1b allele to keep upstream regulatory elements functional. Also, the 3′-untranslated region and polyadenylation signals were conserved to maintain potential mRNA stabilization signals functional.
In our targeting strategy, the GFP reporter allele inactivates the Gfi1b gene. In agreement with previously published data,11 Gfi1b+/GFP mice are indistinguishable from their wt littermates with regard to all hematopoietic parameters analyzed so far. Because we were also unable to detect lower than wt levels of Gfi1b protein in Gfi1b+/GFP mice (data not shown), it is conceivable that the absence of a phenotype in Gfi1b+/GFP animals is a consequence of the autoregulatory features that have been previously described to occur at the Gfi1b locus.15 Quantitative real-time PCR demonstrated that mRNA expression of the GFP and Gfi1b alleles followed the same regulation. Moreover, GFP fluorescence was detected in all cell types where Gfi1b expression has already been demonstrated before, suggesting that the Gfi1b:GFP knock-in mice are well suited as a reporter mouse to follow Gfi1b expression. Using GFP fluorescence as a readout, we conclude that Gfi1b is differentially expressed at relatively high levels in early and intermediate stages of erythroid and megakaryocyte maturation, at moderate levels in immature B cells and pre-DN2 T cells and in peripheral blood granulocytes and monocytes. The latter finding was most unexpected. Although GFP expression was readily detected in CMPs, it was absent in GMPs, the progenitors that are committed to the myeloid and granulocytic lineages and also in all Mac-1+, that is, myeloid cells from bone marrow and peripheral lymphoid organs, suggesting a silencing of the Gfi1b locus in these lineages. The fact that Gfi1b is reexpressed in circulating monocytes and granulocytes may indicate a so far unsuspected role of Gfi1b in the mobilization of these cells from the bone marrow and by inference in the inflammatory reaction.
The most prominent phenotype of Gfi1b−/− mice that is reproduced in our Gf1bGFP/GFP (also termed KI/KI) mutants is the profound deficiency in mature erythrocytes.11 Consistent with an essential role of Gfi1b in erythroid maturation, we found Gfi1b expression in MEPs and throughout the development of the erythroid lineage already beginning at very early stages. According to the measured GFP levels, Gfi1b expression that rises through erythroid maturation is highest at the erythroblast stage, and steeply decreases in later stages. Because Gfi1b is already present in MEPs, the common progenitor cell for erythrocytes and megakaryocytes, it is possible that Gfi1b is more important for terminal differentiation of these cell types than for the commitment to these lineages.11,22 GFP fluorescence drastically increased in erythroid cells from Gfi1bGFP/GFP compared with Gfi1b+/GFP embryos, suggesting an autoregulatory phenomenon at the Gfi1b locus. However, introduction of a Gfi1b transgene under the control of the pan-hematopoietic Vav promoter15 by cross-breeding did not lead to a reduction of GFP fluorescence in erythroid cells (data not shown) but only in pre-B ells. Either the expression level of the vav-Gfi1b transgene is not sufficient to suppress the strongly active Gfi1b promoter in erythrocytes, or autoregulation is restricted to certain cell types, namely lymphoid cells, as it was previously proposed for the Gfi1 locus.7 Notably, the expression from the vav-Gfi1b transgene was not sufficient to rescue the lethality of the Gfi1bGFP/GFP animals, because Gfi1bGFP/GFP/vav-Gfi1b embryos also died around E15 (data not shown).
It is well established that Gfi1b acts as a transcriptional repressor and that the repressory function is mediated by the N-terminal Snail-Gfi-homology (SNAG) domain, which is also required to induce erythroid differentiation.22 Recent findings suggested that senseless, the drosophila homologue of Gfi1 and Gfi1b, can act as a repressor or activator depending on its expression level. Because very high expression levels of Gfi1b were found during the differentiation of erythroid cells compared with pre-B cells, it is possible that a similar situation as found for senseless may apply to Gfi1b and that functions of Gfi1b other than transcriptional repression prevail in erythroid cells. Recent observations show that in erythroid precursors Gfi1b is a component of a multimeric complex also containing SCL and ETO-2,23 both of which have been shown to interact with transcriptional corepressors,24 or histone-modifying enzymes (histone deacetylases, HDACs).25,26 Interestingly, Gfi1b and SCL can both bind to pericentric heterochromatin, and, in addition, Gfi1 as well as Gfi1b were shown to be involved in epigenetic gene silencing through interaction with HDAC-1 and the histone methyltransferase G9A.27,28 Because DNA compaction by heterochromatinization is an important step in erythroid maturation that occurs prior to enucleation of terminally differentiated erythrocytes, a role of Gfi1b in this process appears likely and represents an attractive explanation for the failure of Gfi1b-deficient erythroid cells to develop to definitive erythrocytes. We have recently reported that a profound lack of heterochromatinization occurs in fetal liver cells of Gfi1bGFP/GFP mice which strongly supports this hypothesis.28
The function of Gfi1b during megakaryocyte differentiation is still unclear and remains to be elucidated. Our data confirm Gfi1b expression in megakaryocytes and their precursors but also suggest a differential regulation of Gfi1b expression in this lineage possibly during the maturation process very similar to erythroid cells. Interestingly, megakaryocytes are polyploid and produce platelets that have no nucleus similar to erythroid differentiation and red blood cells pointing to a role of Gfi1b in chromatin compaction during platelet formation as proposed for erythroid maturation.
Transplantation experiments with Gfi1b−/− fetal liver cells showed that Gfi1b may be dispensable for B-cell development29 or for lymphoid differentiation in general. These experiments also suggested that Gfi1b, other than Gfi1,29,30 is not required for normal stem-cell development, despite that both proteins are expressed in HSCs. Both Gfi1 and Gfi1b are expressed in HSCs, but only the deletion of Gfi1 leads to a stem-cell defect.29,30 A function for Gfi1b in HSCs appeared unlikely, which is concluded from serial transplantation experiments, starting with fetal liver cells from Gfi1b-deficient embryos.20,29 We have previously shown that Gfi1b can replace Gfi1 in hematopoietic differentiation and have observed more recently that the expression of Gfi1b under the control of the Gfi1 locus in a knock-in mouse model can rescue at least the loss of HSCs observed in Gfi1-deficient mice (Katharina Fiolka and T.M., unpublished data, June 2006). Why the endogenous Gfi1b apparently fails to make up for the loss of Gfi1 in Gfi1-deficient HSCs remains an open question.
Following GFP fluorescence in Gfi1b+/GFP heterozygous mice strongly suggests that Gfi1b expression is tightly controlled during B-cell development, because it is regulated from undetectable levels in very early B-cell precursors (B220vlow, CD43+), to intermediate levels in pro-B cells and decreases again in mature B cells, suggesting a function of Gfi1b during this process. This new finding broadens the spectrum of cells where the expression of Gfi1b and Gfi1 overlap, because Gfi1 has been described to be expressed with very similar kinetics in early B-cell progenitors and to decline during B-cell maturation.9 In contrast to Gfi1, however, Gfi1b expression cannot be induced by activating B cells, for instance, with LPS according to GFP fluorescence. A similar situation is found in T cells where Gfi1 but not Gfi1b is up-regulated on activation. Because Gfi1b can compensate for the loss of Gfi1 in T-cell development16 and T-cell activation (Katharina Fiolka and T.M., unpublished data, June 2006), it is conceivable that one Gfi protein may be sufficient for a given hematopoietic compartment. However, as for HSCs, the question remains why both Gfi proteins are coexpressed during B-cell development and what their respective roles during this process are.
In contrast to Gfi1, which is expressed in T cells of all developmental stages,9 the only T cells expressing Gfi1b were found in a very small subset within the early thymic pre-T cells. Our analysis based on monitoring GFP fluorescence suggested that Gfi1b is expressed mainly in a very small, pre-DN2 population, in a heterogeneous pattern in the DN2 and at very low levels in the DN3 population. The significance of this very distinct and short up-regulation of Gfi1b expression during T-cell development is unclear. However, because Gfi1 potentiates the response to T-cell receptor (TCR) and IL2 stimulation, whereas Gfi1b transgenic T cells are defective in T-cell activation,31 the balance between Gfi1b and Gfi1 expression at the time of TCRβ rearrangements (ie, in DN2 and DN3 subsets) may play an important role in pre–T-cell differentiation that has yet to be clarified.
It has been well established that both the Gfi1 and the Gfi1b locus are under autoregulatory and cross-regulatory control at least in some cell types.9,15,32 The near complementary expression pattern of Gfi1 and Gfi1b that we observed in the different lineage committed early hematopoietic progenitor cells9 (present study) could be interpreted as the consequence of cross-regulation. If this is the case, the coexpression of both Gfi1 and Gfi1b in HSCs and early B cells must be assured by a mechanism that inhibits cross-regulation in particular cell types. How this is achieved remains to be elucidated. Because the promoters of both Gfi1 genes have cognate binding sites for Gfi1 and Gfi1b that have been shown to be occupied by Gfi1 and Gfi1b in particular in lymphoid cells,9,15,32 the abolition of cross-regulation is likely to involve the availability of cofactors that bind to Gfi1 proteins or, alternatively, is achieved via a regulation of a threshold that inhibit cross-regulation below a certain expression level. Also, alternative promoters could exist in both Gfi1 and Gfi1b loci which are not under autoregulatory control but under the control of other transcription factors for instance in HSCs or B cells.
Authorship
Contribution: L.V., T.O., and T.M. were involved in designing and performing the research, data analysis, and writing the paper,
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tarik Möröy, Institut de recherches cliniques de Montreal, I R C M, 110, Avenue des Pins West, Montreal, QC, H2W 1R7, Canada; e-mail: Tarik.Moroy@ircm.qc.ca or moeroey@uni-essen.de.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Angelika Warda, Wojciech Wegrzyn, Adriane Parchatka, Nadine Esser, and Inge Spratte for technical assistance and Petra Plessow and Tomas Civela for excellent animal care.
This work was supported by the Deutsche Forschungsgemeinschaft, DFG, (grant Mo 435/18-1), the Fonds der chemischen Industrie, and the IFORES Program of the University of Essen Medical School.

![Figure 1. Generation of Gfi1b:GFP knock-in mice. (A) Schematic representation of the targeting construct design and simplified restriction maps of the wt and targeted alleles after CRE recombinase excision of the thymidine kinase-neomycin cassette (TK-neo) by crossing of heterozygous mice with CRE-transgenic mice. All coding sequences of Gfi1b are replaced by the GFP open reading frame. lxP indicates lox-P site; H, HindIII; E, EcoRI; N, NotI; Sl, SalI; B, BamHI; DTA, diphtheria toxin; 1-7, exon 1 to 7; GFP, green fluorescent protein. (B) Southern blotting to detect excision of the thymidine kinase-neomycin resistance gene fusion protein (TK-neo) cassette in mice after gene targeting and CRE-recombination showed the expected 8-kb fragment using a 5′-external probe. (C) PCR strategy and results to detect the insertion of the GFP cDNA into the Gfi1b locus in mouse embryo tail DNA representing the embryos shown in panel D. Because all 5′ sequences are retained, but intron 2 to intron 6 were deleted in the targeted allele, the wt, wt/KI (Gfi1b+/GFP), and KI/KI (Gfi1bGFP/GFP) mice were analyzed by multiplex PCR covering either intron 1 to 2 or intron 1 to GFP (Figure 1C) to demonstrate proper targeting. (D) Appearance of (top panel) and detection of GFP fluorescence in day 13.5 postconception (pc) embryos which were wt, heterozygous, or homozygous for the GFI1b:GFP allele (wt, wt/KI [Gfi1b+/GFP] and KI/KI [Gfi1bGFP/GFP], bottom panel). In heterozygous embryos, GFP expression is restricted to the fetal liver, where erythropoiesis takes place. (Bottom panel) CD31 whole-mount staining showing normal vascularization in wt and mutant embryos. Embryos were analyzed with the Leica MZ/FLIII stereomicroscope (Leica, Wetzlar, Germany) and photographed with the KAPPA CF 15/4 MC camera (KAPPA Opto-Electronics, Gleichen, Germany) using 0.5× magnification (0.125 NA). (E) Western blot analysis of whole-cell extracts from fetal liver cells of wt or homozygous GFP knock-in embryos at day 13.5 pc showing the loss of Gfi1b expression (top) and gain of GFP expression (bottom) in Gfi1b:GFP knock-in mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/6/10.1182_blood-2006-06-030031/4/m_zh80060709530001.jpeg?Expires=1765937494&Signature=rVNdGLKcYNRxxM1hJJxwE3fRmI-A82AcPCjHDPtKzANR25Y90QaILi6AfXOpod54V-VseHfJuSflYE3jt4QXJpXcA8M3J2PZGj8czHrxbGOJ~kb8gDv~xJ3FcWKG2NoLsrcJv7fYOFppCIHdrM1Sf3diRpH1SxbXyrIT19ZWgIuPwidtVVF0v16PuagIcohhmhrEQbMQM4J47~qqoNIEaDZe0GWTwld4-HqTgpQxi9vlEWFiIqbd58PdW4bpfpSNIUtjfppST4DivXUrE6qU2RdgvN93cgINieg7vws34dO3U5YITP~YMg9sPOwvwCmeS-roH6UJxArjHQOvfPQQrQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Gfi1b expression in early precursor and hematopoietic stem cells. (A-C) Bone marrow cells from wt or wt/KI (Gfi1b+/GFP) mice, respectively, were depleted for lineage-positive cells (Lin: CD3, CD4, CD8, Mac-1, B220, Ter119, Ly6G, Pan natural killer [NK] plus Sca1, IL7Rα for CMP-GMP-MEP analysis), and negative cells were restained for surface markers as indicated and analyzed by flow cytometry (A-C, left panels). HSCs (A), MEPs, CMPs, GMPs (B), or CLPs (C) were electronically gated as previously described (see “Material and methods”) and analyzed for GFP expression (right panels).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/6/10.1182_blood-2006-06-030031/4/m_zh80060709530004.jpeg?Expires=1765937494&Signature=ldmsLA1k64dPFEeO03mvv1ivU7R-eYL7dc2YwiAhuvMORZPPadw3Zh6PQg2r0SCWee7s~X57Uzb9UfER6QB22VUG540iEh0-yVIPFUt7pUnmdAzqrCtQupwF-KYqIGwkxL7xZAq~XxQap16W8WGWeGmIY35urkbCFib8jxQTUBHwFZVqU-SyiPt2ffe~ZDVW9iSqs68FAucHl7z4R3boicncHgamE8GvRQ9128VFT3YCnKF-2lIDTQK1bRjDPgCXLnk0f4vescdsB8orRnrzeolgN~1dOpkHMCiLgylL72GHnyD5Ya0ms3~9I1XDv7ZdyLacwOysyQYt3adnTzQr6A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal