Abstract
The bone morphogenetic protein (BMP) family of proteins participates in regulation of angiogenesis in physiologic and pathologic conditions. To investigate the molecular mechanisms that contribute to BMP-dependent angiogenic signaling, we performed gene expression profiling of BMP6-treated mouse endothelial cells. We detected 77 mRNAs that were differentially regulated after BMP6 stimulation. Of these, cyclooxygenase 2 (Cox2) was among the most highly up-regulated by BMP stimulation, suggesting a role for Cox2 as a downstream regulator of BMP-induced angiogenesis. Up-regulation of Cox2 by BMP6 was detected at both mRNA and protein levels in endothelial cells, and BMP6 increased production of prostaglandins in a Cox2-dependent fashion. BMP6 up-regulated Cox2 at the transcriptional level through upstream SMAD-binding sites in the Cox2 promoter. Pharmacologic inhibition of Cox2, but not Cox1, blocked BMP6-induced endothelial cell proliferation, migration, and network assembly. BMP6-dependent microvessel outgrowth was markedly attenuated in aortic rings from Cox2−/− mice or after pharmacologic inhibition of Cox2 in aortas from wild-type mice. These results support a necessary role for Cox2 in mediating proangiogenic activities of BMP6. These data indicate that Cox2 may serve as a unifying component downstream from disparate pathways to modulate angiogenic responses in diseases in which neovascularization plays an underlying pathophysiologic role.
Introduction
During the tightly regulated process of angiogenesis, a balance between pro- and antiangiogenic factors results in the sequential steps required for new blood vessel formation. Several mitogens are considered proangiogenic, including members of the basic fibroblast growth factor, platelet-derived growth factor, vascular endothelial growth factor (VEGF), and transforming growth factor beta families. Since endothelial cells (ECs) are essential components of angiogenesis, the abilities of these growth factors to stimulate angiogenesis are tightly associated with their effects on ECs.
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-β (TGF-β) superfamily, and more than 30 members of the BMP family have been identified from different species.1,2 BMPs are synthesized as precursor proteins, then are proteolytically cleaved at the N-terminus and secreted into the extracellular matrix. BMPs dimerize through disulfide bonds and initiate signaling by binding cooperatively to both type I and type II membrane–bound serine/threonine kinase receptors.3,4 The constitutively active type II receptors trans-phosphorylate type I receptors, which then activate intracellular substrates by phosphorylation, and thus determine the specificity of intracellular signals. In vertebrates, small mothers against decapentaplegic proteins (Smads) are downstream signaling molecules for the receptors. Receptor-regulated Smads (R-Smads) bind to the common partner Smad (Smad4), and these heteromeric complexes translocate to the nucleus and regulate the transcription of target genes by interacting with other transcription factors at cis-acting elements defined by the sequence CAGAC[A].5,6
BMPs have important but incompletely understood effects on angiogenesis by affecting EC differentiation, migration, and proliferation. BMP6 is a potent agonist for EC migration and tube formation in vitro. 7 BMP2 enhances neovascularization in tumors, and this activity may involve the stimulation of EC proliferation, migration, and tube formation by BMP2.8 BMP4 induces EC differentiation of mouse embryonic stem cells, and this induction is blocked by the expression of a dominant-negative BMP receptor.9,10 On the other hand, the transcorneal injection of BMP4 in rats promotes apoptosis of capillary ECs,11 suggesting that BMP4 contributes to EC apoptosis during rat papillary membrane regression. In developing Xenopus embryos, progenitor cells in dorsal and ventral marginal zones fail to differentiate into ECs in the ventral blood island region when BMP signaling is blocked by injection of RNA coding for a dominant-negative, truncated form of the BMP type I receptor.12 Taken together, these studies demonstrate an important yet complex role for BMPs in angiogenesis.
Although many lines of evidence indicate that BMPs are important in angiogenesis, there are still unresolved issues, in particular the delineation of downstream effectors that respond to BMP signaling during angiogenesis. This study has taken advantage of microarray technology to analyze the BMP6-mediated signaling network in ECs. Among the list of differentially expressed genes, we have selected cyclooxygenase 2 (Cox2) for further analysis based on the sustained change in transcription levels. Cox2, which catalyses the conversion of arachidonic acid to prostaglandins (PGs), is linked to several physiologic and pathogenetic pathways that participate in angiogenesis, inflammation, and invasiveness.13 The increased risk of heart attack and stroke among patients taking Cox2 inhibitors14-18 indicates a complicated role for Cox2 activity in maintaining vascular homeostasis. Our expression and EC functional studies indicate that Cox2 is an essential downstream component mediating BMP6-dependent EC activation, which could be one of the mechanisms underlying its cardioprotective effects.
Materials and methods
Plasmids and reagents
Recombinant human BMP6 protein was obtained from R&D Systems (Minneapolis, MN). A selective Cox2 inhibitor, NS398, the Cox1 inhibitor SC560, and immunoassay kits for PGE2 and 6-keto-PGF1α were obtained from Cayman (Ann Arbor, MI). Cox2 promoter regions from −8653 to +53 bp and −7444 to +53 bp were inserted into pGL3-Basic vector (Promega, Madison, WI). Two similar Cox2 reporter constructs (−1500 to +1 bp and −371 to +70 bp) were kindly provided by Dr Curtis C. Harris (National Institutes of Health, Bethesda, MD)19 and Dr Carol C. Pilbeam (University of Connecticut Farmington).20
Microarray analysis
Mouse intraembryonic endothelial cells (MECs) were cultured as previously described.9 Total RNA was extracted from MECs treated with BMP6 for 4, 12, or 24 hours, or from mock-treated controls. Universal mouse reference RNA was used as a reference,21 and was labeled with Cy3 florescence dye using the Fluorescent Linear Amplification Kit (Agilent Technologies, Santa Clara, CA). Samples of MEC RNA were labeled with Cy5, and equimolar mixtures of the 2 florescence dye–labeled probes were hybridized to Agilent 22k Mouse Development Oligo Microarrays (G4120A; Agilent Technologies). Microarrays were scanned with a GenePix 4000B scanner and images were quantified with GenePix Pro 5.0 Software (Molecular Devices, Sunnyvale, CA). Each experimental group included independent triplicate biological replications. Raw expression data were normalized via a modified quantiles local algorithm (Xiaorui He and C.P., manuscript in preparation). Differentially expressed genes were identified with a 2-tailed type 2 Student t test. Cluster analysis and visualization using Java-TreeView (Stanford University, Stanford, CA) were accomplished as previously described.22 Selected genes were validated by reverse transcriptase–polymerase chain reaction (RT-PCR). The complete, Minimum Information About a Microarray Experiment (MIAME)–compliant dataset is available at the Gene Expression Omnibus23 (reference no. GSE4909).
Reporter gene analysis and gel mobility shift assays
Luciferase reporter assays were performed as previously described.9 Electrophoretic gel mobility assays (EMSAs) were performed essentially as described24 with 3 μg of nuclear extract. DNA and protein complexes were resolved on a nondenaturing 4% polyacrylamide gel and visualized by autoradiography. The probes used were −8147—AGG CAG ACA GAC AGA CAA CCA GAT AGA TA— −8119 (sense) and −8119—TCC GTC TGT CTG TCT GTT GGT CTA TCT AT— −8147 (antisense).
Cell proliferation, migration, and tube formation assays
Cell proliferation assays were performed as described previously.25 The migration assays were prepared using a 48-well chamber apparatus (NeuroProbe, Cabin John, MD). The lower chambers of the apparatus were filled with DMEM with or without BMP6 and then covered with the gelatin-coated filter and the upper chambers. Cell suspensions in DMEM with or without Cox inhibitors were then added to the upper chambers. After incubating for 6 hours at 37°C, cells present on the lower surface were identified using a 10 × objective on a Nikon Eclipse TS100 inverted microscope (Nikon, Melville, NY).
EC tube formation was analyzed with the Matrigel-based tube formation assay.7 Chilled 24-well plates were coated with Matrigel (Becton Dickinson, San Jose, CA) that was polymerized at 37°C for 30 minutes. MECs were serum-starved overnight, trypsinized, and plated at equal numbers into each Matrigel-coated well. After 16 hours of incubation in the absence or presence of BMP6 and/or Cox inhibitors, the formation of tubes was photographed. Images were processed with Adobe Photoshop CS (Adobe, San Jose, CA).
Mouse aortic ring angiogenesis assays
Mouse aortic ring assays were performed with modifications from a previously reported method.26 Thoracic aortas were removed from 2-month-old mice, aortic rings were sectioned, and then embedded in rat tail collagen gel (1.5 mg/mL) prepared by mixing 7.5 vol of 2 mg/mL collagen, 1 vol of 10 × MEM, 1.5 vol of NaHCO3 (15.6 mg/mL), and 0.1 vol of 1M NaOH. Each well containing the aortic rings was incubated in 250 μL MCDB131 supplemented with 25 mM NaHCO3 and 1% mouse serum. Images of aortic rings were taken using an Olympus BX41 microscope (Olympus, Center Valley, PA) at day 6.
Results
Characterization of the BMP-responsive transcriptome in ECs
We used MECs to characterize BMP6-responsive transcriptional events in ECs for 2 reasons. First, MECs express all the components of the BMP signaling pathway from cell-surface receptors to Smads (data not shown). Second, Id1, a well-characterized target of BMP6 signaling in ECs,7 is faithfully up-regulated in MECs (Figure 1). To characterize the BMP6-dependent transcriptome in ECs, RNA samples extracted from MECs with or without BMP6 treatment for 4, 12, or 24 hours were analyzed with microarrays. Differentially expressed genes were identified by Student t test (P < .05) and then filtered for fold change. As expected, Id1 was up-regulated at each time point in this analysis, confirming the validity of our dataset and the sensitivity of this approach for identifying BMP-responsive genes (Table 2).
BMP6 induces the up-regulation of Id1 in a time-dependent fashion. RT-PCR analysis of Id1 in MECs treated with or without BMP6. Total RNA was extracted from MECs collected at 4, 12, and 24 hours after BMP6 treatment. GAPDH was used as a loading control.
BMP6 induces the up-regulation of Id1 in a time-dependent fashion. RT-PCR analysis of Id1 in MECs treated with or without BMP6. Total RNA was extracted from MECs collected at 4, 12, and 24 hours after BMP6 treatment. GAPDH was used as a loading control.
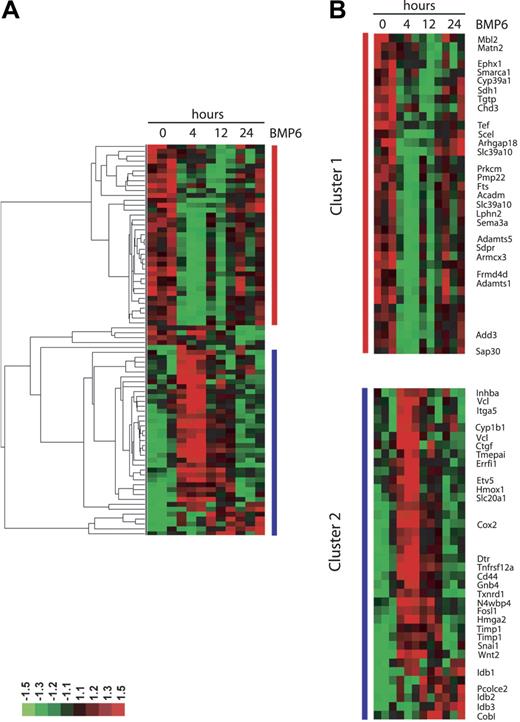
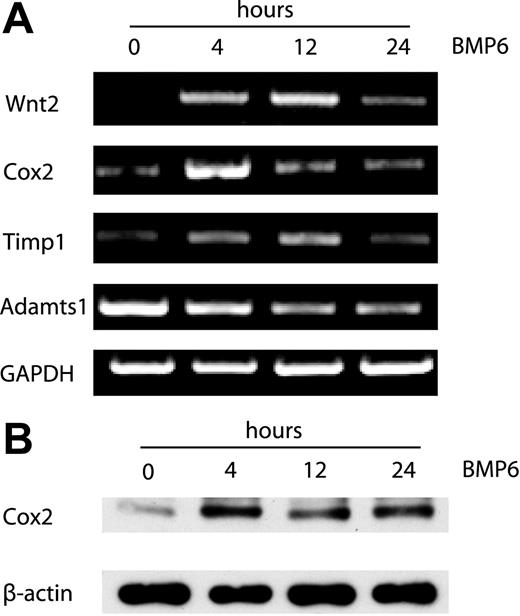
Out of 20 000 genes surveyed, a total of 37 genes were up-regulated and 40 genes were down-regulated by treatment with BMP-6 at any time point using relatively stringent criteria to reduce false-positive results (representative genes shown in Tables 1–2). Two groups of differentially expressed genes were identified by hierarchic clustering analysis (Figure 2). The first group comprised genes down-regulated by BMP6, including two members of the metalloproteinase family (Adamts1 and Adamts5) that are known inhibitors of neovascularization.28 The second group contains genes up-regulated by BMP6. Wnt2, Cox2, Id1, two other members of the Id family (Id2 and Id3), CD44, Tnfrsf12a (tumor necrosis factor receptor superfamily), Snail1 (a Wnt-regulated transcriptional factor), and Timp1 (an inhibitor of metalloproteinases) are present in this cluster. To establish the validity of the microarray dataset, we performed RT-PCR analysis for several selected genes, confirming that Cox2, Wnt2, Timp1, and Adamts1 were indeed differentially regulated by BMP6 at the mRNA level (Figure 3A). In addition, the up-regulation of Cox2 after BMP6 treatment was also demonstrated at the protein level by Western blotting (Figure 3B).
Genes down-regulated by BMP6 in MECs
| Gene name . | NCBI Genebank accession no. . | Fold change . | ||
|---|---|---|---|---|
| 4 h . | 12 h . | 24 h . | ||
| Mbl2 | NM_010776.1 | 0.81 | 0.61 | 0.68 |
| Matn2 | NM_016762.1 | 0.86 | 0.66 | 0.80 |
| Ephx1 | BC029636.1 | 0.83 | 0.63 | 0.77 |
| Smarca1 | NM_053123.1 | 0.77 | 0.64 | 0.93 |
| Cyp39a1 | NM_018887.1 | 0.65 | 0.54 | 0.93 |
| Sdh1 | NM_146126.1 | 0.67 | 0.67 | 0.82 |
| Tgtp | NM_011579.1 | 0.65 | 0.44 | 0.63 |
| Chd3 | NM_146019.1 | 0.67 | 0.65 | 0.84 |
| Tef | NM_153484.1 | 0.60 | 0.72 | 0.92 |
| Scel | AE014176.1 | 0.55 | 0.84 | 1.24 |
| Arhgap18 | NM_176837 | 0.64 | 0.74 | 0.85 |
| Slc39a10 | NM_172653.1 | 0.63 | 0.82 | 0.84 |
| Prkcm | NM_002742.1 | 0.67 | 0.77 | 0.93 |
| Pmp22 | NM_008885.1 | 0.62 | 0.77 | 0.87 |
| Fts | NM_010241.1 | 0.63 | 0.79 | 0.90 |
| Acadm | NM_007382.1 | 0.66 | 0.81 | 0.91 |
| Slc39a10 | NM_172653.1 | 0.61 | 0.85 | 0.90 |
| Lphn2 | XM_131258.1 | 0.67 | 0.85 | 0.90 |
| Sema3a | XM_131903.1 | 0.55 | 0.75 | 0.85 |
| Adamts5 | NM_007038.1 | 0.41 | 0.60 | 0.71 |
| Sdpr | NM_138741.1 | 0.42 | 0.66 | 0.76 |
| Armcx3 | NM_027870.1 | 0.60 | 0.76 | 0.98 |
| Frmd4b | BC031369.1 | 0.61 | 0.67 | 0.91 |
| Adamts1 | D67076.1 | 0.57 | 0.65 | 0.75 |
| Add3 | BC037116.1 | 0.65 | 0.77 | 0.96 |
| Sap30 | NM_021788.1 | 0.66 | 0.91 | 1.01 |
| Sap30 | NM_021788.1 | 0.63 | 0.82 | 1.01 |
| Gene name . | NCBI Genebank accession no. . | Fold change . | ||
|---|---|---|---|---|
| 4 h . | 12 h . | 24 h . | ||
| Mbl2 | NM_010776.1 | 0.81 | 0.61 | 0.68 |
| Matn2 | NM_016762.1 | 0.86 | 0.66 | 0.80 |
| Ephx1 | BC029636.1 | 0.83 | 0.63 | 0.77 |
| Smarca1 | NM_053123.1 | 0.77 | 0.64 | 0.93 |
| Cyp39a1 | NM_018887.1 | 0.65 | 0.54 | 0.93 |
| Sdh1 | NM_146126.1 | 0.67 | 0.67 | 0.82 |
| Tgtp | NM_011579.1 | 0.65 | 0.44 | 0.63 |
| Chd3 | NM_146019.1 | 0.67 | 0.65 | 0.84 |
| Tef | NM_153484.1 | 0.60 | 0.72 | 0.92 |
| Scel | AE014176.1 | 0.55 | 0.84 | 1.24 |
| Arhgap18 | NM_176837 | 0.64 | 0.74 | 0.85 |
| Slc39a10 | NM_172653.1 | 0.63 | 0.82 | 0.84 |
| Prkcm | NM_002742.1 | 0.67 | 0.77 | 0.93 |
| Pmp22 | NM_008885.1 | 0.62 | 0.77 | 0.87 |
| Fts | NM_010241.1 | 0.63 | 0.79 | 0.90 |
| Acadm | NM_007382.1 | 0.66 | 0.81 | 0.91 |
| Slc39a10 | NM_172653.1 | 0.61 | 0.85 | 0.90 |
| Lphn2 | XM_131258.1 | 0.67 | 0.85 | 0.90 |
| Sema3a | XM_131903.1 | 0.55 | 0.75 | 0.85 |
| Adamts5 | NM_007038.1 | 0.41 | 0.60 | 0.71 |
| Sdpr | NM_138741.1 | 0.42 | 0.66 | 0.76 |
| Armcx3 | NM_027870.1 | 0.60 | 0.76 | 0.98 |
| Frmd4b | BC031369.1 | 0.61 | 0.67 | 0.91 |
| Adamts1 | D67076.1 | 0.57 | 0.65 | 0.75 |
| Add3 | BC037116.1 | 0.65 | 0.77 | 0.96 |
| Sap30 | NM_021788.1 | 0.66 | 0.91 | 1.01 |
| Sap30 | NM_021788.1 | 0.63 | 0.82 | 1.01 |
Genes up-regulated by BMP6 in MECs
| Gene name . | NCBI Genebank accession no. . | Fold change . | ||
|---|---|---|---|---|
| 4 h . | 12 h . | 24 h . | ||
| Inhba | NM_008380.1 | 1.56 | 1.28 | 1.16 |
| Vcl | NM_009502.2 | 1.68 | 1.02 | 1.08 |
| Itga5 | NM_010577.1 | 1.60 | 1.08 | 1.03 |
| Cyp1b1 | NM_009994.1 | 1.66 | 1.24 | 1.27 |
| Vcl | NM_009502.2 | 1.70 | 0.98 | 0.98 |
| Ctgf | NM_010217.1 | 2.64 | 1.03 | 0.88 |
| Tmepai | BC036995.1 | 1.85 | 1.36 | 1.05 |
| Errfi1 | NM_133753.1 | 1.62 | 0.97 | 0.87 |
| Etv5 | NM_023794.2 | 1.62 | 1.00 | 0.88 |
| Hmox1 | NM_010442.1 | 1.56 | 0.99 | 0.94 |
| Slc20a1 | NM_015747.1 | 1.52 | 1.16 | 0.95 |
| Cox2 | X98792.1 | 2.48 | 1.52 | 1.19 |
| Dtr | NM_010415.1 | 1.98 | 1.38 | 1.16 |
| Tnfrsf12a | NM_013749.1 | 2.19 | 1.56 | 1.11 |
| Cd44 | XM_130536.1 | 1.85 | 1.21 | 1.05 |
| Gnb4 | NM_013531.1 | 1.55 | 1.24 | 1.04 |
| Txnrd1 | NM_015762.1 | 1.58 | 1.31 | 0.95 |
| N4wbp4 | BC022122.1 | 1.54 | 1.21 | 1.03 |
| Fosl1 | NM_010235.1 | 1.54 | 1.30 | 1.11 |
| Hmga2 | NM_003483.3 | 1.94 | 1.53 | 1.08 |
| Timp1 | NM_011593.1 | 1.65 | 1.66 | 1.12 |
| Timp1 | M59906.1 | 1.45 | 1.56 | 1.17 |
| Snai1 | NM_011427.1 | 1.72 | 1.53 | 1.32 |
| Wnt2 | BC026373.1 | 2.71 | 1.97 | 1.45 |
| Id1 | U43884.1 | 1.94 | 1.91 | 2.06 |
| Pcolce2 | NM_029620.1 | 1.21 | 1.64 | 1.42 |
| Id2 | XM_176747.1 | 1.76 | 2.34 | 2.23 |
| Id3 | NM_008321.1 | 1.54 | 1.72 | 1.88 |
| Gene name . | NCBI Genebank accession no. . | Fold change . | ||
|---|---|---|---|---|
| 4 h . | 12 h . | 24 h . | ||
| Inhba | NM_008380.1 | 1.56 | 1.28 | 1.16 |
| Vcl | NM_009502.2 | 1.68 | 1.02 | 1.08 |
| Itga5 | NM_010577.1 | 1.60 | 1.08 | 1.03 |
| Cyp1b1 | NM_009994.1 | 1.66 | 1.24 | 1.27 |
| Vcl | NM_009502.2 | 1.70 | 0.98 | 0.98 |
| Ctgf | NM_010217.1 | 2.64 | 1.03 | 0.88 |
| Tmepai | BC036995.1 | 1.85 | 1.36 | 1.05 |
| Errfi1 | NM_133753.1 | 1.62 | 0.97 | 0.87 |
| Etv5 | NM_023794.2 | 1.62 | 1.00 | 0.88 |
| Hmox1 | NM_010442.1 | 1.56 | 0.99 | 0.94 |
| Slc20a1 | NM_015747.1 | 1.52 | 1.16 | 0.95 |
| Cox2 | X98792.1 | 2.48 | 1.52 | 1.19 |
| Dtr | NM_010415.1 | 1.98 | 1.38 | 1.16 |
| Tnfrsf12a | NM_013749.1 | 2.19 | 1.56 | 1.11 |
| Cd44 | XM_130536.1 | 1.85 | 1.21 | 1.05 |
| Gnb4 | NM_013531.1 | 1.55 | 1.24 | 1.04 |
| Txnrd1 | NM_015762.1 | 1.58 | 1.31 | 0.95 |
| N4wbp4 | BC022122.1 | 1.54 | 1.21 | 1.03 |
| Fosl1 | NM_010235.1 | 1.54 | 1.30 | 1.11 |
| Hmga2 | NM_003483.3 | 1.94 | 1.53 | 1.08 |
| Timp1 | NM_011593.1 | 1.65 | 1.66 | 1.12 |
| Timp1 | M59906.1 | 1.45 | 1.56 | 1.17 |
| Snai1 | NM_011427.1 | 1.72 | 1.53 | 1.32 |
| Wnt2 | BC026373.1 | 2.71 | 1.97 | 1.45 |
| Id1 | U43884.1 | 1.94 | 1.91 | 2.06 |
| Pcolce2 | NM_029620.1 | 1.21 | 1.64 | 1.42 |
| Id2 | XM_176747.1 | 1.76 | 2.34 | 2.23 |
| Id3 | NM_008321.1 | 1.54 | 1.72 | 1.88 |
Hierarchic clustering of BMP6 target genes. Differentially expressed genes were selected with a P value of .05 or less and ratio fold change of 1.5 or more, and were subjected to hierarchic cluster analysis. Median-centered clusters were viewed with Java-TreeView. Fold change relative to common reference is indicated by red (+1.5; full scale) and green (−1.5; full scale) intensity. (A) Global view of the clustered genes. (B) Detail of the clustered genes with selected genes labeled. Cluster 1 corresponds to genes in Table 1, and cluster 2 corresponds to genes in Table 2. The complete data set is accessible online through the University of North Carolina Microarray Database.27
Hierarchic clustering of BMP6 target genes. Differentially expressed genes were selected with a P value of .05 or less and ratio fold change of 1.5 or more, and were subjected to hierarchic cluster analysis. Median-centered clusters were viewed with Java-TreeView. Fold change relative to common reference is indicated by red (+1.5; full scale) and green (−1.5; full scale) intensity. (A) Global view of the clustered genes. (B) Detail of the clustered genes with selected genes labeled. Cluster 1 corresponds to genes in Table 1, and cluster 2 corresponds to genes in Table 2. The complete data set is accessible online through the University of North Carolina Microarray Database.27
RT-PCR and Western blotting analysis of gene expression in BMP6-induced EC activation. (A) RT-PCR analysis of Wnt2, Cox2, Timp1 and Adamts1 expression using total RNA from MECs collected at 0, 4, 12, and 24 hours after BMP6 treatment. GAPDH was used as a loading control. (B) Western blot analysis of Cox2 protein expression after BMP6 stimulation for the indicated times. β-actin was used as a loading control.
RT-PCR and Western blotting analysis of gene expression in BMP6-induced EC activation. (A) RT-PCR analysis of Wnt2, Cox2, Timp1 and Adamts1 expression using total RNA from MECs collected at 0, 4, 12, and 24 hours after BMP6 treatment. GAPDH was used as a loading control. (B) Western blot analysis of Cox2 protein expression after BMP6 stimulation for the indicated times. β-actin was used as a loading control.
Enhanced production of prostanoids through the up-regulation of Cox2 by BMP6
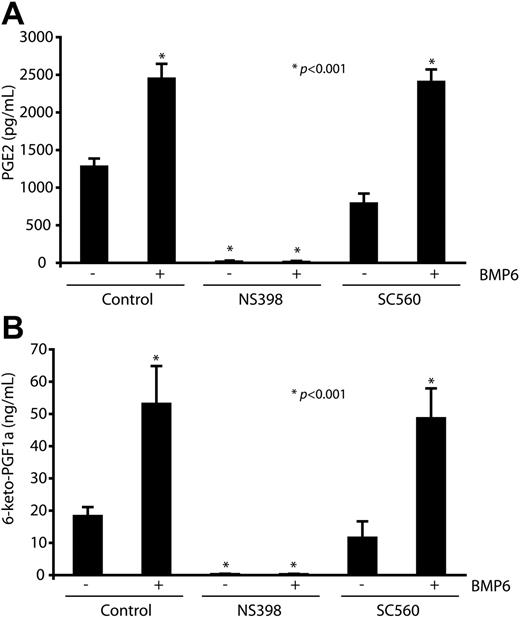
Because cyclooxygenases catalyze the formation of PGs from arachidonic acid, we next investigated whether stimulation of ECs by BMP6 also leads to increased PG production by MECs. These experiments were performed in the presence of arachidonic acid concentrations that favored PG generation by Cox2 over Cox1 and measured the 2 functional important PGs in ECs, PGE2 and the dominant form PGI2.29 BMP6 potently increased generation of PGE2 and 6-keto-PGF1α (a stable metabolism of PGI2) in MECs, as determined by accumulation of these prostanoids in the culture media after BMP stimulation (Figure 4A-B). Generation of PGE2 and 6-keto-PGF1α was abolished by pretreatment with the Cox2 inhibitor NS398 but not the Cox1 inhibitor SC560, indicating their derivation via a Cox2-dependent mechanism.
Elevated levels of secreting prostaglandins derived from Cox2 after BMP6 stimulation. MECs were pretreated with NS398 (10 μM), SC560 (100 nM), or vehicle before BMP6 (100 ng/mL) treatment. Cox2-dependent prostanoid generation was determined by measurement of the major forms of PGs generated by ECs, PGE2 (A), and 6-keto-PGF1α (B) after 4 hours of stimulation in the presence of exogenous arachidonic acid (2.5 μM). Data are expressed as means ± SEM of 6 replicates. *P < .001 compared with control.
Elevated levels of secreting prostaglandins derived from Cox2 after BMP6 stimulation. MECs were pretreated with NS398 (10 μM), SC560 (100 nM), or vehicle before BMP6 (100 ng/mL) treatment. Cox2-dependent prostanoid generation was determined by measurement of the major forms of PGs generated by ECs, PGE2 (A), and 6-keto-PGF1α (B) after 4 hours of stimulation in the presence of exogenous arachidonic acid (2.5 μM). Data are expressed as means ± SEM of 6 replicates. *P < .001 compared with control.
Transcriptional activation of the Cox2 promoter by BMP6
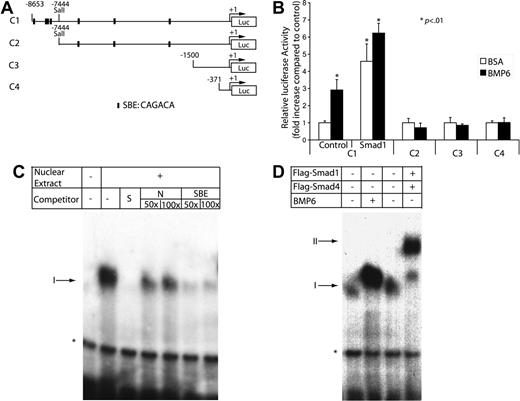
Because Cox2 is primarily regulated at the transcriptional level by factors such as interleukin-1β (IL-1β)29,30 and tumor necrosis factor-α,31 we reasoned that the rapid up-regulation of Cox2 mRNA by BMP6 might also occur at the transcriptional level. To investigate the mechanism of transactivation of the Cox2 promoter by BMP6, we tested four Cox2 promoter–luciferase reporter constructs containing various lengths of Cox2 promoter regions upstream from its transcription start site (Figure 5A). These constructs were transiently transfected into MECs, which were then treated with or without BMP6. Only the −8653 to +53 bp promoter construct, but not those containing smaller fragments of the Cox2 promoter, could be stimulated by BMP6 (Figure 5B). In addition, we observed a 5-fold activation of this 8.7-kb reporter when Smad1 was cotransfected into MECs, which is consistent with previous observations that overexpressed Smad proteins can transactivate BMP target genes in a ligand-independent manner.32,33 These data indicate that necessary BMP-responsive elements in the Cox2 promoter are located between base pairs −8653 and −7444.
Activation of the Cox2 promoter by BMP signaling. (A) Depiction of 4 luciferase reporters of the Cox2 promoter, −8653 to +53 bp, −7444 to +53bp, −1500 to +1 bp, and −371 to +70 bp. Putative Smad-binding sites are denoted. (B) MECs were transfected with luciferase reporter plasmids carrying different lengths of the Cox2 promoter in the presence or absence of BMP6 treatment or the cotransfection of an active Smad1 expression vector. After 48 hours, cells were lysed to measure luciferase activity. Luciferase activity was normalized using Renilla luciferase activity. Data shown are the means ± SD of triplicates. (C) An end-labeled fragment containing the 29-bp Smad-responsive region was incubated with nuclear extracts from MECs and with different nonlabeled competitors. Complexes were resolved on a nondenaturing 4% polyacrylamide gel followed by autoradiography of the dried gel. The addition of nuclear extract produced a specific shifting band denoted as I. *Nonspecific band. S indicates specific; NS, nonspecific; and SBE, Smad-binding element. (D) Nuclear extracts were extracted from MECs treated with BSA or BMP6 or cotransfected with Flag-Smad1 and Flag-Smad4. Overexpression of Smads induced a slower-migrating complex labeled as II.
Activation of the Cox2 promoter by BMP signaling. (A) Depiction of 4 luciferase reporters of the Cox2 promoter, −8653 to +53 bp, −7444 to +53bp, −1500 to +1 bp, and −371 to +70 bp. Putative Smad-binding sites are denoted. (B) MECs were transfected with luciferase reporter plasmids carrying different lengths of the Cox2 promoter in the presence or absence of BMP6 treatment or the cotransfection of an active Smad1 expression vector. After 48 hours, cells were lysed to measure luciferase activity. Luciferase activity was normalized using Renilla luciferase activity. Data shown are the means ± SD of triplicates. (C) An end-labeled fragment containing the 29-bp Smad-responsive region was incubated with nuclear extracts from MECs and with different nonlabeled competitors. Complexes were resolved on a nondenaturing 4% polyacrylamide gel followed by autoradiography of the dried gel. The addition of nuclear extract produced a specific shifting band denoted as I. *Nonspecific band. S indicates specific; NS, nonspecific; and SBE, Smad-binding element. (D) Nuclear extracts were extracted from MECs treated with BSA or BMP6 or cotransfected with Flag-Smad1 and Flag-Smad4. Overexpression of Smads induced a slower-migrating complex labeled as II.
Interestingly, this interval in the Cox2 promoter contains a tight cluster of putative Smad-binding elements (identified using the Vector NTI algorithm v.9.1; Invitrogen, Carlsbad, CA) between base pairs −8147 and −8119 (Figure 5A). To further characterize the BMP6-responsive element in the Cox2 promoter, we used this 29-bp sequence as a probe in gel shift assays after performing pilot experiments to exclude the possibility that other potential Smad-binding sites within the promoter had nuclear protein-binding activity. Using this 29-bp fragment as a probe, we found that extracts from MECs contained a DNA-binding activity that migrates with slower mobility than the probe alone (Figure 5C). This binding was competed away by addition of unlabeled specific probe at 50-fold excess but not by 100-fold excess of unlabeled nonspecific competitor, indicating its specificity. To analyze whether this binding is related to Smad-binding activity, we tested a consensus Smad-binding element sequence in competition assays.34 This sequence efficiently inhibited binding of the labeled probe to nuclear proteins, which strongly suggested that Smads are required nuclear proteins in the extract for binding to the 29-bp Cox2 element. Stimulation of MECs with BMP6 for 4 hours retarded the mobility and enhanced the intensity of the DNA-protein interaction (Figure 5D), suggesting that BMP6 remodels or posttranslationally modifies the protein complex that binds to this element. In addition, the coexpression of Flag-tagged Smad1 and Smad4 caused an additional DNA-protein complex of slightly slower mobility. Known difficulties in supershifting Smad proteins preclude definitive testing that the endogenous complexes contains proteins within this family, but our data are most consistent with a necessary role for Smad binding within the Cox2 promoter in mediating BMP6-dependent trans-activation.
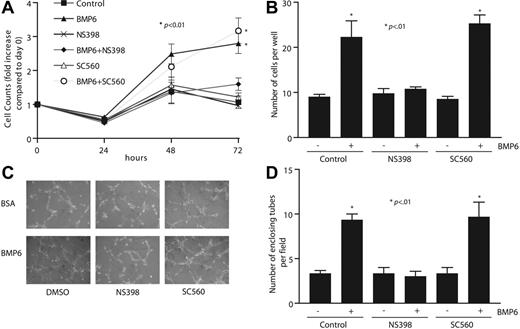
Inhibition of Cox2 blocks BMP6-induced EC proliferation, migration, and tube formation
To determine whether the up-regulation of Cox2 plays a necessary intermediary role in BMP6-dependent endothelial functions, we examined whether changes in BMP6-mediated MEC proliferation and migration occur after inhibition of Cox2. At the concentrations used here, NS398 exerted specific inhibition of Cox2 over Cox1, as determined by measurement of PGE2 production (data not shown). In cell-counting assays as an index of proliferation, BMP6 stimulated cell growth at both 48 and 72 hours after treatment (Figure 6A). Inhibition of Cox2 blocked BMP6-dependent MEC proliferation almost totally, whereas a specific Cox1 inhibitor, SC560, had no effect. We next measured cell migration activity using 2 assays: wound healing and Boyden chambers. BMP6 potently enhanced migration through wells, as has been previously described,7 and the addition of NS398 prevented BMP6-stimulated migration, whereas SC560 had no effect (Figure 6B). Similar results were observed in wound-healing assays (data not shown). We also tested the role of BMP6-dependent Cox2 activation on EC tube formation using Matrigel tube formation assays. The addition of BMP6 alone increased the number of enclosing tubes by more than 2-fold, and this increase could only be blocked by cotreatment with NS398, but not SC560 (Figure 6C-D). The results from these functional assays strongly support a model in which Cox2 is a necessary mediator of the effects of BMP6 on EC function.
Inhibiting Cox2 activity attenuates BMP6-induced MEC proliferation, migration, and tube formation. (A) Time-dependent response of MEC proliferation to BMP6 (100 ng/mL) treatment. MECs were pretreated with NS398 (10 μM), SC560 (100 nM), or vehicle before treatment with BMP6. Cells were counted at each time point. (B) MEC cell migration on gelatincoated filters to BMP6 was assayed after pretreatment with NS398, SC560, or vehicle. After 6 hours, transmigrated cells were counted. (C) MECs were plated on Matrigel-coated 24-well plates, followed by treatment with BMP6, NS398, SC560, and/or vehicle. Images were taken 6 hours after incubation. Original magnification, × 10. A Nikon 10×/0.25 NA objective lens (Nikon, Melville, NY) was used along with QCapture 2.6.0 software (Quantitative Imaging, Burnaby, BC, Canada). (D) The mean and SD of 3 replicate wells of a representative experiment is shown. The p-value was determined by Student t test.
Inhibiting Cox2 activity attenuates BMP6-induced MEC proliferation, migration, and tube formation. (A) Time-dependent response of MEC proliferation to BMP6 (100 ng/mL) treatment. MECs were pretreated with NS398 (10 μM), SC560 (100 nM), or vehicle before treatment with BMP6. Cells were counted at each time point. (B) MEC cell migration on gelatincoated filters to BMP6 was assayed after pretreatment with NS398, SC560, or vehicle. After 6 hours, transmigrated cells were counted. (C) MECs were plated on Matrigel-coated 24-well plates, followed by treatment with BMP6, NS398, SC560, and/or vehicle. Images were taken 6 hours after incubation. Original magnification, × 10. A Nikon 10×/0.25 NA objective lens (Nikon, Melville, NY) was used along with QCapture 2.6.0 software (Quantitative Imaging, Burnaby, BC, Canada). (D) The mean and SD of 3 replicate wells of a representative experiment is shown. The p-value was determined by Student t test.
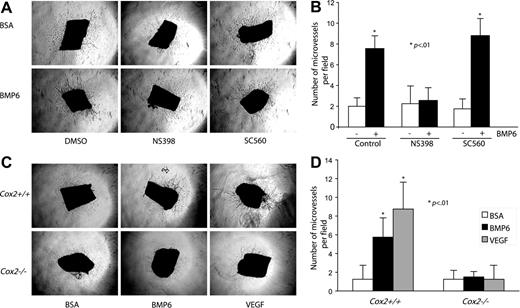
Requirement of Cox2 activity in BMP6-induced microvessel outgrowth
To better understand the effect of Cox2 activity in BMP6-induced angiogenesis, mouse aortic ring assays were selected to mimic an in vivo environment.26 Mouse thoracic aortas were sectioned, embedded in collagen gel, and incubated with or without BMP6 and/or Cox inhibitors. To minimize the background of microvessel outgrowth under control conditions, only 1% mouse serum was added to media. After a 6-day incubation, BMP6 treatment increased the total number of outgrowing microvessels from aortic rings (Figure 7A-B). Cotreatment with NS398 reduced BMP6-activated neovascular outgrowth to control levels, whereas SC560 had no effect. We also used genetic deletion of Cox2 to provide more definitive evidence of a role for endogenous Cox2 in mediating BMP6-dependent responses. Aortas from Cox2−/− mice and Cox2+/+ littermates were used in these experiments. The activation of microvessel outgrowth by BMP6 was abolished in aortas from Cox2−/− mice (Figure 7C-D). Although Cox2 activity may not be necessary for all VEGF-mediated events,35 VEGF induces Cox2 expression in ECs,36 and this induction is required for VEGF-mediated angiogenesis.37 Therefore, we also tested the effects of Cox2 deletion on VEGF-induced neovascularization. The addition of recombinant VEGF (25 ng/mL) significantly increased microvessel outgrowth in wild-type aortas, but not in Cox2−/− aortas (Figure 7C-D). These data indicate that BMP6 and VEGF signal pathways unexpectedly share common requisite downstream mediators in the regulation of angiogenesis.
The proangiogenic activity of BMP6 is blocked by Cox2 inhibition in a mouse aorta ring assay. (A) Mouse aorta rings (n = 4) were embedded in collagen gel in the presence of vehicle, BMP6, NS398, or SC560. Neovessel sprouts were blindly counted at day 6. Original magnification, × 4. (B) Quantitative analysis of aortic ring assays. Columns indicate mean of triplicates. Bars indicate SD. *P < .01 compared with control. Similar results were obtained in a second independent experiment. (C) Aortas from Cox2+/+ and Cox2−/− mice were tested in this neoangiogenic assay. Images of microvessel outgrowth from aorta with or without BMP6 treatment are presented. An Olympus 4×/0.16 NA objective lens was used. (D) Statistical data of aortic ring assay using Cox2−/− mice. Error bars indicate SD of triplicates; *P < .01 compared with Cox2+/+ BSA-treated groups.
The proangiogenic activity of BMP6 is blocked by Cox2 inhibition in a mouse aorta ring assay. (A) Mouse aorta rings (n = 4) were embedded in collagen gel in the presence of vehicle, BMP6, NS398, or SC560. Neovessel sprouts were blindly counted at day 6. Original magnification, × 4. (B) Quantitative analysis of aortic ring assays. Columns indicate mean of triplicates. Bars indicate SD. *P < .01 compared with control. Similar results were obtained in a second independent experiment. (C) Aortas from Cox2+/+ and Cox2−/− mice were tested in this neoangiogenic assay. Images of microvessel outgrowth from aorta with or without BMP6 treatment are presented. An Olympus 4×/0.16 NA objective lens was used. (D) Statistical data of aortic ring assay using Cox2−/− mice. Error bars indicate SD of triplicates; *P < .01 compared with Cox2+/+ BSA-treated groups.
Discussion
The BMP signaling pathway has an important but incompletely characterized role in mediating vascular development and angiogenesis. Mechanistically, little is known about downstream transcriptional targets of BMPs other than Id1 in ECs.7 In the present study, we have been able to generate a comprehensive list of BMP6 target genes in ECs using microarray technology. The accuracy of our dataset has been confirmed by both RT-PCR and Western blotting (Figure 3A-B; data not shown). Three major clusters of BMP-responsive genes were identified based on their patterns of expression (Figure 2), suggesting that independent pathways for gene activation by BMPs exist in ECs. In this screen, we found angiogenesis-associated genes that participate in intracellular and extracellular signaling and matrix reorganization, indicating a coordinated program to allow vessel assembly and maturation, including Cox2, Wnt2, Timp1, Adamts1, and Adamts5. An independent microarray study in our laboratory also found that Wnt2 is enriched in flk1+ differentiated embryonic stem cells, and that this Wnt signaling pathway plays a critical role in the regulation of angiogenesis.38 The present studies indicate that BMP6 is likely to be upstream from Wnt2 expression in the cascade of blood vessel growth, an observation that will be helpful to establish network relationships of different signaling pathways in angiogenesis.
Our analyses indicate that Cox2 is indeed a bona fide transcriptional target of the BMP-responsive signaling pathway. Cox2 mediates the biosynthesis of PGs from arachidonic acid, and the expression of Cox2 is up-regulated by cytokines, including VEGF and IL-1β.39 Increased Cox2-dependent PG generation plays diverse roles in increasing cell proliferation,40 inhibiting apoptosis,41 and stimulating angiogenesis.42,43 The present results suggest that the role of Cox2 in angiogenesis in vivo may be due in part to its induction by BMPs acting on the vascular endothelium to alter prostanoid generation.
We have identified in these studies a heretofore-uncharacterized BMP-responsive enhancer located at −8147 bp upstream from the Cox2 transcription start site (Figure 5). A number of transcription factors and signaling events have been previously linked to Cox2 transcriptional activation; for example, VEGF induces Cox2 up-regulation through p38 MAPK and JNK signaling pathways,37 and Wnt signaling increases Cox2 transcription via a β-catenin–dependent pathway that activates the Cox2 promoter through a Tcf-4–binding element.19 In osteoblasts, BMP2 activates Cox2 transcription via a specific binding site on the Cox2 promoter for core-binding factor activity 1;20 however, our data indicate that the induction of Cox2 by BMP6 follows different rules, at least in ECs, and these experiments provide the first link between Smad activation and Cox2 transcriptional regulation. Based on our analyses, we also predict the involvement of the EC-specific transcriptional factor GATA2 in BMP6-mediated Cox2 induction, since a GATA-rich sequence is adjacent to the Smad-binding site (data not shown). Additional reporter studies are being performed to clarify the relationship among these angiogenic transcription factors in the regulation of Cox2 in ECs.
Results from in vitro EC function assays, including proliferation, migration, and tube formation, clearly show that Cox2 activity is required for BMP6-induced EC activation (Figure 6). In addition, pharmacologic and genetic disruptions of Cox2 activity indicate that BMP6-activated microvessel outgrowth also requires Cox2 activity (Figure 7). At the same time, the inhibition of Cox1 does not attenuate angiogenesis stimulated by BMP6. Given the remarkable structural and kinetic similarity between these 2 enzymes, our findings are consistent with other observations that Cox2, but not Cox1, plays an important role in mediating angiogenesis under both physiologic and pathologic situations.13,35 Coupled with our own conclusions, this suggests that angiogenesis-promoting prostanoid generation in ECs must occur in a precise spatiotemporal fashion to activate the program of blood vessel growth. Remarkably, inhibition of Cox2 by NS398 reduces BMP6 mRNA and protein expression in bone cells.44 This suggests that positive feedback loops may exist in the BMP6 and Cox2 pathway, and such an interaction could play an important role in amplifying the activation of ECs by BMP6. Taken together, these studies provide further support for the consideration of pharmacologic Cox2-targeting strategies to modulate new vessel formation in diseases in which angiogenesis is a major component, including cancer and cardiovascular disease.
Authorship
Contribution: R.R., H.W., and C.P. designed research; R.R., C.Z., Y.W., and H.W. performed research; R.R., P.C.C., and C.P. analyzed data; and R.R. and C.P. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cam Patterson, Director, Division of Cardiology and Carolina Cardiovascular Biology Center, 8200 Medical Biomolecular Research Building, Chapel Hill, NC 27599-7126; e-mail: cpatters@med.unc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to Dr Da-Zhi Wang for providing Flag-tagged Smad1 and Smad4 constructs, and to Beverly Koller and Alysia Kern Lovgren for assistance with cyclooxygenase-deficient mice. We thank Xiaorui He for technical advice with microarray analysis.
This work was supported by National Institutes of Health grants HL 61656, HL 03658, and HL 072347 (C.P.). C.P. is an Established Investigator of the American Heart Association and a Burroughs Wellcome Fund Clinician Scientist in Translational Research.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal