Abstract
Standard chemotherapy fails in 40% to 50% of patients with diffuse large B-cell lymphoma (DLBCL). Some of these failures can be salvaged with high-dose regimens, suggesting a role for drug resistance in this disease. We examined the expression of genes in the glutathione (GSH) and ATP-dependent transporter (ABC) families in 2 independent tissue-based expression microarray datasets obtained prior to therapy from patients with DLBCL. Among genes in the GSH family, glutathione peroxidase 1 (GPX1) had the most significant adverse effect on disease-specific overall survival (dOS) in the primary dataset (n = 130) (HR: 1.68; 95% CI: 1.26-2.22; P < .001). This effect remained statistically significant after controlling for biologic signature, LLMPP cell-of-origin signature, and IPI score, and was confirmed in the validation dataset (n = 39) (HR: 1.7; 95% CI: 1.05-2.8; P = .033). Recursive partitioning identified a group of patients with low-level expression of GPX1 and multidrug resistance 1 (MDR1; ABCB1) without early treatment failures and with superior dOS (P < .001). Overall, our findings suggest an important association of oxidative-stress defense and drug elimination with treatment failure in DLBCL and identify GPX1 and ABCB1 as potentially powerful biomarkers of early failure and disease-specific survival.
Introduction
Anthracycline and alkylator combination chemotherapy, such as CHOP (cyclophosphamide, adriamycin, vincristine, and prednisone),1 has remained the standard first-line treatment for patients with diffuse large B-cell lymphoma (DLBCL) for almost 3 decades,2 with the recent addition of rituximab. However, only about 50% to 60% of patients are cured with this approach. An additional 10% to 20% of patients can be salvaged with high-dose chemotherapy followed by stem cell transplantation,3 implicating drug resistance as a significant cause of treatment failure in this disease. Several mechanisms of drug resistance, mostly arising from work in epithelial cancer cell lines, have been put forward to account for this variability
Anthracyclines are lipophilic compounds that enter the cell by free diffusion across the plasma membrane.4 ATP-dependent drug efflux via transmembrane proteins (ABC transporters) is a major route of anthracycline elimination and thus, a major determinant of intracellular drug levels (Figure 1). Since the identification of MDR1 glycoprotein (ABCB1) in multidrug-resistant cell lines, several additional ABC transporters have been identified, among them MRP1 (ABCC1), MRP2 (ABCC2), and breast cancer–resistance protein (BCRP; ABCG2). Although several lines of work suggest an association between ABC-transporter expression and inferior clinical outcome in various malignancies, studies in previously untreated patients with lymphomas, and DLBCL in particular, have produced controversial findings.5-8
Role of the glutathione pathway and ABC-transporter family in drug elimination and defense from ROS-mediated oxidative stress. Shaded diamonds represent intracellular drug and drug metabolites. Glutathione (GSH) is represented by clear pentagons. The rate-limiting step in its de novo biosynthesis is the enzyme glutamate-cysteine ligase (GCL). The balance of drug/drug metabolites and intracellular GSH is a significant determinant of drug levels and ROS stress. Glutathione peroxidase 1 (GPx1) reduces hydrogen peroxide generated after exposure to certain drugs, by oxidizing GSH to its disulfide form GSSG. Glutathione S-transferases (GST) conjugate GSH to drugs and drug metabolites, thus facilitating drug-conjugate elimination and efflux. GSTs also function as peroxidases to a lesser extent than GPx1 (not shown for simplicity). The ABC transporters ABCB1 (MDR1), ABCC1 (MRP1), and ABCC2 (MRP2) among others are involved in ATP-dependent efflux of drugs or drug-GS conjugates.
Role of the glutathione pathway and ABC-transporter family in drug elimination and defense from ROS-mediated oxidative stress. Shaded diamonds represent intracellular drug and drug metabolites. Glutathione (GSH) is represented by clear pentagons. The rate-limiting step in its de novo biosynthesis is the enzyme glutamate-cysteine ligase (GCL). The balance of drug/drug metabolites and intracellular GSH is a significant determinant of drug levels and ROS stress. Glutathione peroxidase 1 (GPx1) reduces hydrogen peroxide generated after exposure to certain drugs, by oxidizing GSH to its disulfide form GSSG. Glutathione S-transferases (GST) conjugate GSH to drugs and drug metabolites, thus facilitating drug-conjugate elimination and efflux. GSTs also function as peroxidases to a lesser extent than GPx1 (not shown for simplicity). The ABC transporters ABCB1 (MDR1), ABCC1 (MRP1), and ABCC2 (MRP2) among others are involved in ATP-dependent efflux of drugs or drug-GS conjugates.
Once inside the cell, anthracyclines exert their cytocidal action by several mechanisms, including DNA intercalation, topoisomerase II inhibition, and reactive-oxygen-species (ROS) formation that can cause widespread damage to DNA, phospholipids, and thiol-containing transport proteins.4 Several anthracycline-resistant cancer cell lines exhibit changes in critical aspects of the ROS defense system.9-12 Glutathione (GSH) is the most abundant intracellular nonprotein thiol13 and a key contributor to ROS defense. Its intracellular levels are primarily regulated by the enzyme glutamate-cysteine ligase (GCL), which catalyzes the rate-limiting step in overall GSH biosynthesis (Figure 1). The major downstream effector of GSH-mediated ROS defense is the enzyme glutathione peroxidase 1 (GPX1), which reduces hydrogen peroxide at the expense of oxidizing GSH to its disulfide form GSSG. Overexpression of GPX1 and other antioxidant enzymes, as well as depletion of intracellular GSH, have been shown to suppress anthracycline-induced apoptosis in several experimental systems, including cancer cell lines and tumor xenografts.14-18 Previous clinical studies have examined GPX1 function19 and genetic polymorphisms20 as they relate to susceptibility to lymphoma but not response to therapy. A cDNA microarray-based study of tumor samples from a patient population with DLBCL21 found increased expression of antioxidant defense enzymes, including GPX1, to be associated with a better prognosis in this disease.
Alkylating agents act through the covalent binding of alkyl groups to various cellular molecules.22 The glutathione family has also been implicated in alkylator resistance in cell lines as well as primary patient tissues (Figure 1).13,23-27 Specifically, glutathione-S-transferases (GSTs) comprise a family of related proteins that can enzymatically conjugate GSH to electrophilic chemotherapeutic agents and even facilitate their elimination through the ABC transporter MRP1 (ABCC1).28 Besides GSH conjugation activity, GSTs have organic peroxidase activity and may also play a role in protection from ROS induced by anticancer drugs.13 Moreover, recent studies have demonstrated that GST subclasses π and μ have a regulatory role in the cell by binding to key members of the survival and death-signaling pathway, such as c-Jun N-terminal kinase 1 (JNK1) and apoptosis signal-regulating kinase 1 (ASK1).29,30 Overexpression of GSTs at the protein and mRNA level has been noted in lymphomas. Although all classes have been studied, GSTP1 seems to be the one most consistently associated with inferior clinical outcomes.31,32 More recently, Ribrag et al showed that 50% or higher immunostaining of DLBCL cells for GSTP1 in 69 previously untreated patients was associated with inferior complete response rates and progression-free survival, but not overall survival.33
Based on the previous experimental observations implicating members of the ABC-transporter family and GSH pathway in resistance to anthracyclines and alkylating agents in several malignancies, we initiated this study in DLBCL. We thought that the complex interaction between these gene families at the molecular level could best be delineated with an approach that measures coordinate tissue expression of critical genes. Our experimental hypothesis was that elevated expression of members of the ABC-transporter and GSH families would be associated with inferior clinical outcomes in previously untreated patients with DLBCL. Additionally, coordinate expression of members of these pathways assessed via classification and regression tree analysis (CART) could identify subgroups with divergent risk for early treatment failure.
Materials and methods
Datasets and data
We used the publicly available dataset reported on by Monti et al34 for our primary analysis (http://www.broad.mit.edu/cgi-bin/cancer/datasets.cgi). This set includes pretreatment patient-level tumor-specimen expression data from the Affymetrix HG-U133 A and B microarray set (Affymetrix, Santa Clara, CA). Tumor specimens were nodal biopsies from newly diagnosed, previously untreated patients with DLBCL seen at Dana Farber/Partners Cancer Care and confirmed by expert hematopathology review. Expression signals were obtained using Affymetrix Microarray Suite 5.0 (MAS 5.0) software with the scaling factor (SF) and normalization factor (NF) set to 1, indicating no scaling or normalization.
Patients in this dataset (n = 130) received full-dose CHOP-based therapy (eg, 3-4 cycles + radiotherapy for localized disease or a minimum of 6 cycles for advanced disease) and had long-term clinical follow-up.34 Baseline clinical characteristics have been previously reported in Table 1 of the supplementary information.34 The primary clinical outcome in this expression dataset is consistent with disease-specific overall survival (dOS). DOS was measured from the date of diagnosis to the date of death or last follow-up without disease.
Based on gene expression in this dataset and using consensus clustering, Monti et al have identified 3 distinct biologic subtypes of DLBCL: oxidative phosphorylation (OxPhos), B-cell receptor/proliferation (BCR), and host response (HR).34 These subtypes have distinct genetic features and association with the tumor microenvironment but have not been found to be predictive of response to conventional chemotherapy. Since they provide important biologic information, they were used to stratify patients in our analysis.
Additionally, the “cell-of-origin” classification method (activated B-cell like, germinal center B-cell like, and type 3), developed by the Lymphoma/Leukemia Molecular Profiling Project (LLMPP)35 and later defined by the expression of 27 genes,36 was calculated for this dataset and used as a predictor/stratifying variable in our analysis. Finally, the international prognostic index (IPI), a 4-level index derived from baseline clinical variables, was made available on 116 patients and was included as a predictor/stratifying variable in this study.
An independent dataset previously reported on by Houldsworth et al37 was updated and used to validate findings from our primary analysis. This set includes pretreatment tumor-specimen expression data from 39 patients with untreated DLBCL from Memorial Sloan-Kettering Cancer Center. Expression-level data were generated on the Affymetrix HG-U95Av2 microarray (Affymetrix) and reanalyzed through MAS 5.0, with SF and NF set to 1. The survival end point evaluated in this dataset is disease-specific overall survival after anthracycline-based chemotherapy.
Derivation of expression level
Genes of interest in the ABC-transporter family and GSH pathway were mapped to unique probe sets in the primary dataset using the Netaffx tool from Affymetrix. When several probe sets were available for the same target gene, the signals across these sets were averaged, as long as there was a high correlation between them (Pearson coefficient > 0.8). When correlation was poor, a representative probe set was selected for each target gene based on Affymetrix probe set suffix designation (#_at preferred over #_s_at, #_s_at preferred over #_x_at) and target sequence characteristics (exemplar sequence preferred over consensus sequence; mRNA support preferred over EST support). The absolute expression signal of each gene was adjusted by the median expression signal of each subject along a set of 1798 one-to-one match genes across all Affymetrix platform generations. This set of matches across platforms (HU 6800, HG-U95Av2, and HG-U133) was derived using the Affymetrix good-match spreadsheets and eliminating one-to-many and many-to-one associations. Unless otherwise noted, all expression values reported are median-adjusted.
Gene expression in the secondary dataset was derived by mapping the selected probe sets from the HG-U133 chip to probe sets from the HG-U95Av2 chip using the Affymetrix best-match spreadsheets. Previous research suggests that correlation coefficients for replicates analyzed with different Affymetrix platforms are uniformly more than 0.8 for best-match probe sets with median adjustment.38 The absolute expression signal of each gene was again adjusted by the median expression signal of each subject along the set of 1798 one-to-one match genes across all Affymetrix generations.
The “redox signature score” recently described by Tome et al21 was calculated in the primary dataset as follows: The probe sets corresponding to the genes in the signature were identified through Netaffx. Specifically, these genes include: thioredoxin interacting protein, superoxide dismutases 1, 2, 3, glutathione peroxidases 1, 3, 4, thioredoxin reductases 1, 2, catalase, GSTAs, GSTOs, thioredoxin, and microsomal GSTs. Absolute expression values were derived as detailed in the first paragraph and adjusted by the median expression signal of the 1798 one-to-one match genes. The adjusted signals were then combined as described by Tome et al, and the signature was generated.
Statistical analysis
The correlation between genes was calculated using the Pearson correlation coefficient. Comparisons between categoric and ordinal variables were performed using the Fisher exact test. The nonparametric Kruskal-Wallis test was used to compare GPX1 levels across stratifying variables. All rates were estimated from survival distributions that were computed using the Kaplan-Meier method. Univariate hazard ratios for dOS were derived from the Cox proportional hazards (CPH) model, and P values were calculated based on the Wald test to assess the significance of factors in the model. The stricter P value cutoff of .01 was used to account for multiple comparisons. The proportional hazards assumption was tested using “log-log plots” (ie, ln(-ln(survival)) versus ln(time)) for each statistically significant variable. Interaction terms were incorporated in the regression model for each IPI, LLMPP, and biologic signature subgroup before generating stratified estimates of effect size.
Recursive partitioning using CART (Salford Systems, San Diego, CA) was used to develop a classification tree for the 2-year disease-specific overall survival rate (dOS2) based on all the genes in Table 1. This algorithm sequentially divides a group of patients into subgroups that become progressively more homogeneous with respect to clinical outcome than the original group. At the first step, all factors entered are examined at every possible cutoff (number of patients minus one) to select the factor that best splits the entire group of patients into 2 subgroups. At the second, as well as subsequent steps, a factor is selected (again from all potential factors and cutoffs) using the same process to subdivide the current group. The process stops when no further improvement in homogeneity can be made by splitting using the available factors. To minimize overfitting the data, groups with fewer than 5 patients were not further split. The tree was then pruned to develop final risk groups, between which the 2-year dOS rates differed significantly (P < .01). The 95% confidence intervals for dOS2 were calculated using the binomial distribution. All P values reported are 2 tailed. Statistical analysis was performed with the STATA software (v.9, 2005; STATA, Research Park, TX).
Association of gene expression with disease-specific overall survival in the primary dataset
| Gene . | Probe set ID* . | IQR range† . | Coefficient‡ . | 95% CI . | P . |
|---|---|---|---|---|---|
| ABCB1 (MDR1) | 209993_at | 0.50-0.88 | 0.01 | −0.7-0.8 | .9 |
| ABCC1 (MRP1) | 202804_at | 1.2-2.3 | 0.01 | −0.2-0.3 | .9 |
| 202805_s_at | |||||
| ABCC2 (MRP2) | 206155_at | 0.07-0.24 | 0.34 | −0.9-1.5 | .6 |
| ABCG2 (BCRP) | 209735_at | 0.7-1.2 | −0.11 | −0.7-0.5 | .7 |
| GCLM | 203925_at | 0.61-0.99 | 0.31 | −0.4-1.0 | .4 |
| GCLC | 202922_at | 1.3-2.0 | 0.24 | 0.008-0.5 | .04 |
| 202923_s_at | |||||
| GPx1 | 200736_s_at | 8.5-17 | 0.05 | 0.02-0.08 | < .001 |
| GPx2 | 202831_at | 0.94-1.6 | 0.20 | −0.1-0.7 | .1 |
| GPx3 | 201348_at | 1.7-3.5 | 0.07 | −0.02-0.2 | .1 |
| 214091_s_at | |||||
| GPx4 | 201106_at | 7.4-13 | −0.02 | −0.07-0.04 | .5 |
| GSTA1 | 203924_at | 0.17-0.55 | −0.74 | −1.6-0.1 | .1 |
| GSTA2 | 202478_at | 0.57-1.1 | 0.09 | 0.4-0.5 | .7 |
| GSTA3 | 222102_at | 0.11-0.24 | −0.28 | −1.8-1.2 | .7 |
| GSTA4 | 202967_at | 1.4-2.2 | −0.34 | −0.7-0.01 | .04 |
| GSTM1 | 204550_x_at | 1.8-2.5 | −0.02 | −0.1-0.06 | .6 |
| 215333_x_at | |||||
| GSTM4 | 204149_s_at | 0.05-0.19 | −0.02 | −0.4-0.4 | .9 |
| GSTM5 | 205752_s_at | 1.4-2.1 | −0.30 | −0.7-0.1 | .2 |
| GSTP1 | 200824_at | 5.9-14 | −0.007 | −0.04-0.02 | .6 |
| GSTT1 | 203815_at | 0.09-0.18 | −0.08 | −1.7-1.5 | .9 |
| Gene . | Probe set ID* . | IQR range† . | Coefficient‡ . | 95% CI . | P . |
|---|---|---|---|---|---|
| ABCB1 (MDR1) | 209993_at | 0.50-0.88 | 0.01 | −0.7-0.8 | .9 |
| ABCC1 (MRP1) | 202804_at | 1.2-2.3 | 0.01 | −0.2-0.3 | .9 |
| 202805_s_at | |||||
| ABCC2 (MRP2) | 206155_at | 0.07-0.24 | 0.34 | −0.9-1.5 | .6 |
| ABCG2 (BCRP) | 209735_at | 0.7-1.2 | −0.11 | −0.7-0.5 | .7 |
| GCLM | 203925_at | 0.61-0.99 | 0.31 | −0.4-1.0 | .4 |
| GCLC | 202922_at | 1.3-2.0 | 0.24 | 0.008-0.5 | .04 |
| 202923_s_at | |||||
| GPx1 | 200736_s_at | 8.5-17 | 0.05 | 0.02-0.08 | < .001 |
| GPx2 | 202831_at | 0.94-1.6 | 0.20 | −0.1-0.7 | .1 |
| GPx3 | 201348_at | 1.7-3.5 | 0.07 | −0.02-0.2 | .1 |
| 214091_s_at | |||||
| GPx4 | 201106_at | 7.4-13 | −0.02 | −0.07-0.04 | .5 |
| GSTA1 | 203924_at | 0.17-0.55 | −0.74 | −1.6-0.1 | .1 |
| GSTA2 | 202478_at | 0.57-1.1 | 0.09 | 0.4-0.5 | .7 |
| GSTA3 | 222102_at | 0.11-0.24 | −0.28 | −1.8-1.2 | .7 |
| GSTA4 | 202967_at | 1.4-2.2 | −0.34 | −0.7-0.01 | .04 |
| GSTM1 | 204550_x_at | 1.8-2.5 | −0.02 | −0.1-0.06 | .6 |
| 215333_x_at | |||||
| GSTM4 | 204149_s_at | 0.05-0.19 | −0.02 | −0.4-0.4 | .9 |
| GSTM5 | 205752_s_at | 1.4-2.1 | −0.30 | −0.7-0.1 | .2 |
| GSTP1 | 200824_at | 5.9-14 | −0.007 | −0.04-0.02 | .6 |
| GSTT1 | 203815_at | 0.09-0.18 | −0.08 | −1.7-1.5 | .9 |
IQR indicates interquartile range
ID from the Affymetrix HG-U133 A and B microarray set (Affymetrix).
Median-adjusted expression signal.
βx coefficient from Cox proportional hazards model.
Results
The median-adjusted expression of genes in the ABC-transporter and GSH families in previously untreated patients with DLBCL from the primary dataset was calculated and is displayed in Table 1. In univariate analysis, none of the ABC transporters evaluated was associated with dOS. Among members of the glutathione pathway, GPX1 was found to have a highly significant association with dOS. The hazard ratio for a 10-fold increase in GPX1 expression was 1.68 (95% CI: 1.26-2.22; P < .001).
CART analysis
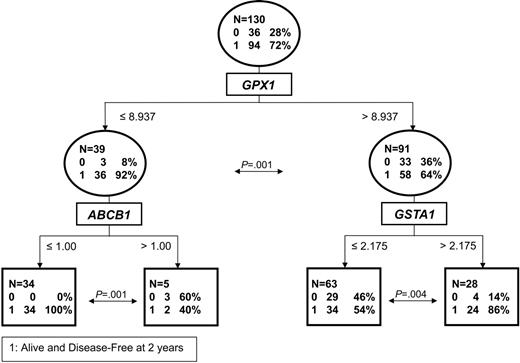
In order to study early resistance to treatment rather than late relapse, we evaluated the 31 failures (59% of total) occurring in the first 2 years and developed a classification tree for the rate of dOS at 2 years based on the median-adjusted expression of genes in Table 1. CART analysis identified the most powerful predictor of the 2-year dOS rate to be GPX1 expression with a cutoff at 8.9 (Figure 2). Specifically, 36 (92%) of 39 subjects (95% CI: 80%-97%) with low GPX1 expression (median: 6.1; range: 1.7-8.9) were alive and disease free at 2 years, compared with 58 (64%) of 91 (95% CI: 53%-73%) with high GPX1 expression (median: 14.5; range: 8.9-50.6) (P = .001). This analysis was also performed with IPI included for the 116 patients with a known IPI value. Of interest, CART identified GPX1 and not IPI as the best predictor of outcome in that approach.
CART for the dOS rate at 2 years. The cutoffs represent median-adjusted expression values of the genes indicated. Circles represent branch points, and squares are terminal nodes. The P values at each split are calculated for the binomial distribution.
CART for the dOS rate at 2 years. The cutoffs represent median-adjusted expression values of the genes indicated. Circles represent branch points, and squares are terminal nodes. The P values at each split are calculated for the binomial distribution.
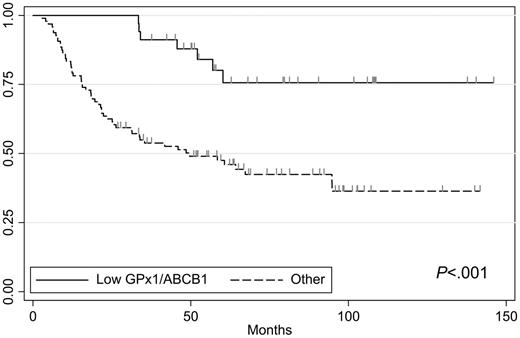
In the low GPX1 subgroup, MDR1 (ABCB1) emerged as the best predictor of outcome, at an expression cutoff of 1. ABCB1 expression above the cutoff (median: 1.4; range: 1.0-2.0) was thus associated with a 2-year dOS rate of 40% in 5 patients (95% CI: 12%-78%). In contrast, low expression of ABCB1 (median: 0.6; range: 0.1-1.0) identified a subgroup of 34 patients with no failures (100%; 95% CI: 90%-100%). The difference in 2-year dOS between the 2 subgroups defined by ABCB1 was statistically significant (P = .001). With an additional median follow-up of 3.1 years, only 7 late relapses occurred in the subgroup (n = 34) characterized by low levels of expression of both ABCB1 and GPX1 (Figure 3). Thus, dOS was highly statistically significantly different with long-term follow-up in this patient cohort compared with the others (P < .001). Moreover, this cohort with excellent prognosis was composed of patients in all IPI and LLMPP signature subgroups (Table 2).
Long-term outcome by CART classification. The group with low GPX1 and low ABCB1 expression levels was defined by the 2-year dOS rate and has no early failures. Tick marks reflect censoring.
Long-term outcome by CART classification. The group with low GPX1 and low ABCB1 expression levels was defined by the 2-year dOS rate and has no early failures. Tick marks reflect censoring.
Comparative distribution of IPI and LLMPP signature score among patients in the low- and high-risk groups identified by CART
| . | Study cohort, no. (%) . | Low GPX1/ABCB1, no. (%) . | High GPX1/GSTA1, no. (%) . |
|---|---|---|---|
| IPI | |||
| 1 | 45 (39) | 11 (37) | 13 (52) |
| 2 | 27 (23) | 8 (27) | 6 (24) |
| 3 | 32 (28) | 8 (27) | 2 (8)* |
| 4 | 12 (10) | 3 (10) | 4 (16) |
| LLMPP | |||
| GCB | 64 (49) | 20 (59) | 12 (43) |
| ABC | 24 (18) | 7 (21) | 4 (14) |
| Type 3 | 42 (32) | 7 (21) | 12 (43) |
| . | Study cohort, no. (%) . | Low GPX1/ABCB1, no. (%) . | High GPX1/GSTA1, no. (%) . |
|---|---|---|---|
| IPI | |||
| 1 | 45 (39) | 11 (37) | 13 (52) |
| 2 | 27 (23) | 8 (27) | 6 (24) |
| 3 | 32 (28) | 8 (27) | 2 (8)* |
| 4 | 12 (10) | 3 (10) | 4 (16) |
| LLMPP | |||
| GCB | 64 (49) | 20 (59) | 12 (43) |
| ABC | 24 (18) | 7 (21) | 4 (14) |
| Type 3 | 42 (32) | 7 (21) | 12 (43) |
Data available on 116 subjects for IPI and 130 subjects for LLMPP signature. Percentages reflect distribution of patients in each cohort.
Significantly different from entire cohort; Fisher exact for heterogeneity (P = .048).
In the high GPX1 expression subgroup, the best predictor of 2-year dOS was expression of glutathione-S-transferase GSTA1 with a cutoff of 2.2. The group with low GSTA1 expression (n = 63) had a poor outcome (dOS2 = 54%; 95% CI: 42%-66%; P = .004). In contrast, high levels of GSTA1 were favorably associated with outcome, with 24 (86%) of 28 subjects (95% CI: 68%-94%) being alive and disease-free at 2 years. In this subgroup, 7 additional failures occurred (11 total), with an additional median follow-up of 2.8 years (Table 2). There was no statistically significant difference in long-term outcome in this subgroup compared with the others (P = .30).
Glutathione peroxidase 1
As noted, GPX1 expression was found to be significantly associated with both early failure by recursive partitioning and dOS by survival analysis in the primary dataset. We thus chose to characterize GPX1 expression further in this patient cohort. Median GPX1 expression was 12.3 (range: 1.7-50.6). We observed no correlation between GPX1 expression and IPI score (Kruskal-Wallis test for heterogeneity, P = .15) or LLMPP subgroup (P = .24). Of interest, we observed a significant association of GPX1 expression with the BCR biologic subtype identified by Monti et al34 (median: 8.9) compared with the OxPhos (median: 15) and HR (median: 16) subgroups (P < .001).
As reported above, GPX1 was significantly associated with dOS in the primary dataset, with a hazard ratio for a 10-fold increase in GPX1 expression of 1.68 (95% CI: 1.26-2.22; P < .001). The association of GPX1 expression and dOS persisted when adjusted for IPI score, with a stratified hazard ratio of 1.72 (95% CI: 1.26-2.34; P = .001), or biologic subtype (stratified hazard ratio: 1.63; 95% CI: 1.17-2.28; P = .004). When adjusted for LLMPP signature subgroup, GPX1 expression also remained significantly associated with dOS, albeit with an effect modification (P = .022) (Figure 4). Specifically, in the activated B-cell–like subgroup (n = 24) high GPX1 expression had a pronounced adverse effect on dOS, with a hazard ratio of 7.7 (95% CI: 1.9-31; P = .004). The association with non-ABC subgroups was still significant but at a lower hazard ratio of 1.79 (95% CI: 1.30-2.47; P < .001). These results suggest that there may be a synergistic association of GPX1 expression and the activated B-cell–like phenotype with clinical outcome.
Effect of GPX1 expression and the LLMPP signature score on dOS. Patients are grouped in quartiles by GPX1 expression, with quartile 1 having the lowest and quartile 4 the highest level. The association with dOS is presented for the entire cohort (A), patients with the activated B-cell (ABC) subtype (B), and patients with non-ABC disease subtypes (C). Censoring not shown for simplicity.
Effect of GPX1 expression and the LLMPP signature score on dOS. Patients are grouped in quartiles by GPX1 expression, with quartile 1 having the lowest and quartile 4 the highest level. The association with dOS is presented for the entire cohort (A), patients with the activated B-cell (ABC) subtype (B), and patients with non-ABC disease subtypes (C). Censoring not shown for simplicity.
A recent cDNA microarray-based study in patients with DLBCL generated a redox signature score21 that was found to correlate with overall survival in this lymphoma. Given the significant role of the GSH pathway, and GPX1 in particular, in ROS defense, we wanted to test whether the redox signature score had any prognostic significance independently of GPX1 in our dataset. We found no significant association of the redox score with dOS (hazard ratio for a 10-fold change: 1.1; 95% CI: 0.99-1.2; P = .070). There was a borderline significant difference in dOS between the highest and lowest quartiles of the score (hazard ratio: 2.1; 95% CI: 0.99-4.6; P = .05) with the highest level of the score being associated with a poor clinical outcome, in contrast to the observation by Tome et al.21 This result is consistent with the association we found, since elevated levels of GPX1 are associated with an elevated redox score and with a poor clinical outcome. In fact, GPX1 expression appears to be the most significant component of that score. When GPX1 expression was removed from the redox signature, no association of the latter with clinical outcome was observed.
Evaluation of GPX1 in an independent dataset
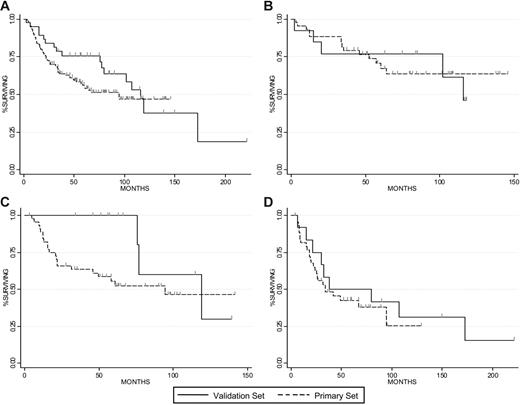
In the independent dataset, there are 39 subjects with DLBCL that were evaluated prior to the initiation of chemotherapy. The median dOS was similar in the primary and validation datasets (95 months and 116 months, respectively, P = .22, Figure 5A). In order to compare the 2 different patient populations, we examined the distribution of subjects across the IPI score (for the 116 patients with available data in the primary dataset) and we found them to be equivalent (P = .12 for heterogeneity). GPX1 expression in this dataset was also found to be adversely associated with dOS with a hazard ratio for a 10-fold change in expression at 1.7 (95% CI: 1.05-2.8; P = .033). Moreover, the survival outcome among subjects by tertile of GPX1 expression was comparable between the 2 datasets (Figure 5B–D). Due to the limited number of patients in the second dataset, we could not study the low-risk group identified by low GPX1 and ABCB1 expression in this patient cohort (no subjects met the cutoff criteria).
Clinical outcome among patients in the primary and validation datasets. DOS is presented for the primary and validation datasets (A). DOS is compared among patients in the lowest (B), middle (C), and highest (D) tertile of GPX1 expression in each dataset. Tick marks reflect censoring.
Clinical outcome among patients in the primary and validation datasets. DOS is presented for the primary and validation datasets (A). DOS is compared among patients in the lowest (B), middle (C), and highest (D) tertile of GPX1 expression in each dataset. Tick marks reflect censoring.
Discussion
In an effort to identify the molecular basis for the differential response to anthracycline- and alkylator-based chemotherapy among patients with DLBCL, we have analyzed the expression of genes in the ABC-transporter family and the GSH pathway in tumor tissue samples obtained prior to therapy. Our study demonstrates that high expression of GPX1, the major effector of GSH-dependent ROS defense, is adversely associated with early failure as well as disease-specific overall survival. This effect is observed even after adjusting for biologic subtype, IPI score, and LLMPP cell-of-origin signature, the most widely used prognostic tools in this disease. Moreover, using CART analysis, we have identified a subgroup of patients with low levels of GPX1 expression that has superior short-term and long-term clinical outcomes. GPX1 expression was also significantly associated with time to treatment failure in an independent dataset composed of 39 DLBCL patients from a different institution. We have made the assumption that microarray-derived expression levels reflect the expression of these genes in cancer cells. They may, however, also reflect gene expression in the organism as a whole, or expression in tumor-infiltrating cells, such as macrophages and T cells. To that effect, it is interesting that GPX1 levels were not significantly different in the HR biologic subtype, which is characterized by infiltrating host cells, compared with the others.
Assuming that the derived expression values primarily reflect mRNA levels in the tumor cells, our results suggest that augmented ROS and oxidative-stress defense may protect DLBCL cells from chemotherapy-induced death. This hypothesis is supported by several lines of investigation. Oxidative stress can induce apoptosis through numerous mechanisms.39-41 Several anthracycline- and alkylator-resistant cancer cell lines exhibit increased levels of enzymes within the free-radical defense system, including increases in GSH, GCL, and GPX1.9-13,23,26 l-buthionine-S-sulphoximine (BSO), a GCL inhibitor, which depletes intracellular GSH levels, has been shown to sensitize cells to treatment with alkylators and anthracyclines.13 Overexpression of GPX1 and other antioxidant enzymes has been shown to suppress apoptosis in cancer cell lines and tumor xenograft models,14-18 including in the setting of doxorubicin treatment.14-16 In lymphoma, the oncogene BCL2, overexpressed in 50% of DLBCLs, is known to inhibit apoptosis, and has been shown to do so in part through the antioxidant pathway and GPX1.18 To our knowledge, only one prior study has suggested that GPX1 overexpression did not have a significant protective effect on doxorubicin-induced apoptosis.42
Our results challenge the findings of a previous cDNA microarray-based study of tumor samples from a patient population with DLBCL,21 which found increased expression of antioxidant defense enzymes, including GPX1, to be associated with better prognosis. When we actually applied the redox signature score generated by these authors to our primary dataset, we found no significant association with dOS, except for the lowest and highest quartiles. Even then, the association was in the opposite direction of what was previously described, and in favor of our hypothesis that a high oxidative-stress defense, especially through high levels of GPX1, is associated with inferior clinical outcome. This adverse association was confirmed by us in an independent oligonucleotide dataset. There are several key differences in our approach, which can account for the different conclusions. The dataset on which the Tome et al21 study is based includes patients seen across decades at the NCI and treated with a variety of anthracycline-based chemotherapy regimens. The dataset we have used is composed of a more homogeneous group of patients from a single institution treated with CHOP chemotherapy. Of most importance, the study by Tome et al21 is based on a cDNA platform, while our dataset is derived from an oligonucleotide platform. Comparative microarray studies have shown generally moderate correlation between expression levels among these 2 inherently different platforms.43,44 Moreover, oligonucleotide-based arrays exhibit higher probe set specificity and are considered more reliable for assessing differential gene expression among samples.45,46
In previous reports, several ABC transporters have been implicated in drug resistance in cancer cell lines and patient samples from a variety of malignancies. The association between ABC-transporter expression and clinical outcomes in patients with lymphoma and DLBCL has been controversial. The ABC transporter MDR1 (ABCB1) is present by immunohistochemistry (IHC) in as many as 49% of newly diagnosed lymphomas.47 Several studies have documented inferior complete response rates among patients with MDR1 overexpression at the protein or mRNA level.5,7,8 However, a recently reported large IHC-based series of 207 patients showed no such association.6 Similarly, MRP1 (ABCC1) is present by IHC in 44% to 63% of previously untreated lymphomas.5,48 MRP1 had no impact on response to chemotherapy or survival in 48 patients examined by Filipits et al,48 but when coexpressed with MDR1 it was shown to correlate with inferior response rates but not overall survival in a similar patient population.5 In our study, we observed a significant association between MDR1 (ABCB1) expression and outcome by CART analysis but not by the univariate linear-effects Cox model. ABCB1 expression was associated with outcome only in the subgroup of patients with low-level GPX1 expression. The nature of this association (confounded by GPX1 and possibly other untested genes) may explain why it is seen in certain lymphoma patient populations and not in others. We did not observe an association between BCRP (ABCG2), MRP1 (ABCC1), or MRP2 (ABCC2) with outcomes by either analysis, confirming the prior IHC-based studies. Our study is based on gene expression rather than protein detection by IHC. Even though mRNA profiling does not account for posttranslational modification and protein localization, it allows a quantitative comparison between patient samples compared with IHC, which is semiquantitative.
GSTs have been associated with drug resistance to several chemotherapeutic agents in cells lines as well as primary patient tissues,25 especially in association with low GSH levels or inhibition of GSH synthesis.13,24 Besides GSH conjugation activity, GSTs have organic peroxidase activity49 and may also regulate key components of the cell survival and cell death signaling pathways.29,30 In our study, GSTA1 was favorably associated with outcome in the subgroup of patients with high GPX1 levels. However, the effect of GSTA1 did not persist with longer follow up.
In conclusion, we have used 2 independent expression microarray datasets derived from untreated patients with DLBCL to evaluate expression of genes in the ABC-transporter and GSH families and how it relates to clinical outcome after CHOP chemotherapy. In a disease such as DLBCL, with a well-established role of drug resistance in treatment failure in vivo, we have identified several members of the GSH pathway, and most notably GPX1, to be significantly adversely associated with early failure as well as disease-specific overall survival, in accordance with most published in vitro studies. We demonstrated that GPX1 expression had a robust association with outcome across clinically and biologically defined risk groups, as well as across datasets. Moreover, we were able to identify a group of patients with low-level expression of GPX1 and MDR1 (ABCB1) that had superior short-term and long-term clinical outcomes. The generalizability of our observations needs to be assessed in ongoing studies using other anthracycline-containing regimens or chemotherapy and monoclonal antibody treatment combinations in patients with DLBCL. Nevertheless, our findings provide the rationale for prospective intervention-based studies that target patients with a high likelihood of chemotherapy resistance for alternative treatment strategies.
Authorship
Contribution: C.A., P.A.G., S.J.S., and T.R.R. designed and contributed to the research; C.A. and P.A.G. performed the research; P.W. and R.H. assisted with the analysis; J.H. contributed the independent dataset; C.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charalambos Andreadis, Abramson Cancer Center, University of Pennsylvania, 16 Penn Tower, 3400 Spruce St, Philadelphia, PA 19104; e-mail: babis.andreadis@uphs.upenn.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grant number 5-K12-CA076931 from the National Cancer Institute (Bethesda, MD). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
We thank Drs Margaret A. Shipp and Donna Neuberg for providing supplemental patient information on the primary dataset. We also thank Dr Georgia Hatzivassiliou for her insightful comments and editorial support.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal