Abstract
Pathogenic mutations in α and β spectrin result in a variety of syndromes, including hereditary elliptocytosis (HE), hereditary pyropoikilocytosis (HPP), and hereditary spherocytosis (HS). Although some mutations clearly lie at sites of interaction, such as the sites of spectrin α-βtetramer formation, a surprising number of HE-causing mutations have been identified within linker regions between distal spectrin repeats. Here we apply solution structural and single molecule methods to the folding and stability of recombinant proteins consisting of the first 5 spectrin repeats of α-spectrin, comparing normal spectrin with a pathogenic linker mutation, Q471P, between repeats R4 and R5. Results show that the linker mutation destabilizes a significant fraction of the 5-repeat construct at 37°C, whereas the WT remains fully folded well above body temperature. In WT protein, helical linkers propagate stability from one repeat to the next, but the mutation disrupts the stabilizing influence of adjacent repeats. The results suggest a molecular mechanism for the high frequency of disease caused by proline mutations in spectrin linkers.
Introduction
Spectrin superfamily proteins have tandem arrays of 3-helix bundle spectrin repeats and are generally found in regions of cells that are exposed to mechanical stress.1 Within erythrocytes, spectrin crosslinks F-actin into a resilient membrane network that normally allows these cells to distort reversibly for months under the high fluid shears of blood circulation.2 Spectrin crosslinks consist of antiparallel heterotetramers of α-spectrin and β-spectrin that form staggered head-to-tail interactions between pairs of lateral α-β heterodimers (Figure 1A, inset). Crystal structures that have emerged in the past decade for a half-dozen tandem arrays of erythroid3-6 and nonerythroid spectrin7 repeats have been surprising and consistent in showing that linkers between spectrin repeats are continuous helices. Although such structures are obtained from static and dense-packed crystals that favor ordered states, solution studies8 and single-molecule measurements of forced extension9 also suggest cooperative coupling between erythroid spectrin repeats.
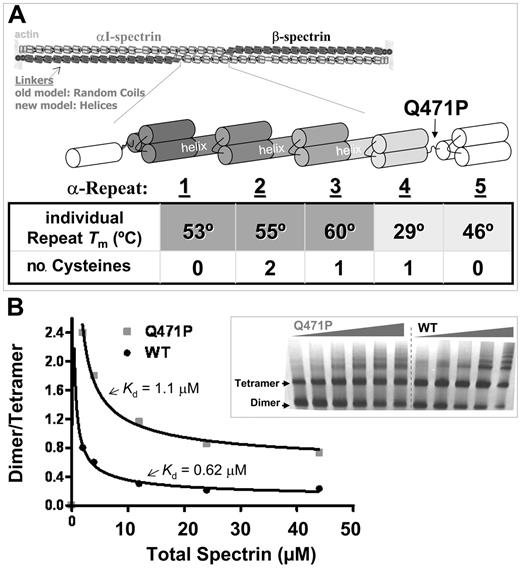
Spectrin structures and interactions and the anemia-causing linker mutant. (A) Antiparallel heterodimers of αI-spectrin (light) and β-spectrin (dark) associate head to tail to form a tetramer that crosslinks F-actin at the red blood cell membrane. Linkers between repeats were originally thought to be random coils, but recent evidence suggests many are helical. Normal and mutant constructs of the αI-spectrin N-terminus allow study of the effects of the pathogenic mutation Q471P, which is located in the linker between repeats α4 and 5. The table lists the melting temperature of the individually expressed repeats2 and the number of cysteines within each repeat. (B) Dimer-tetramer ratio for spectrin purified from control and patient blood, with model fits that yield a weaker dissociation constant Kd-tetramer for the Q471P mutant (data not shown). (inset) A 2% to 4% gradient nondenaturing polyacrylamide gel of spectrin from control and patient erythrocytes. Lanes 1 to 5 contain WT sample spectrin concentrations of 11, 6, 3, 1, and 0.5 mg/dL. Lanes 6 to 11 contain hereditary elliptocytosis patient spectrin concentrations of 15, 11, 6, 3, 1, and 0.5 mg/dL.
Spectrin structures and interactions and the anemia-causing linker mutant. (A) Antiparallel heterodimers of αI-spectrin (light) and β-spectrin (dark) associate head to tail to form a tetramer that crosslinks F-actin at the red blood cell membrane. Linkers between repeats were originally thought to be random coils, but recent evidence suggests many are helical. Normal and mutant constructs of the αI-spectrin N-terminus allow study of the effects of the pathogenic mutation Q471P, which is located in the linker between repeats α4 and 5. The table lists the melting temperature of the individually expressed repeats2 and the number of cysteines within each repeat. (B) Dimer-tetramer ratio for spectrin purified from control and patient blood, with model fits that yield a weaker dissociation constant Kd-tetramer for the Q471P mutant (data not shown). (inset) A 2% to 4% gradient nondenaturing polyacrylamide gel of spectrin from control and patient erythrocytes. Lanes 1 to 5 contain WT sample spectrin concentrations of 11, 6, 3, 1, and 0.5 mg/dL. Lanes 6 to 11 contain hereditary elliptocytosis patient spectrin concentrations of 15, 11, 6, 3, 1, and 0.5 mg/dL.
A wide range of mutations within erythrocyte spectrin perturbs cell structure and compromises the ability of the red blood cell to withstand mechanical forces, resulting in hereditary spherocytosis (HS), hereditary elliptocytosis (HE), or hereditary pyropoikilocytosis (HPP).10,11 Many of these mutations are in the N-terminal tetramerization site of the α-subunit or the β spectrin–ankyrin interaction. However, more than a dozen mutations distal to these sites have been linked to HE/HPP, and, of these, 7 mutations have been mapped to the linkers between repeats of α-spectrin. Five of these known mutations are prolines.11 Given that it has recently become clear from studies of individual recombinant spectrin repeats that the melting temperatures of almost one third of the repeats are at or below 37°C,12 we hypothesized that proline-induced interruption of a helical linker would result in local nonhelicity within the linker,13 decoupling and destabilizing some spectrin repeats even at physiological temperatures and, thus, contributing to disease. Therefore, the continuity of helical linkers between tandem repeats may be critical to the overall mechanical properties of the erythrocyte membrane.
Here, the first 5 repeats of α-spectrin (aa 1-584) were expressed with either the wild-type (WT) sequence or the pathologic proline mutation Q471P. Although previous reports emphasize a disruption in RBC spectrin tetramers, we show that the mutant has equal affinity to WT in stable head-to-tail interactions with monomeric β-spectrin. However, thermal unfolding measured by several methods, including single-molecule forced extension and a cysteine-labeling technique, indicate a loss in structural stability of the Q471P mutant, even at 37°C. This is also predicted by temperature-dependent molecular dynamic simulations. The results thus demonstrate that erythroid spectrin with proline mutations in linkers between repeats can destabilize adjacent helical repeats and contribute directly to disease.
Materials and methods
Analysis of spectrin oligomers
Spectrin extracts were prepared at 4°C, as described,14 then concentrated to 12 to 14 mg/mL by vacuum dialysis against 10 mM Tris, 20 mM NaC1, 130 mM KC1, 1 mM EDTA, and 0.1 mM NaN3, pH 7.4. Serial dilutions of spectrin in the same buffer were incubated for 3 hours at 30°C before electrophoresis. Spectrin oligomer formation was studied by electrophoresis of extracted spectrin on 2% to 4% gradient nondenaturing polyacrylamide slab gels at 4°C. Gels were electrophoresed at 50 V for 48 hours in buffer containing 40 mM Tris, 20 mM Na acetate, and 2 mM EDTA, pH 7.4. Quantitation of spectrin oligomers and peptides from Coomassie blue–stained acrylamide gels was performed by elution of excised gel slices with 1.0 mL of 25% pyridine for 3 to 4 days until the gel slice was colorless. Optical density of the eluted material at 700 nm was measured in a spectrophotometer.
Protein expression and purification of recombinant spectrin proteins
Residues 1-584 of αI spectrin and 1902-2084 of βI spectrin (repeats 16-17) were amplified by polymerase chain reaction (PCR) from erythrocyte spectrin cDNA clones.15,16 Site-directed mutagenesis was used to introduce Q471P in the 5-repeat α-spectrin construct (α1-5). All constructs were expressed as glutathione S-transferase (GST) fusions using a pGEX-2T vector and DH5α Escherichia coli. GST moiety was cleaved and purified from the supernatant by affinity chromatography, as described previously.17-19 The protein was concentrated using Centriprep concentrators (Millipore, Billerica, MA) and was further purified on G3000SW and G2000SW columns connected in series (60 × 2.5 cm; Tosohaus, Montgomeryville, AL) equilibrated with PBS plus 1 mM β-ME, 1 mM EDTA, and 0.15 mM PMSF, pH 7.4. Protein concentrations were determined by measuring OD280 with ϵ280 calculated using the GPMAW program.
Isothermal titration calorimetry
A VP-ITC isothermal titration calorimeter (MicroCal, Northampton, MA) was used for thermodynamic analysis of the binding between the α1-5 WT and mutant and β16-17 (residues 1902-2084). Samples were dialyzed in 2 steps (3 hours and overnight) against PBS. All samples and buffer were degassed under vacuum for 5 minutes before loading in the instrument. The reference cell was filled with the dialysate buffer, and the sample cell was filled with the dialysate for the control or with β16-17 recombinant spectrin, used as titrated protein, at a concentration of 20 μM; α1-5 constructs were used as titrants in the syringe for control and sample experiments. Titration experiments were performed with stirring at 300 rpm and 25 to 29 injections of 10 μL protein over 40 to 50 minutes. The area under each spike, representing the Δ heat for each injection, was integrated, and the control was subtracted from experimental data to correct for the dilution heat of the titrant protein into buffer. Integrated injections were then fit to a simple binding model with the use of nonlinear least square regression analysis. Measurements reported are for 25°C; measurements at 37°C gave weaker associations, as expected from past work, but mutant and normal controls were statistically the same.
Circular dichroism
Circular dichroism spectra were measured with the use of a spectrophotometer (J715; Jasco, Sydney, Australia) and a 1-mm path-length cell in PBS buffer (pH 7.5). The instrument was calibrated before sample measurement with d-10-camphorsulfonic acid. Three consecutive measurements at each temperature were made after allowing for thermoequilibrium and were averaged.
Differential scanning calorimetry
An MCS differential scanning calorimeter (DSC; MicroCal) was used to perform thermal denaturation studies on the α1-5 and Q471P mutant. All samples and buffer were degassed for 5 minutes before they were loaded into the calorimeter. The dialysate buffer was used to fill the reference cell in buffer control and denaturation experiments, and the sample cell was filled with the dialysate buffer in control or protein sample at 0.5 to 1.0 mg/mL (6.5-13 μM). All scans were taken from 20° to 90°C at a scanning rate of 90°C/h. Data analysis was performed using software provided with the instrument by subtracting the buffer control, subtracting the baseline fitted to the ends of the transitions, normalizing for the protein concentration, and curve-fitting using nonlinear least square regression analysis.
Molecular dynamics simulations
Atomistic-level molecular dynamics (MD) simulations were performed for the WT and the Q471P mutations using the 3D model of repeats 4-5, obtained from the Robetta website.20-27 The sequence for the spectrin repeats was obtained from the UniProtKB/Swiss-Prot Protein Knowledgebase, accession number P02549. Constructs were equilibrated in a box of explicit water for 10 nanoseconds at temperatures of 17°C, 27°C, or 37°C using the package NAMD28 with the CHARMM27 force field.29 Measurements of the root-mean-square-deviation (rmsd) of the positions of the backbone atoms show that after 5 nanoseconds, the protein structure reached equilibrium for all temperatures sampled; therefore, data were collected for the last 3 nanoseconds of simulation. Average helical content of the residues was calculated using the Lifson-Roig model,30 by which a residue, i, was marked helical if, and only if, its backbone dihedral angle pair (φi, ψi) and its adjacent residue angle pairs (φi-1, ψi-1 and φi+1, ψi+1) lay in the helical region φ = −65(± 35)° and ψ = − 37(± 30)°. The average solvent-exposed area of the cysteine was calculated using the software MSMS31 for integration of molecular surfaces using a probe radius of 1.4 Å, a value typically used for water.
Cysteine labeling
Labeling of accessible cysteines was achieved by adding 13 μL of 6 mM IAEDANS to a solution consisting of 0.5 mM TCEP and 200 μL of 1 mg/mL protein in PBS, pH 7.5. Samples were allowed to react at 23°C, 30°C, 37°C, 45°C, 60°C, and 75°C for 45 minutes, after which the reaction was quenched by the addition of 10 μL of 282 mM β-mercaptoethanol. After IAEDANS labeling, protein samples were run on a 12% Bis-Tris SDS gel, and the fluorescence intensity of the protein bands was imaged with a UV transilluminator. To account for disparities in sample loading, the gel was stained using Simply Blue colloidal stain and was digitally scanned. Fluorescence and blue stain intensities of each protein band were quantitated using Scion Image for Windows (http://www.scioncorp.com), and the fluorescence intensities were normalized by the blue stain intensity (sample loaded).
Atomic force microscopy measurements
Collection and analysis of single molecule unfolding measurements were carried out as described.9 Atomic force microscopy (AFM) was carried out on a Nanoscope IIIa Multimode (Digital Instruments, Santa Barbara, CA) equipped with a liquid cell and heater for temperature control. Protein density was adjusted so that less than 10% of tip-to-surface contact resulted in a protein unfolding event, and 5000 surface-to-tip measurements were made for each sample to ensure good statistics. Analysis of the force curves was conducted using a semiautomated visual analysis program written in C++.
Results
α-β Spectrin head-to-tail heterodimer interaction is unaffected by mutation
Effects of the Q471P mutation on head-to-tail interactions between the N-terminal α1-5 construct (Figure 1A) and a C-terminal β-spectrin construct were first examined with the use of isothermal titration calorimetry (ITC). ITC measures protein binding through measurement of the heat given off during protein interaction at constant temperature. From the measured addition of heat as protein was added, the equilibrium dissociation constant, Kd, was determined, together with the heat or enthalpy of binding ΔH from a nonlinear curve fit. For the WT α1-5 construct, Kd = 0.44 μM (Figure S2, available on the Blood website; see the Supplemental Figures link at the top of the online article), and ΔH = −30 kcal/mol. Additionally, from the relationship RTln Kd = ΔG = ΔH − TΔS, the interaction entropy ΔS = −73 cal/mol K was calculated and indicated a significant and understandable increase in system order (water, protein, salt) upon association. These thermodynamic measures of interaction are close to those reported or estimated for full-length spectrin.32 The Q471P mutant also gave Kd = 0.44 μM, with similar ΔH and ΔS values (−35 kcal/mol and −87 cal/mol K). No significant difference in binding was thus detected between normal and mutant monomer.
Heterodimer association into full-length spectrin tetramers has been studied for decades, with one of the earliest studies by equilibrium ultracentrifugation indicating Kd-tetramer = 0.5 μM.32 This is similar to the dissociation constant for monomeric proteins, at least within the usual 10% to 15% error for protein concentration. Early work on the association of spectrin dimer extracted from human erythrocytes with the Q471P mutation suggested a weaker interaction of mutant full-length heterodimers, but no quantitative analysis was attempted.33 Nondenaturing PAGE study of association was, therefore, repeated for normal and mutant spectrin from mutant and WT red cells, showing a 2-fold weaker association of mutant heterodimers into tetramers (Figure 1B). The control Kd-tetramer = 0.62 μM was remarkably close to results cited, whereas the mutant gave Kd-tetramer* = 1.1 μM. Because the defect was not in the monomeric association sites, we hypothesized that conformational perturbation within the mutant heterodimers might be the basis for weaker tetramer association.
Thermal denaturation in solution shows the mutant is less folded at 37°C
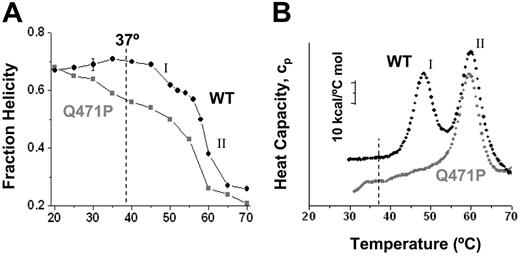
To examine the structural properties of the WT and mutant α1-5 proteins, circular dichroism, tryptophan fluorescence, and calorimetric studies were conducted. CD spectra of the secondary structure for both constructs showed approximately 68% helical content at 19°C, which was consistent with past measurements on a range of spectrin constructs.12 For the WT protein, this helicity increased slightly with temperature to more than 70%, up to 40°C. The Q471P mutant showed the opposite behavior: helicity decreased to approximately 55% at 37°C. The clear difference provided the first definitive evidence of destabilization resulting from a pathogenic linker mutation.
For both constructs, helicity decreased for temperatures higher than 40°C, with multistate unfolding. For the WT, a minor transition was seen first at TmI ≈ 48°C, followed by the major transition at TmII ≈ 59°C. Helicity loss, Δ, in each transition occurred in the ratio of ΔI:ΔII = 13%:34% ≈ 0.3:0.7. This ratio suggested unfolding of one or 2 of the 5 WT repeats in the first transition. Two distinct melting temperatures might have been anticipated from past results34 for the relevant tandem repeat constructs: α4-5 had a Tm = 41°C, whereas α1-2 and α2-3 had similar Tm = 53°C or 57°C. Figure 1 also tabulates Tm values that were measured recently14 for individual repeats, and again these were low for R4 and R5 (29°C and 46°C, respectively) and high for R1, R2, and R3 (53°C, 55°C, and 60°C, respectively). The comparisons clearly showed that placing repeats in tandem increased the stability above the simple average (eg, ½ [TmR4 + TmR5) < Tm,α4-5]), which was consistent with cooperative stabilization by the linking helix.9,34 A tentative mechanism of unfolding the WT construct is, therefore, (I) for initial unfolding of the C-terminal α4-5 repeats followed at higher temperature by (II) for cooperative unfolding of the N-terminal α1-3 repeats. Destabilizing the linking helix between R4 and R5 with Q471P should primarily, as shown, have decreased and broadened transition-I for the mutant.
To assess the energetics of these transitions, differential scanning calorimetry (DSC) studies were also conducted on both constructs (Figure 2B). DSC on the WT recombinant protein showed 2 peaks in the heat capacity at similar transition temperatures (48°C and 60°C). By fitting a 2-state model to each peak, transition enthalpies of ΔHI = 123 kcal/mol and ΔHII = 215 kcal/mol were determined and appeared in the approximate proportion ΔHI:ΔHII ≈ 0.4:0.6. This ratio provided perhaps the best evidence that the WT protein denatured first (I) through unfolding of the smaller α4-5 domain followed by (II) unfolding of the larger α1-3. For Q471P, the first transition appeared lost in a broad first peak. For the second transition with the mutant, however, estimates of TmII and ΔHII were only slightly lower than those of WT (59°C and 188 kcal/mol, respectively), suggesting that the α1-3 in the mutant was relatively well folded. Solution structural studies were thus collectively clear in identifying a folding defect that arose with the pathogenic mutation Q471P.
Thermal denaturation studies of the WT and Q471P mutant. WT and Q471P recombinant spectrin fraction folded as a function of temperature based on (A) circular dichroism and (B) DSC measurements all show a significant thermal destabilization of the Q471P mutant relative to the WT construct at or below 37°C.
Thermal denaturation studies of the WT and Q471P mutant. WT and Q471P recombinant spectrin fraction folded as a function of temperature based on (A) circular dichroism and (B) DSC measurements all show a significant thermal destabilization of the Q471P mutant relative to the WT construct at or below 37°C.
Molecular dynamics demonstrates thermal unfolding of the mutant linker
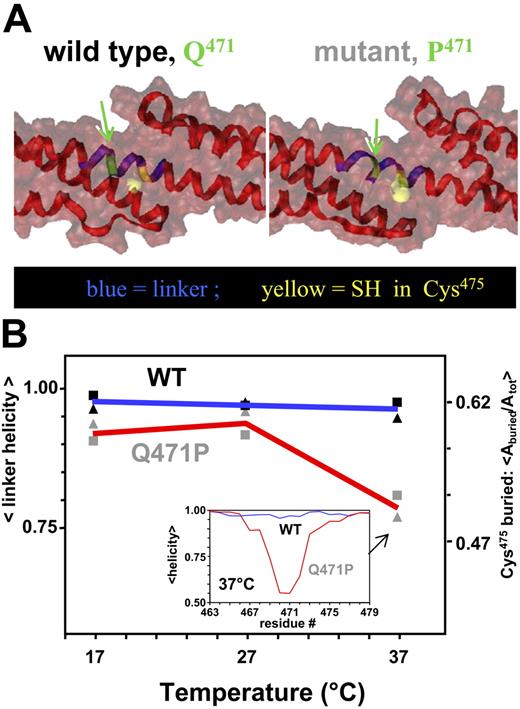
Based on the structural evidence for coupling within α4-5, the linker between repeats R4 and R5 was likely to be helical. For a better picture of this and the expected proline-induced effects in the putative helical linker, molecular dynamics simulations were performed on homology models of α4-5 WT and the Q471P mutant. The linker for WT was indeed a stable helix after 8-nanosecond equilibration. For the mutant, in contrast, the helix was locally disordered immediately near the proline substitution. Shown in opaque yellow and in space-filling representation was the thiol in Cys475, which was approximately one helical turn away from the site of mutation and was more clearly exposed in the mutant.
At temperatures of 17°C, 27°C, and 37°C, helicity of the linker and solvent exposure of the thiol were quantified (Figure 3B). The relevant part of the linker on which to focus was identified based on the perturbation to helicity at 37°C in Q471P (Figure 3B, inset; blue residues in Figure 3A). Pro471 was at the center of the perturbation, as expected, and the loss in helical content extended several residues either side of it. At the low temperatures in simulation, the perturbation was minimal, consistent with solution studies (Figure 2). As with helicity at 37°C, however, Cys475 was less buried in the mutant. This increased solvent accessibility was directly assessed with the novel use of small thiol-reactive reagents to probe thermal unfolding.
Molecular dynamics simulations show α4-5 linker destabilization. (A) Snapshots of the α4-5 linker (blue) for WT and mutant at 37°C. The mutated site and Cys475 are represented in green and yellow, respectively. Ribbon representations show a disruption of the helix in the linker of the mutant, whereas the molecular surface representation shows an increase in solvent-exposed area of the Cys. (B) Quantitation of Cys475 exposure (▴) and linker helical content (▪) from molecular dynamics simulation. (inset) Normalized fraction of helical content along the backbones at 37°C. Note that compared with WT (blue line), the Q471P mutant (red line) shows considerable loss in helicity (approximately 40%) in the linker region. Arrows denote area of helix altered by proline mutation.
Molecular dynamics simulations show α4-5 linker destabilization. (A) Snapshots of the α4-5 linker (blue) for WT and mutant at 37°C. The mutated site and Cys475 are represented in green and yellow, respectively. Ribbon representations show a disruption of the helix in the linker of the mutant, whereas the molecular surface representation shows an increase in solvent-exposed area of the Cys. (B) Quantitation of Cys475 exposure (▴) and linker helical content (▪) from molecular dynamics simulation. (inset) Normalized fraction of helical content along the backbones at 37°C. Note that compared with WT (blue line), the Q471P mutant (red line) shows considerable loss in helicity (approximately 40%) in the linker region. Arrows denote area of helix altered by proline mutation.
Cysteine labeling increases with thermal unfolding and more so for the mutant
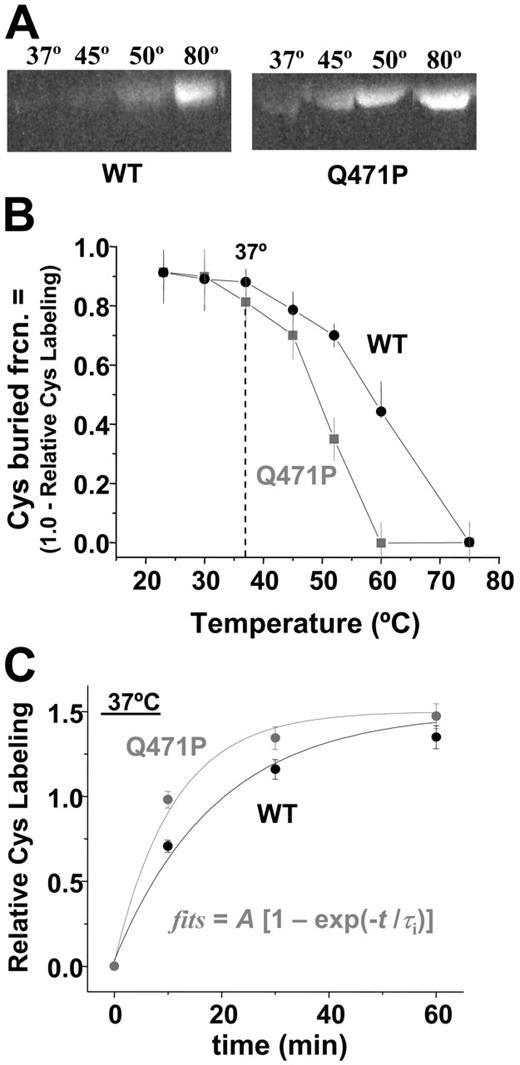
Cysteines are relatively hydrophobic amino acids, and at least some of the 4 Cys residues of the α1-5 construct were evidently buried. Unfolding is expected to increase cysteine exposure and, thus, to increase reactivity to thiol-reactive compounds, such as the fluorescent dye IAEDANS. Because the reaction exhibited only weak temperature dependence in denaturing solutions such as urea, changes in fluorescence labeling of the constructs with temperature (Figure 4A–B) largely reflected changes in the Cys-buried fraction. However, the first-order kinetics of labeling (Figure 4C) means that reaction rate differences resulting in differences in buried Cys generally decreased with time as label accumulated. Labeling of either construct was relatively low—up to 30°C (for 60 minutes)—but for temperatures of 37°C and higher, Cys was increasingly exposed and labeled, with greater labeling of Q471P than of WT up to full labeling of Cys at approximately 75°C (Figure 4B). Fits of labeling kinetics also revealed a 1.9-fold increased rate of labeling for the Q471P mutant (0.052 min−1) relative to WT (0.099 min−1), which was again indicative of increased Cys accessibility (Figure 4C). Because the only difference between the 2 recombinant constructs was the proline mutation localized near Cys, we hypothesized that the mutation locally disrupted linker helicity and exposed the nearby Cys. Mass spectrometry on 30-minute samples indeed detected considerable IAEDANS labeling of Cys475 in the Q471P mutant, but comparatively little labeling of WT was detected.
Cysteine labeling compared with temperature for mutant and WT. (A) SDS-PAGE followed by fluorescence imaging of IAEDANS-labeled proteins illustrates an increase in Cys labeling at lower temperatures for the mutant. (B) Unlabeled Cys compared with temperature after 60-minute reaction. Splayed curves resemble the unfolding curves of Figure 2, with significant differences in labeling at temperatures higher than 30°C. (C) IAEDANS reaction kinetics of WT and Q471P with associated fits. First-order reaction rate fits to the data demonstrate a 1.9-fold faster rate of reaction for Q471P than for WT.
Cysteine labeling compared with temperature for mutant and WT. (A) SDS-PAGE followed by fluorescence imaging of IAEDANS-labeled proteins illustrates an increase in Cys labeling at lower temperatures for the mutant. (B) Unlabeled Cys compared with temperature after 60-minute reaction. Splayed curves resemble the unfolding curves of Figure 2, with significant differences in labeling at temperatures higher than 30°C. (C) IAEDANS reaction kinetics of WT and Q471P with associated fits. First-order reaction rate fits to the data demonstrate a 1.9-fold faster rate of reaction for Q471P than for WT.
Just as Cys exposure increased with temperature, so did Trp hydration. Moreover, the mutant showed lower Tm than the WT (Figure S1A), yielding a picture similar to that of Cys labeling, but the latter appeared more definitive.
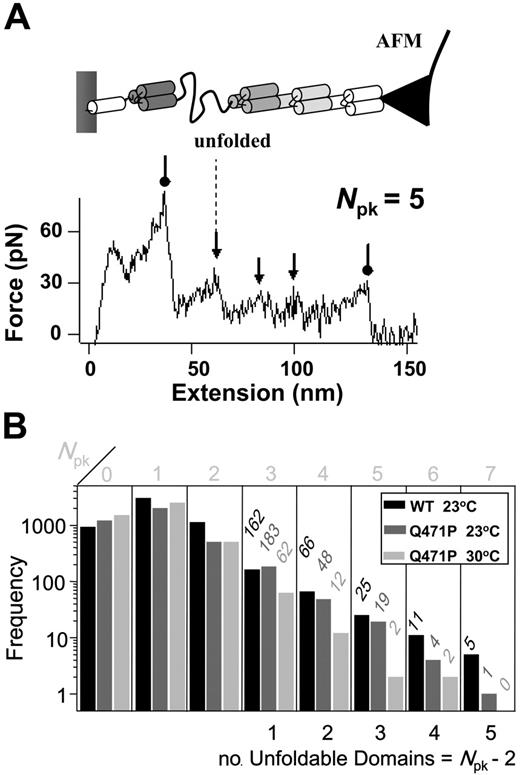
Single-molecule forced unfolding shows the mutant has fewer stable domains
Given that spectrin is a mechanically important protein to the red blood cell and that linker mutants, such as Q471P, lead to less stable membranes, we also sought to assess the mechanical effects of the Q471P linker mutation. Forced extension of single molecules by AFM was performed on both constructs. The characteristic sawtooth patterns of forced unfolding for the WT and the Q471P mutants at 23°C showed a maximum of 7 peaks (Npk = 1-7), and this maximum was equivalent to the number of mechanically resilient spectrin domains plus initial and final desorption events (Figure 5A). The first and last peaks are not caused by protein unfolding.9 Therefore, we determined the number of unfoldable domains (Npk − 2). The sawtooth thus implied up to 5 unfoldable domains for the WT and the Q471P mutants at 23°C, but the mutant showed 80% less such 5-domain events, and the mutant showed no such events at 30°C. Trends for 4-domain events, 3-domain events, and so on were similar. This simple, first analysis of forced extension thus implies that the proline linker mutant has fewer resilient domains under mechanical stress. Results are collectively consistent with the proline mutation destabilizing spectrin repeats.
Domain numbers under mechanical stress. (A) Schematic diagram of AFM-forced unfolding with a single repeat shown unfolded. Such unfolding yields a single peak (arrows) in the sawtooth-shaped force-extension plot; the first and last peaks (circles) correspond to desorption of the protein from either the surface or the AFM tip. (B) Frequency of occurrence for unfolding spectrin domains (Npk-2). The mutant shows fewer high Npk sawtooth patterns, indicating a relative loss of domain structure.
Domain numbers under mechanical stress. (A) Schematic diagram of AFM-forced unfolding with a single repeat shown unfolded. Such unfolding yields a single peak (arrows) in the sawtooth-shaped force-extension plot; the first and last peaks (circles) correspond to desorption of the protein from either the surface or the AFM tip. (B) Frequency of occurrence for unfolding spectrin domains (Npk-2). The mutant shows fewer high Npk sawtooth patterns, indicating a relative loss of domain structure.
Discussion
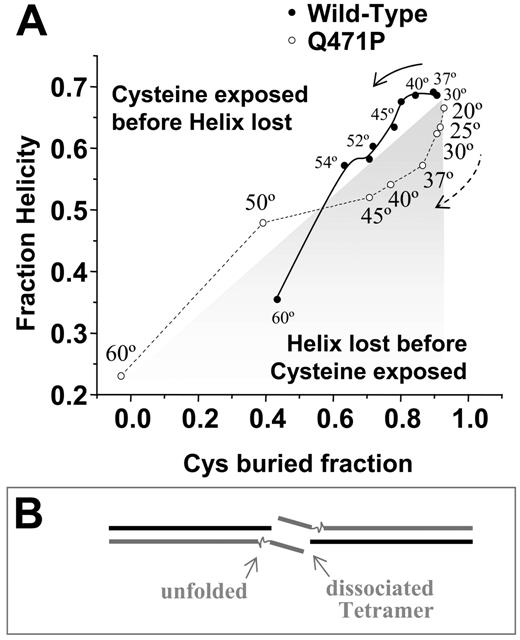
Thermal denaturation studies here suggest that the WT α1-5 construct unfolds in a partially cooperative 3-state manner. Cooperativity between other spectrin repeats is increasingly clear in solution studies and single-molecule force measurements of multirepeat stability.9,12 The single aa mutation at Q471P uncoupled repeats (Figures 1A, 3), limiting the cooperativity between repeats and leading to a divergence in unfolding pathways (Figure 6A). By plotting helicity versus Cys labeling for both constructs and using temperature to create parameters for the pathways, the WT construct showed no significant perturbation of structure up to approximately 37°C, after which Cys exposure increased (above the diagonal) before significant loss of helical secondary structure occurred greater than approximately 42°C. The Q471P showed clearly opposite behavior (below the diagonal) to approximately 37°C. Although the helicity measurements were equilibrium measurements and the temperature-dependent Cys exposure results (Figure 4B) were kinetic snapshots (Figure 4C), the clear differences in initial unfolding suggest independent and distinct multistate unfolding pathways. Additional analyses of fractional helicity compared with Trp exposure showed a similar divergence (Figure S1B) and highlighted an initial loss in helicity for the mutant that preceded other changes.
Proline mutation alters folding state. (A) Plot of helicity compared with fraction unlabeled cysteine at temperatures ranging from 20°C to 60°C reveals divergent unfolding pathways on mutation (B) The reduced structural stability of the mutant is speculated to add flexibility and to increase entropy in destabilizing the αβassociation in the tetramer.
Proline mutation alters folding state. (A) Plot of helicity compared with fraction unlabeled cysteine at temperatures ranging from 20°C to 60°C reveals divergent unfolding pathways on mutation (B) The reduced structural stability of the mutant is speculated to add flexibility and to increase entropy in destabilizing the αβassociation in the tetramer.
Based on the localized thermal instability seen in molecular dynamics simulations and the weaker mechanical stability detected in force measurements, the proline mutation lowered the thermal stability of the α4-5 linker, resulting in a loss of structure at physiological temperature (Figure 3). This loss in linker helicity limited the stabilizing effects of R5 on R4 and led to independent unfolding of R4 (Tm = 29°C11 ). Linker coupling generally appears to be sequence specific rather than based on helical content alone,35 and steered molecular dynamics studies on related repeats have found this coupling to be largely based on the sequestration of linker residues from water.36 Exposure of the hydrophobic residue side-chains within this region lead to a loss in the tertiary structure of the linker and the repeats.
Human erythrocytes with the Q471P mutation show a shift in the tetramer–dimer equilibrium toward heterodimers, with almost a 2-fold weaker Kd-tetramer for the mutant. However, our ITC experiments on dimers demonstrated no effect on head-to-tail dimerization of monomeric α1-5 and β16-17 recombinant peptides. We propose that unfolding of repeats affects association into tetramer because of the staggered nature of dimerization (Figure 1A, inset). In the tetramer, α4-5 repeats are proximal to the dimerization site in the adjacent spectrin chain so that unfolded, flexible chains could enhance the configuration space of the dissociated state or could sterically inhibit association, just as sterics limits thiol reactivity (Figure 4). The flexibility would shift the tetramer–dimer distribution toward dissociation by decreasing the on-rate (Figure 6B). With this in mind, it becomes clear why a single amino acid mutation in a normally ordered linker can result in the shape changes and reduced erythrocyte membrane integrity associated with hemolytic elliptocytosis.
Authorship
Contribution: D.E.D., P.G., and D.W.S. designed the research, analyzed the data, and wrote the paper. C.P.J., G.M., V.O., N.B., and S.H. performed the research and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
C.P.J. and G.M contributed equally to this article.
This article is dedicated to the memory of Dr Sally Marchesi for her pioneering investigations into the molecular basis of spectrin pathologies.
Correspondence: Patrick G. Gallagher, Department of Medicine, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06520; e-mail: patrick.gallagher@yale.edu; David W. Speicher, The Wistar Institute, 3601 Spruce St, Philadelphia, PA 19104; e-mail: speicher@wistar.org; and Dennis E. Discher, Molecular and Cell Biophysics Laboratory, University of Pennsylvania, 3699 Market St, Philadelphia, PA19104; e-mail: discher@seas.upenn.edu.
The online version of this manuscript contains a data supplement.
An Inside Blood analysis of this article appears at the front of this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supported by funding from National Institutes of Health grants HL65448 (P.G.G.) and HL38794, the National Institutes of Health–National Heart, Lung and Blood Institute (D.E.D.), NCI Cancer Core grant CA10815, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (D.W.S.).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal