Abstract
We report a novel t(7;9)(q11;p13) translocation in 2 patients with B-cell acute lymphoblastic leukemia (B-ALL). By fluorescent in situ hybridization and 3′ rapid amplification of cDNA ends, we showed that the paired box domain of PAX5 was fused with the elastin (ELN) gene. After cloning the full-length cDNA of the chimeric gene, confocal microscopy of transfected NIH3T3 cells and Burkitt lymphoma cells (DG75) demonstrated that PAX5-ELN was localized in the nucleus. Chromatin immunoprecipitation clearly indicated that PAX5-ELN retained the capability to bind CD19 and BLK promoter sequences. To analyze the functions of the chimeric protein, HeLa cells were cotransfected with a luc-CD19 construct, pcDNA3-PAX5, and with increasing amounts of pcDNA3-PAX5-ELN. Thus, in vitro, PAX5-ELN was able to block CD19 transcription. Furthermore, real-time quantitative polymerase chain reaction (RQ-PCR) experiments showed that PAX5-ELN was able to affect the transcription of endogenous PAX5 target genes. Since PAX5 is essential for B-cell differentiation, this translocation may account for the blockage of leukemic cells at the pre–B-cell stage. The mechanism involved in this process appears to be, at least in part, through a dominant-negative effect of PAX5-ELN on the wild-type PAX5 in a setting ofPAX5 haploinsufficiency.
Introduction
PAX5 is a member of the highly conserved paired-box (PAX) domain family of transcription factors. The PAX5 gene plays an important role in both cell differentiation as well as in embryonic development, and is located on chromosome 9p13.1 This locus contains 2 distinct promoters, resulting in 2 alternative 5′ exons (1a and 1b).2 PAX5b is transcribed in the central nervous system and testis as well as in the B-lymphoid lineage, while PAX5a, also named B-cell–specific activator protein (BSAP), is specifically transcribed in the lymphoid lineage. PAX5a/BSAP is expressed from early stages of B-cell development up to mature B cells and is down-regulated during terminal differentiation into plasma cells.3 In bone marrow of Pax5−/− mice, B lymphopoiesis is completely arrested at the pro-B stage, revealing the essential role of this gene in early B-cell lymphopoiesis.4
PAX5-binding sites have been identified in the regulatory sequences of a number of genes, including CD19,5 BLK,6 BLNK,7 MB1 (Igα), N-myc, LEF1,8 and RAG2.9 PAX5 can act as an activator or a repressor of transcription. It has been shown to activate CD19, N-myc, MB1, and LEF1 expression and to repress PD-1 transcription.8 PAX5 plays an essential role in B-cell lineage commitment by suppressing alternative lineage choices.10-12 Indeed, it represses the transcription of Notch1, which is necessary for the commitment of lymphoid progenitors to the T-cell pathway,13 and the transcription of M-CSFR, thus rendering B-cell precursors unresponsive to the myeloid cytokines such as macrophage colony-stimulating factor (M-CSF).12
Chromosomal translocations involving the PAX5 gene have been described in different types of B-cell malignancies. The t(9;14)(p13;q32) translocation in B-cell non-Hodgkin lymphoma results in juxtaposition of the complete coding sequence of PAX5 to the IGH gene cluster, and leads to elevated PAX5 expression.14,15 Moreover, PAX5 is involved in the chimeric transcript PAX5-ETV6 in patients with B-cell acute lymphoblastic leukemia (B-ALL) who have a dic(9;12)(p13;p13) translocation.16,17 The latter fusion protein retains the paired-box domain of PAX5 fused to the helix-loop-helix and DNA-binding domain of ETV6. In this case, the chimeric protein most probably acts as an aberrant transcription factor.17 The paired box is a DNA-binding domain that is highly conserved during evolution in all PAX genes.18-20 This domain is conserved in all fusion transcripts involving a PAX gene, including PAX5-ETV6 and others described, such as PAX3-FKHR21 and PAX8-PPARG1.22 PAX5 also contains a conserved octapeptide motif and a partial homeodomain. The C-terminal region contains a transcriptional activation domain, and the extreme C-terminal region acts as a repression domain.23 Furthermore, PAX5 displays aberrant hypermutation in more than 50% of diffuse large B-cell lymphomas.24
In this study, we report the molecular characterization of a new chromosomal t(7;9)(q11;p13) translocation in 2 patients with B-ALL. This translocation juxtaposes the 5′ region of PAX5 (at 9p13), including its paired-box domain (PBD) and almost the entire sequence of elastin (ELN) (at 7q11.3).
Patients, materials, and methods
Patients
Patient 1 is a 38-year-old male who presented with a B-ALL of pre-B3 phenotype (CD10+ Cμ+). Cells were obtained from a bone marrow aspiration. The karyotype showed an isolated t(7;9)(q11;p13) translocation: 46, XY, t(7;9)(q11;p13)[17].
Patient 2 is a 16-year-old male with B-ALL of pre-B2 phenotype (CD10+ Cμ−). Cells were obtained from a bone marrow aspiration. The karyotype displayed a balanced t(7;9)(q11;p13) translocation associated with a deletion 9p on the other chromosome 9: 46, XY, t(7;9)(q11;p13), del(9)(p11p13)[14]/46, XY[1].
In parallel, 6 other patients with B-ALL were selected to serve as controls. All these patients had a pre–B-cell phenotype (pre-B1 to pre-B4).25 Their karyotypes were as follows: C1 (pre-B4): 46,XY, del(6)(q13q23),der(19)t(1;19)(q23;p13)[10]/46, idem,der(21)t(1;21)(q12;p11)[2]/46, XY[4]; C2 (pre-B2): 46, XY, t(9;22)(q34;q11)[11]; C3 (pre-B1): 46, XX, t(4;11)(q21;q23)[19]/46, XX[1]; C4 (pre-B3): 46, XY, del(11)(q21), dup(21)(q?)[2]/46, XY[10]; C5 (pre-B2): 46, XX, t(1;22)(q11;p11)[10]/46, XX, t(1;22)(q11;p11), del(11)(q14q23)[1]/ 46, XX[8]; and C6 (pre-B2/3): 46, XY, t(9;22)(q34;q11)[3]/45, idem,−7[7].
The approval of the French equivalent of the Institutional Review Board (Comité Consultatif de Préservation des Personnes en Recherche Biologique [CCPPRB]) is not required for investigations based on archived materials that have been routinely used for diagnostic purposes.
Cell lines
DG75, HeLa, and NIH3T3 cells were grown at 37°C, 5% CO2, in RPMI medium or Dulbecco modified Eagle medium (DMEM; Invitrogen-Gibco, Carlsbad, CA) with 10% decomplemented fetal calf serum (FCS; Invitrogen-Gibco), 2 mM l-glutamine, 1 mM sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin.
FISH
The bacterial artificial chromosome (BAC) clones spanning the PAX5 gene (RP11-243F8, RP11-297B17, and RP11-344B23) and the ELN gene (CTD-2278A11) were obtained from the Welcome Trust Sanger Institute (http://www.sanger.ac.uk). DNA was labeled with biotin-16-dUTP or digoxigenin-11-dUTP (Roche, Indianapolis, IN) using a nick-translation kit (Amersham Biosciences, Arlington Heights, IL) according to the manufacturer's instructions. Slides were observed under a Zeiss Axioplan 2 microscope (Zeiss, Jena, Germany) equipped with a 100×/1.3 numerical aperture (NA) oil objective. Images were acquired with a high-resolution JAIM3000 camera (Metasystems, Altlussheim, Germany) and were processed using ISIS3 software (Metasystems).
3′ RACE PCR and RT-PCR
Total RNA was extracted according to the TRIZOL method (Invitrogen-Gibco). Total RNA (1 μg) from Patient 1 was reverse-transcribed into cDNA using an oligo dT-anchor primer (5′/3′ rapid amplification of cDNA ends [RACE] kit, 2nd generation; Roche). The cDNA product was used as template in a polymerase chain reaction (PCR) experiment using the BD Advantage 2 PCR enzyme system (BD Biosciences, Palo Alto, CA), a PCR anchor primer (5′-GACCACGCGTATCGATGTCGAC-3′), and a PAX5-specific forward primer, PAXF (5′-CATAGTGTCCACTGGCTCCGTG-3′), corresponding to the nucleotides spanning exons 4 and 5 junction of PAX5 mRNA (NM_016734, National Center for Biotechnology Information [NCBI]). PCR was carried out with a GeneAmp PCR system 9600 (Perkin Elmer, Shelton, CT) according to the following cycling parameters: 95°C initial denaturation for 2 minutes, 5 cycles of 95°C for 5 seconds, 72°C for 3 minutes, 5 cycles of 95°C for 5 seconds, 70°C for 3 minutes, and 25 cycles of 95°C for 5 seconds, 68°C for 3 minutes. A final extension was performed at 72°C for 10 minutes. The PCR product was purified using the QiaQuick PCR Purification kit (Qiagen, Valencia, CA). An internal PAXG primer (5′-GGTATTCAGGAGTCTCCGGTGC-3′) was used to sequence the PCR product using the ABI Prism Dye Terminator kit (Perkin-Elmer; Applied Biosystems, Foster City, CA).
The PAX5a-ELN transcript was amplified using PAXQ (5′-CCCTGTCCATTCCATCAAGTCCTG-3′), and ELN9 (5′-ATGAGGTCGTGAGTCAGGGGTC-3′) primers; and the PAX5b-ELN transcript was amplified using PAXQ and PAXR (5′-CCGGCCTGCAGTCTGGAGCG-3′) primers.
Cloning of PAX5-ELN
Full-length PAX5-ELN and wild-type (wt)–PAX5 cDNA was amplified from patient 1 using PAXQ or ELN9 and PAXQ or PCR5 (5′-GGCTGGGCTGGGGCTGCGGTTATTT-3′) primers, respectively, and was cloned in the pCRII vector (TA cloning kit, Dual Promoter; Invitrogen-Gibco). After digestion with EcoRI, PAX5-ELN was cloned into a pcDNA3 vector (Invitrogen-Gibco). PAX5 was also cloned in pcDNA3 after digestion using HindIII and NotI. The constructs were sequenced and tested using the Transcend Non-Radioactive Translation Detection Systems (Promega, Madison, WI).
Cell transfections
Transient transfections were performed on DG75 with 15 μg pcDNA3-PAX5-ELN by electroporation as described previously.26 HeLa and NIH3T3 cells were transfected using FuGENE 6 Transfection Reagent (Roche).
For real-time quantitative (RQ)–PCR experiments, 5 × 106 DG75 cells were cotransfected with 30 μg pcDNA3 or pcDNA3-PAX5-ELN constructs and 1 μg of a green fluorescence protein (GFP) expression construct (pmaxGFP; Amaxa Biosystems, Cologne, Germany). GFP positive cells were sorted, 1 day after transfection, using an EPICS ALTRA cell sorter (Beckman Coulter, Hialeah, FL).
Western blotting
Cells were lysed with Passive Lysis Buffer (PLB; Promega) for 30 minutes at 4°C on a rocking platform and then sonicated 3 times for 10 seconds. Total proteins were separated by 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then electrotransferred onto nitrocellulose membranes. The membranes were blocked with a solution of 1% milk and 1% bovine serum albumin (BSA) in TBST (0.1% Tween 20 in TBS) and probed with appropriate primary and secondary antibodies diluted in TBST. The immunoreactive bands were visualized using the enhanced chemiluminescence (ECL) lighting system (Perkin Elmer).
Confocal microscopy
Cells were labeled as described previously,27 24 hours after transfection, using the anti-PAX5 N-terminal rabbit polyclonal antibody (ab12000; Abcam, Cambridge, MA) or the anti-PAX5 mouse monoclonal antibody (sc-13146; Santa Cruz Biotechnology, Santa Cruz, CA) and the anti-ELN rabbit polyclonal antibody (sc-25736; Santa Cruz Biotechnology). They were then incubated with the appropriate secondary antibody (Alexa Fluor 488 anti-rabbit antibody and Alexa Fluor 594 goat anti-mouse antibody). Slides were observed under a Zeiss LSM 510 confocal inverted microscope equipped with a 63×/1.4 NA oil objective. Images were processed using Zeiss LSM 510 software.
ChIP
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's instructions. Briefly, DG75 cells were transiently transfected with 15 μg of pcDNA3-PAX5-ELN, pcDNA3-PAX5, or pcDNA3 alone (negative control). After 40 hours of growth, 1% formaldehyde was added directly to the culture medium to cross-link proteins to DNA. The cell pellet was lysed with SDS lysis buffer and sonicated to shear DNA to a size range between 200 and 1000 bp. After centrifugation, the supernatant was diluted 10-fold in ChIP dilution buffer and incubated and rotated overnight at 4°C with anti-ELN, anti-PAX5, mouse IgG2a (DAKO, Carpinteria, CA), normal rabbit IgG (Upstate Biotechnology), or without antibody. Protein A agarose/salmon sperm DNA were then used to immunoprecipitate the antibody/protein/DNA complex. After washing, the complex was incubated at 65°C for 4 hours to reverse the protein/DNA cross-links. The DNA was then purified by phenol/chloroform extraction and used as a template for PCR amplification. PCR was performed using the BD Advantage 2 PCR enzyme system (BD Biosciences), CD19F (5′-GTGGTAGTGAGAGCTGGGATGAA-3′), and CD19R (5′-CAGCTTCGCGCAGGGGATGCT-3′) primers to amplify the region spanning the PAX5-binding site of the CD19 promoter, BLKPrF (5′-GAGGCTCTGAGATCCAAGTCAAC-3′) and BLKPrR (5′-TGTGCCATCTGTGTGCACCAGG-3′) primers for the BLK promoter, and SERPINA1PrF (5′-AGCCCAGATGCTGCAATAGA-3′) and SERPINA1PrR (5-AGCAGGTGAGGCAAGTAAGG-3′) for the SERPINA1 promoter. PCR products were resolved on 2% agarose gels, visualized after ethidium bromide staining, and subsequently sequenced.
Luciferase assay
HeLa cells were transfected with 5 μg luc-CD19, 1 μg pcDNA3-PAX5, increasing amounts of pcDNA3-PAX-ELN (0.3-9.0 μg), and 0.4 μg pRL-CMV (Promega). Cells were lysed, 40 hours after transfection, with 1 mg/mL BSA in PLB. Firefly luciferase gene expression normalized for Renilla luciferase activity was analyzed using the Dual-luciferase Reporter assay system (Promega). Additional experiments using 5 μg luc-CD19, increasing amounts of pcDNA3-PAX5 (1.3-10.0 μg), and 0.4 μg pRL-CMV were performed as a control of specificity (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article).
Quantitative RT-PCR
cDNA was synthesized using Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT; Invitrogen) according to the manufacturer's instructions. Quantitative PCR was performed on an ABI7000 (Applied Biosystems) using qPCR MasterMix Plus for SYBR green (Eurogentec, Herstal, Belgium). CD19A (5′-CTTCA TCTTCAAAGAGCCCTGG-3′) and CD19B (5′-CTTTGAAGAATCTCCTGGTGGG-3′) primers were used to amplify the CD19 cDNA. BLKF (5′-CCTGGAAATGGAAAGGTGG TTC-3′) and BLKR (5′-GGAGAAGGCACCTTTGTTGGTT-3′) amplified the BLK cDNA. BLNKA (5′-CTGCCAAGATTTACAGAAGGGG-3′) and BLNKB (5′-AACGCCAGCTTC CTGTTCTGAA-3′) amplified the BLNK cDNA. MB1A (5′-CTGCTGCTGTTCAGGAAA CG-3′) and MB1B (5′-GTCCAGGTTCAGGCCTTCAT-3′) amplified the MB1 cDNA. LEF1A (5′-ACACCCGTCACACATCCCAT-3′) and LEF1B (5′-CACCCGGAGACAA GGGATAA-3′) amplified the LEF1 cDNA. The presented data correspond to the mean of 2−ΔΔCt from 3 independent reactions, normalized to the MLN51 and S14 reference genes.
Results
Rearrangement of PAX5
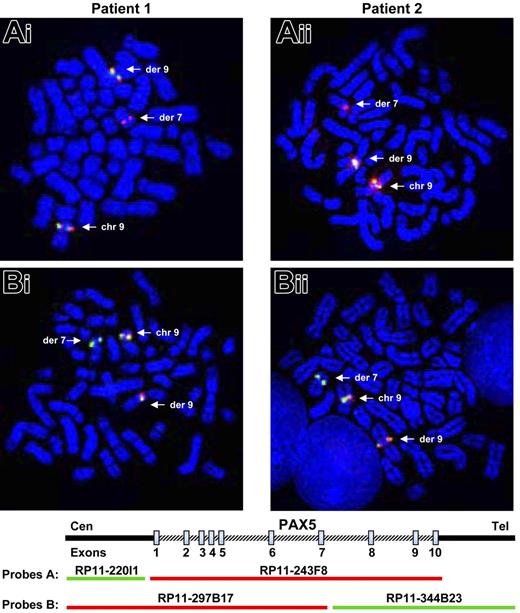
The investigations were based on the assumption that PAX5, located at 9p13, was rearranged in the 2 patients. As PAX5 plays an important role in the B-cell lineage commitment, its deregulation could explain the differentiation blockage of the leukemic cells. We therefore tested metaphase chromosomes of the 2 patients by FISH (fluorescent in situ hybridization) with PAX5 BAC probes. When we used the probe RP11-243F8 corresponding to the entire sequence of PAX5 gene, we observed a split signal in both patients: one part on chromosome 9p13 and the other part on chromosome 7q11 (Figure 1Ai-ii). With additional probes (RP11-297B17 and RP11-344B23), we observed that PAX5 was split between exons 7 and 10 in both patients (Figure 1Bi-ii).
Rearrangement of the PAX5 gene detected by FISH. FISH with PAX5 probes on both patients. (Ai-ii) FISH with PAX5 probe A: the RP11-243F8 BAC probe corresponds to the entire sequence of PAX5 gene. The signal of this probe is split between derived chromosomes 7 and 9 (arrows). (Bi-ii) FISH with PAX5 probe B: the BAC RP11-297B17 and RP11-344B23 correspond respectively to the 5′ part of the PAX5 gene up to intron 7 and the 3′ part of PAX5 from intron 7. FISH only shows a split of the RP11-344B23 probe. Thus, PAX5 is rearranged in both patients between exons 7 and 10.
Rearrangement of the PAX5 gene detected by FISH. FISH with PAX5 probes on both patients. (Ai-ii) FISH with PAX5 probe A: the RP11-243F8 BAC probe corresponds to the entire sequence of PAX5 gene. The signal of this probe is split between derived chromosomes 7 and 9 (arrows). (Bi-ii) FISH with PAX5 probe B: the BAC RP11-297B17 and RP11-344B23 correspond respectively to the 5′ part of the PAX5 gene up to intron 7 and the 3′ part of PAX5 from intron 7. FISH only shows a split of the RP11-344B23 probe. Thus, PAX5 is rearranged in both patients between exons 7 and 10.
Identification of the partner of PAX5
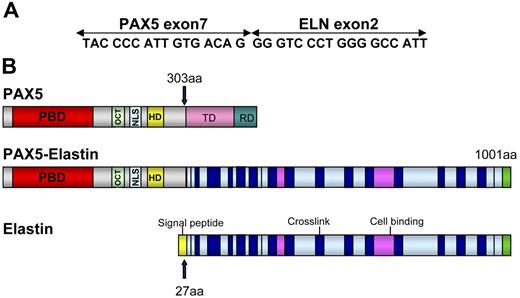
Several lines of evidence indicated that a new chimeric gene was created, composed of the 5′ part of PAX5 containing the PBD domain and the 3′ part of another gene. To identify the partner of PAX5, we performed 3′ RACE PCR using primers located upstream from exon 7. A 1000-bp product was obtained from the RNA of Patient 1 (Figure 2A). Sequencing revealed that this product corresponded to the 5′ part of PAX5 fused to ELN. FISH confirmed involvement of ELN with a specific BAC probe (Figure 2B). For both patients, the structure of the open reading frame of the chimeric PAX5-ELN transcript showed that PAX5 exon 7 (NM_016734, NCBI) was fused in frame with ELN exon 2 (NM_000501) (Figure 3A).
Rearrangement of the ELN gene. (A) 3′ RACE PCR was performed using a PAX5-specific primer and an anchor primer. We obtained a 1000-bp product that corresponds after sequencing to a part of PAX5 fused to the ELN transcript from its exon 2. (B) FISH analysis with the ELN BAC probe (CTD-2278A11). (C) Amplification of the 2 alternative transcripts by RT-PCR: PAX5a-ELN and PAX5b-ELN from the mRNA of patient 1.
Rearrangement of the ELN gene. (A) 3′ RACE PCR was performed using a PAX5-specific primer and an anchor primer. We obtained a 1000-bp product that corresponds after sequencing to a part of PAX5 fused to the ELN transcript from its exon 2. (B) FISH analysis with the ELN BAC probe (CTD-2278A11). (C) Amplification of the 2 alternative transcripts by RT-PCR: PAX5a-ELN and PAX5b-ELN from the mRNA of patient 1.
Schematic representation of PAX5-ELN. (A) Open reading frame of the chimeric PAX5-ELN gene. (B) Structure of PAX5, PAX5-ELN and ELN proteins. OCT indicates octapeptide; TD, transactivation domain; and RD, repressor domain.
Schematic representation of PAX5-ELN. (A) Open reading frame of the chimeric PAX5-ELN gene. (B) Structure of PAX5, PAX5-ELN and ELN proteins. OCT indicates octapeptide; TD, transactivation domain; and RD, repressor domain.
RT-PCR analysis revealed the presence of 2 alternative fusion transcripts, PAX5a-ELN and PAX5b-ELN, as shown in Figure 2C. However, we were unable to detect the reverse fusion transcript ELN-PAX5, which may be explained by differential promoter activity, as we observed the presence of wild-type PAX5 mRNA but not ELN mRNA (M.B., unpublished results, July 2006).
The structure of the chimeric protein therefore comprised the PAX5 PBD (DNA-binding domain) and its nuclear localization sequence (NLS) fused to ELN without its signal peptide (Figure 3B).
Nuclear localization of PAX5-ELN
Due to the structure of PAX5-ELN, we postulated that this fusion protein should be localized to the nucleus. To confirm this hypothesis, we cloned the full-length cDNA amplified from Patient 1's material in a pcDNA3 vector. The pcDNA3-PAX5a-ELN construct was selected for further experiments because PAX5a corresponds to BSAP, specific to the B lineage. The construct was sequenced and successfully tested in in vitro transcription-translation experiments (M.B., unpublished results, October 2005). Transient transfections with this construct were performed in NIH3T3 cells. Because NIH3T3 cells do not contain PAX5, we used an anti-PAX5 N-terminal antibody to visualize the PAX5-ELN protein. As shown in Figure 4A, we observed a nuclear localization of PAX5-ELN. This result was expected since PAX5-ELN retains the nuclear localization signal of wt-PAX5. Similar results were obtained in DG75-transfected cells (Figure 4B). Conversely, DG75 cells do not contain ELN, so we used an anti-ELN antibody to visualize the PAX5-ELN protein. An antibody directed against the C-terminal region of PAX5 was used to visualize the endogenous protein. Confocal microscopy confirmed that PAX5-ELN and PAX5 were both localized in the nucleus. However, although there was an overlap of PAX5 and PAX5-ELN staining, it remains difficult to categorically state that both proteins colocalized (Figure 4B). Transfection experiments on HeLa cells further support the nuclear localization of the PAX5-ELN protein (M.B., unpublished results, April 2006).
Localization of PAX5-ELN by confocal microscopy. (A) Nuclear localization of PAX5-ELN in NIH3T3 cells (anti-PAX5; green) (B) Nuclear localization of PAX5-ELN and endogenous PAX5 in DG75 cells (anti-elastin [H-300] is green; anti–PAX-5 [A-11], red).
Localization of PAX5-ELN by confocal microscopy. (A) Nuclear localization of PAX5-ELN in NIH3T3 cells (anti-PAX5; green) (B) Nuclear localization of PAX5-ELN and endogenous PAX5 in DG75 cells (anti-elastin [H-300] is green; anti–PAX-5 [A-11], red).
Binding of PAX5-ELN on CD19 and BLK gene promoters
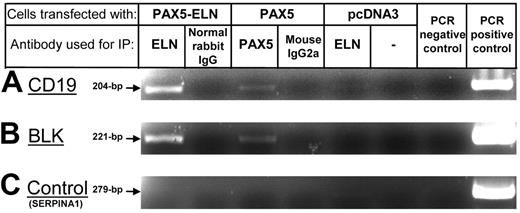
Since PAX5-ELN is localized to the nucleus, we sought to determine whether PAX5-ELN was able to bind the promoter of PAX5 target genes. The CD19 gene was chosen because it is described as a specific target of PAX5. Therefore, we transfected DG75 cells with pcDNA3-PAX5-ELN or pcDNA3-PAX5 constructs. ChIP experiments on transfected DG75 cells were performed using anti-ELN or anti-PAX5 antibodies. PCR reactions with specific primers clearly showed that PAX5-ELN was able to bind endogenous CD19 promoter sequences, as was PAX5 (Figure 5A). The same experiments were performed for BLK promoter sequences, another known target of PAX5. We observed the same results: both PAX5-ELN and PAX5 were able to bind to BLK promoter sequences (Figure 5B).
ChIP experiments on CD19 and BLK promoters. (A) PCR amplification of the CD19 promoter fragments after ChIP: PAX5-ELN binds CD19 promoter sequences as PAX5. (B) PCR amplification of the BLK promoter. PAX5-ELN binds BLK promoter sequences as PAX5. (C) Non-PAX5 target sequence PCR control. PCR amplification of the SERPINA1 promoter.
ChIP experiments on CD19 and BLK promoters. (A) PCR amplification of the CD19 promoter fragments after ChIP: PAX5-ELN binds CD19 promoter sequences as PAX5. (B) PCR amplification of the BLK promoter. PAX5-ELN binds BLK promoter sequences as PAX5. (C) Non-PAX5 target sequence PCR control. PCR amplification of the SERPINA1 promoter.
Dominant-negative effect of PAX5-ELN on PAX5
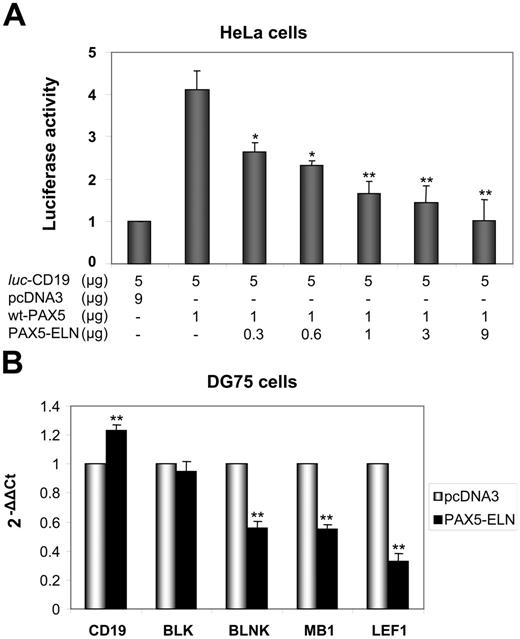
Given the structure of PAX5-ELN (which lacks the transactivation domain of PAX5), we postulated that the chimeric protein might block CD19 transcription. We cotransfected HeLa cells with the luc-CD19 construct,23 pcDNA3-PAX5, and increasing amounts of pcDNA3-PAX5-ELN. PAX5 alone activated the transcription of the luciferase reporter gene (Figure 6A). Concomitant transfection of the pcDNA3-PAX5-ELN led to a down regulation of PAX5-driven CD19 transcription (Figure 6A). Taken together, the results of the ChIP and the luciferase assay indicated that PAX5-ELN could function in a dominant-negative manner over PAX5.
Repression of PAX5-dependent transactivation by PAX5-ELN. (A) The plasmids luc-CD19 (5 μg), pcDNA3-PAX5 (1 μg), and the reference pRL-CMV (0.4 μg; transfection efficiency control) were transfected together into HeLa cells with increasing amounts of pcDNA3-PAX5-ELN. The luc-CD19 plasmid contains 3 copies of the high-affinity PAX5-binding site of CD19 upstream of the TATA box and the luciferase gene. Luciferase activity in 3 independent transfection experiments was normalized to the measured Renilla activities and are shown as average values relative to the basal activity observed with pcDNA3 alone (results are mean ± SD; Student t test; *P < .05, **P < .01 compared with DG75 transfected with pcDNA3-PAX5, without PAX5-ELN). (B) DG75 cells were transfected with pcDNA3-PAX5-ELN and pmaxGFP. GFP-positive cells were sorted 24 hours after transfection. CD19, BLK, BLNK, MB1, and LEF1 mRNA levels were evaluated by RQ-PCR (Results are mean ± SD; Student t test: *P < .05, **P < .01).
Repression of PAX5-dependent transactivation by PAX5-ELN. (A) The plasmids luc-CD19 (5 μg), pcDNA3-PAX5 (1 μg), and the reference pRL-CMV (0.4 μg; transfection efficiency control) were transfected together into HeLa cells with increasing amounts of pcDNA3-PAX5-ELN. The luc-CD19 plasmid contains 3 copies of the high-affinity PAX5-binding site of CD19 upstream of the TATA box and the luciferase gene. Luciferase activity in 3 independent transfection experiments was normalized to the measured Renilla activities and are shown as average values relative to the basal activity observed with pcDNA3 alone (results are mean ± SD; Student t test; *P < .05, **P < .01 compared with DG75 transfected with pcDNA3-PAX5, without PAX5-ELN). (B) DG75 cells were transfected with pcDNA3-PAX5-ELN and pmaxGFP. GFP-positive cells were sorted 24 hours after transfection. CD19, BLK, BLNK, MB1, and LEF1 mRNA levels were evaluated by RQ-PCR (Results are mean ± SD; Student t test: *P < .05, **P < .01).
Furthermore, RQ-PCR experiments on DG75 cells were also carried out to test the role of PAX5-ELN on endogenous target genes. As shown in Figure 6B, PAX5-ELN was able to down regulate BLNK, LEF1, and MB1. Unexpectedly, CD19 and BLK transcription appeared unaffected by the presence of PAX5-ELN (Figure 6B).
Down-regulation of the transcription of PAX5 targets in patients' leukemic cells
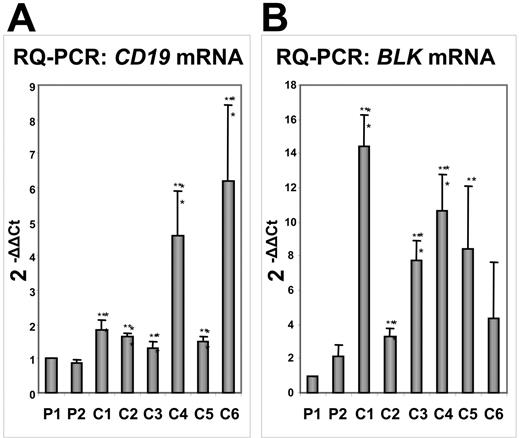
To confirm our results, we extracted RNA from the marrow of 6 patients with B-ALL but with distinct cytogenetic abnormalities. RQ-PCR showed that the levels of CD19 expression were lower in PAX5-ELN–positive tumors compared with those in the other patients with B-ALL (Figure 7A). The study of another target of PAX5, the BLK gene, corroborated our results since its expression was clearly lower in PAX5-ELN–positive patients compared with the 6 other patients (Figure 7B). However, contrary to results obtained from DG75 cells, BLNK, MB1, and LEF1 expression did not significantly vary in PAX5-ELN–positive patients compared with PAX5-ELN–negative cases (M.B., unpublished results, October 2006).
Comparative transcription of the PAX5 target genes CD19 and BLK by RQ-PCR in PAX5-ELN–positive and –negative patients with B-ALL. (A) CD19 mRNA levels: CD19 transcription in Patient 1 (P1) and Patient 2 (P2) is lower compared with the other patients with B-ALL (C1, C2, C3, C4, C5, and C6). (B) BLK mRNA levels: P1 and P2 display lower levels of BLK transcripts than the other patients with B-ALL. (Results are mean ± SD; Student t test: *P < .05, **P < .01 compared with Patient 1).
Comparative transcription of the PAX5 target genes CD19 and BLK by RQ-PCR in PAX5-ELN–positive and –negative patients with B-ALL. (A) CD19 mRNA levels: CD19 transcription in Patient 1 (P1) and Patient 2 (P2) is lower compared with the other patients with B-ALL (C1, C2, C3, C4, C5, and C6). (B) BLK mRNA levels: P1 and P2 display lower levels of BLK transcripts than the other patients with B-ALL. (Results are mean ± SD; Student t test: *P < .05, **P < .01 compared with Patient 1).
Last, we did not find a correlation between gene transcription and CD19 protein expression. We could retrieve pellets of cells from only 1 patient (patient 1 [P1]) carrying the PAX5-ELN fusion gene and 2 patients with B-ALL previously investigated by RQ-PCR (C3 and C5). The levels of CD19 protein (detected by fluorescence-activated cell sorter [FACS]) were not significantly different in all 3 patients (Table S1). However, no definitive conclusion can be drawn from these data since only 3 patients were tested. In addition, it is worth noting that the 2 PAX5-ELN–negative patients had the lowest level of CD19 transcription in the series of control cases. Moreover, one cannot exclude differential posttranscriptional regulation of CD19 in those patients.
Discussion
Chromosomal translocations are frequently observed in cancers, particularly in leukemia. They lead to gene rearrangements and, in some instances, dramatically alter gene expression. Since PAX5 is crucial for B-cell differentiation, we postulated that in the 2 patients with B-ALL carrying a t(7;9)(q11;p13) translocation, the differentiation arrest at the pre-B stage was the result of PAX5 deregulation. By FISH analysis, we confirmed the hypothesis that PAX5 was rearranged: a genomic breakpoint between exons 7 and 10 was observed using PAX5 BAC probes. This indicated that PAX5 was involved in a chimeric gene, a part of which retained sequences coding for the DNA-binding PBD of the protein. Unexpectedly, using a 3′ RACE method, we found that the partner gene on chromosome 7 was ELN, which encodes an extracellular matrix protein. We next demonstrated that the chimeric PAX5-ELN gene was identical in the 2 patients carrying the t(7;9)(q11;p13) translocation. The 2 patients reported in this study were retrieved from a collection of 147 other patients with adult B-ALL in our chromosome bank, suggesting that the t(7;9)(q11;p13) translocation is not an exceptional event. For comparison, in a series of 250 pediatric patients with B-ALL, we could retrieve only 2 patients with a dic(9;12)(p13;p13) translocation, the incidence of which is estimated at about 1%. Larger cohorts of cases are necessary to assess the real incidence of the t(7;9)(q11;p13) translocation. However, we think that this abnormality is underestimated, and that its characterization will favor further reports.
In all previously described chimeras involving PAX genes, the partner is a transcription factor. This is true in solid tumors such as thyroid carcinoma, with the PAX8-PPARG1 oncogene,22 and in rhabdomyosarcoma, with the PAX3-FKHR21 or PAX7-FKHR proteins.28 In all of these disorders, the various chimeras may act both as transdominant-negative proteins and as abnormal and/or ectopic transcription factors. Regarding the only previously reported translocation involving PAX5 in B-ALL that yields a PAX5-ETV6 hybrid gene,16 Stehl et al17 suggested that the corresponding protein could be an aberrant transcription factor. Interestingly, in the patients reported here, PAX5-ELN expression is likely to yield an aberrant transcription factor retaining the DNA-binding properties of wild-type PAX5, but without its transactivation and repressor domains, which are replaced by ELN. The discovery of ELN as the partner gene of PAX5 was somewhat unexpected, as its product is a 72-kDa insoluble extracellular matrix protein. It is the main component of elastic fibers, which are crucial for the elasticity of structures such as skin, blood vessels, heart, lungs, intestines, tendons, and ligaments.29 ELN is composed largely of glycine, proline, and other hydrophobic residues, and contains multiple lysine-derived cross-links, such as desmosines and isodesmosines.30 A defect in elastin is implicated in several genetic disorders: Williams-Beuren syndrome (WBS), supravalvular aortic stenosis (SVAS), and Cutis Laxa. SVAS is a rare obstructive arterial disease characterized by the narrowing of the large elastic vessels.31 Patients with WBS have additional clinical manifestations, including typical facial anomalies, neurodevelopmental abnormalities, growth retardation, and cardiovascular manifestations.32 Mutations or deletions limited to the ELN gene appear to result in SVAS, whereas deletions spanning at least 114-kb around the gene lead to WBS. Moreover, ELN is implicated in other chromosomal translocations. Curran et al described a t(6;7)(p21.1;q11.23) translocation that disrupts the ELN gene with a breakpoint in exon 28 in a patient with SVAS.33 Von Dadelszen et al reported a case of SVAS with a t(6;7)(q27;q11;23) translocation and with the same breakpoint on ELN, but with different breakpoints on chromosome 6.34 In 2002, Duba et al35 described 5 patients with the same balanced translocation t(7;16)(q11.23;q13) associated with a variable expression of WBS. This translocation disrupts the ELN gene within intron 5 and creates a fusion gene with intron 1 of the TM7XN1 gene.35 However, in all these disorders, alterations of the ELN gene are transmitted in the germ line and are frequently accompanied by a defect in the organization of the elastin network. In contrast, in our observations, the rearrangement of ELN occurs in somatic cells and curiously involves cell types that do not express this gene. This may be the reason why the reverse ELN-PAX5 hybrid gene was not transcribed. The fact that the breakpoints observed in the tumors are identical in the 2 patients strongly suggests that this rearrangement is not completely sporadic. Interestingly, the fact that the breakpoints between germinal and somatic alterations of the ELN gene are distinct also suggests that the mechanisms underlying these chromosomal rearrangements are completely different.
Although particularly aberrant, the chimeric protein PAX5-ELN is probably functional in leukemic cells. To confirm this point, we transfected Burkitt lymphoma cells (DG75) with a construct containing the full-length cDNA of the chimeric gene. Thus, we showed that PAX5-ELN was localized to the nucleus, as was PAX5. The same result was obtained in other transfected cell types (eg, HeLa cells, NIH3T3, etc). This was expected since PAX5-ELN retained the NLS of PAX5 and lost the signal peptide of ELN. ChIP experiments clearly demonstrated that PAX5-ELN was able to bind CD19 and BLK promoter sequences, as was wild-type PAX5, suggesting that it could act in a dominant-negative manner. To further demonstrate this, we then cotransfected HeLa cells with a luc-CD19 construct, pcDNA3-PAX5, and pcDNA3-PAX5-ELN. We observed a down-regulation of PAX5-driven CD19 transcription proportional to the amounts of the transfected pcDNA3-PAX5-ELN construct. Thus, PAX5-ELN was able to block PAX5-dependent transactivation by competing on PAX5-binding sites. However, RQ-PCR experiments (on DG75 cells) revealed that the effect of PAX5-ELN on PAX5 endogenous targets is probably more complex than expected. In our hands, PAX5-ELN was able to down-regulate BLNK, LEF1, and MB1, but failed to affect CD19 and BLK transcription. This is in agreement with the data of Nutt et al,8 showing that when transfected in RAC65 embryonic carcinoma cells, the sole DNA-binding domain (paired box) of PAX5 can act in a transdominant negative manner on luc-CD19construct expression.8 Nevertheless, the same domain fails to interfere with the expression of the endogenous CD19 gene in B-cell lines, even if it is expressed at a 50-fold higher level compared with the endogenous PAX5 protein (Dr. Meinrad Busslinger, written personal communication, October 2006).
As B-cell differentiation is not altered in Pax5+/− animals,36 it is probable that the haploinsufficiency observed in such tumors may not alone account for the maturation blockage. Nevertheless, some degree of haploinsufficiency may be involved in this process, since 1 allele is interrupted and the transcription of PAX5 is supposed to be biallelic at this stage.36 Concerning the tumor cells, we observed that both CD19 and BLK genes were expressed at lower levels in PAX5-ELN–positive tumors compared with other patients with B-ALL with different chromosomal abnormalities, whereas BLNK, MB1, and LEF1 genes were not significantly affected (M.B., unpublished results, October 2006). These results do not correlate with the data obtained on DG75 transfected cells, suggesting that the regulation of PAX5 target gene transcription is context dependent, being different in mature (DG75 post–germinal-center B cells) and in leukemic pre-B cells. Further experiments are needed to unravel the subtle mechanisms underlying these differences.
One can assume that PAX5-ELN rearrangement is, to a large extent, an early event in the process of transformation of leukemic cells. The competition of PAX5-ELN with PAX5 may interfere with the process of B-cell differentiation, which is tightly linked to PAX5 expression. Moreover, as this translocation is isolated in 1 of the 2 patients and associated with a chromosomal deletion [del(9)(p11p13)] outside the PAX5 locus in the other patient, it is probable that this phenomenon represents a key event in the process of B-cell transformation. We suspect that additional (secondary) molecular events are involved in the process of transformation of normal B cells into leukemic cells. In this regard, further biochemical experiments are needed to understand the consequences of the presence of ELN in the nucleus, since this protein is usually exclusively located in the extracellular compartment.
In conclusion, we report herein a new chromosomal translocation in patients with B-ALL that yields a chimeric PAX5-ELN protein with a dominant-negative function on PAX5 in a background of haploinsufficiency. To the best of our knowledge, this is only the second description of such an oncogenic process involving a transcription factor and a structural protein in hematologic malignancies,37 and the first in human lymphoid tumors.
Authorship
Author contributions: M.B., C.B., and C.Q. performed most of the experiments described in this study. N.D. performed and interpreted all experiments on chromosome metaphases (karyotypes, FISH). F.M. played an important role in the management of all molecular biology experiments (PCR, RT-PCR, RACE, ChIP). E.K. performed FACS analysis on leukemic cells. G.D. and P.B. designed the study and wrote the manuscript. M.B. and C.B. contributed equally to this work.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: Pierre Brousset, Department of Pathology, CHU Purpan, Place Baylac, 31059 Toulouse Cedex, France; e-mail: brousset.p@chu-toulouse.fr.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Prof Meinrad Busslinger (Vienna, Austria) for providing the luc-CD19 construct and fruitful discussion on our results. Special thanks to Neil Ledger, Charles A. Stewart, Jean-Pierre Jaffrézou, and Brian Huntly for critical reading of the manuscript.
Supported by grants from l'Institut National du Cancer (INCa), France; l'ARC contrat no. 3705; La ligue nationale contre le Cancer (Comités de Haute-Garonne et du Gers); and Cancéropole Grand Sud-Ouest.


![Figure 4. Localization of PAX5-ELN by confocal microscopy. (A) Nuclear localization of PAX5-ELN in NIH3T3 cells (anti-PAX5; green) (B) Nuclear localization of PAX5-ELN and endogenous PAX5 in DG75 cells (anti-elastin [H-300] is green; anti–PAX-5 [A-11], red).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/8/10.1182_blood-2006-05-025221/4/m_zh80080711020004.jpeg?Expires=1767751045&Signature=k3~3pXbTSAO8MZtAypd7~vM7gVVopccdQuzBbNrL1zwEC4L3U8BIpAauIrNuwmy8XucsPkyfT0aw6BM4wHh6gLyjv6fqzY6gcdMqYpwcq-sJCja4EJyuB2aiRHnyZaK89KwdAH~JmKfNTnhlNmsKQ0h3Nu1YGmod5UxcVYa9PqjSOiseRQrqmGtUFL5QzZNfi2D1FgSZ4hZPJsKhKUIP1emHBKzuaVilbm9RPBlKwPUbCS3jdICNs4-oKPdznHozSnWHKfY0ZK-6Pq1LU8aESFgwtLhd~XYT1lrinGbDQ014aWsa9HDxPVcTM4rZuOej3lbcQm56bAdGjq0hk12cMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal