Abstract
Expression of the constitutively activated TEL/PDGFβR fusion protein is associated with the t(5;12)(q33;p13) chromosomal translocation found in a subset of patients with chronic myelomonocytic leukemia. TEL/PDGFβR activates multiple signal transduction pathways in cell-culture systems, and expression of the TEL-PDGFRB fusion gene induces myeloproliferative disease (MPD) in mice. We used gene-targeted mice to characterize the contribution of signal transducer and activator of transcription (Stat) and Src family genes to TEL-PDGFRB–mediated transformation in methylcellulose colony and murine bone marrow transduction/transplantation assays. Fetal liver hematopoietic stem and progenitor cells harboring targeted deletion of both Stat5a and Stat5b (Stat5abnull/null) genes were refractory to transformation by TEL-PDGFRB in methylcellulose colony assays. Notably, these cell populations were maintained in Stat5abnull/null fetal livers and succumbed to transformation by c-Myc. Surprisingly, targeted disruption of either Stat5a or Stat5b alone also impaired TEL-PDGFRB–mediated transformation. Survival of TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− mice was significantly prolonged, demonstrating significant sensitivity of TEL-PDGFRB–induced MPD to the dosage of Stat5a. TEL-PDGFRB–mediated MPD was incompletely penetrant in TPiGFP→Stat5b−/− mice. In contrast, Src family kinases Lyn, Hck, and Fgr and the Stat family member Stat1 were dispensable for TEL-PDGFRB disease. Together, these data demonstrate that Stat5a and Stat5b are dose-limiting mediators of TEL-PDGFRB–induced myeloproliferation.
Introduction
Chronic myelomonocytic leukemia (CMML) is characterized by dysplastic monocytosis, hypercellular bone marrow, splenomegaly, variable bone marrow fibrosis, and progression to acute myelogenous leukemia (AML). The t(5;12) (q33;p13) chromosomal translocation occurs in a subset of CMML patients. The TEL/PDGFβR fusion protein retains the pointed (PNT) domain found in TEL and the split tyrosine kinase (intracellular) portion of platelet-derived growth factor receptor β (PDGFβR).1 TEL/PDGFβR self-associates in the cytoplasm as oligomers via the PNT domain and becomes constitutively activated.2–4 This constitutive tyrosine kinase activity is required for recruitment of SH2-domain signaling intermediates including phosphatidylinositol 3-kinase (PI3K), phospholipase-C γ (PLCγ), SHP2, as well as signal transducers and activators of transcription 1 and 5 (STAT1 and STAT5, respectively).2,5–7
A murine bone marrow transduction/transplantation model of TEL-PDGFRB is marked by a rapidly fatal myeloproliferative disorder (MPD) that recapitulates aspects of human CMML including leukocytosis, splenomegaly, and extramedullary hematopoiesis.8 A TEL/PDGFβR mutant with tyrosine to phenylalanine mutations in the juxtamembrane SH2-domain–binding tyrosine residues (TEL/PDGFβR-F2) retains tyrosine kinase activity but does not induce myeloproliferation in mice.8 TEL/PDGFβR-F2 fails to bind and activate Stat56 and, in the context of native PDGFβR, the F2 tyrosines mediate binding and activation of both Src and Stat proteins.9–11 Therefore Src and Stat are candidate signaling molecules, but their role in mediating disease induced by TEL-PDGFRB is unclear. We used gene-targeted mice to characterize the contribution of signal transducer and activator of transcription (Stat) and Src family genes to TEL-PDGFRB–mediated transformation in methylcellulose colony and murine bone marrow transduction/transplantation assays
Materials and methods
Plasmids
TEL-PDGFRB ires green fluorescent protein (TPiGFP) was constructed in a murine stem cell virus (MSCV)2.2-ires-GFP vector backbone as previously described.8 TEL-PDGFRB-PGK-Neo (TPNeo) was generated by subcloning the TEL-PDGFRB cDNA into MSCV-Neo (Clontech, Mountain View, CA). MSCV-Myc was generated as described.12 TEL-PDGFRB ires c-Src (TPiSrc) was generated by inserting the c-Src cDNA (provided by Sara Courtneidge, Burnham Institute for Medical Research, La Jolla, CA) into MSCV2.2-ires-GFP, replacing GFP with c-Src. The TEL-PDGFRB cDNA was then excised from TPiGFP and placed into MSCV2.2-ires-cSrc. Stat51*6 ires GFP was created by placing the Stat51*6 cDNA (Toshio Kitamura, University of Tokyo, Japan) into MSCV2.2-ires-GFP. TEL-PDGFRB ires Stat5a (TPiStat5a) was created by placing the Stat5a cDNA into MSCV2.2-ires-GFP backbone to create MSCV2.2-ires-mStat5a, into which the TEL-PDGFRB cDNA was subcloned at the EcoRI site. TEL-PDGFRB ires Stat5b (TPiStat5b) was generated by inserting the mStat5b cDNA (courtesy of Lothar Hennighausen, NIDDK, Warren Leonard NHLBI, National Institutes of Health, Bethesda, MD) into MSCV2.2-ires-GFP. The TEL-PDGFRB cDNA was then subcloned into the EcoRI site of MSCV2.2-ires-mStat5b.
Retrovirus production
Retroviral supernatants were generated by transient transfection of Ecopac (Cell Genesys, Foster City, CA) with MSCV-based retroviral construct using Superfect transfection reagent (Qiagen, Chatsworth, CA) per the manufacturer's instructions. Retroviral supernatants were harvested 48 hours after transfection. To evaluate viral titer of constructs containing eGFP, Ba/F3 cells were transduced with retroviral supernatants and GFP expression was determined by flow cytometric detection of GFP positivity. To assess MSCV-Neo, TPiNeo, TPiStat5a, and TPiStat5b viral titer, 3T3 cells were transduced with retroviral supernatants. 3T3 DNA was isolated and quantitative polymerase chain reaction (PCR) amplication was used to detect psi retroviral sequences compared with control genomic sequences.
Mouse strains
Lyn, Hck, Fgr triply deficient mice13 were provided by Clifford Lowell (University of California, San Francisco). Stat1−/− mice14 were provided by Robert Schreiber (Washington University, St Louis). Lothar Hennighausen (NIDDK, National Institutes of Health) contributed Stat5a−/−15 and Stat5ab+/null mice.16 Stat5ab+/null mice were crossed with NIH Black Swiss female mice (Taconic Farms, Hudson, NY). The resulting outbred Stat5ab+/null male and female mice were used to generate Stat5abnull/null, Stat5abnull/null, and Stat5abnull/null fetuses. Stat5b−/− mice17 were acquired from Helen Davey (AgResearch, Hamilton, New Zealand). Assisted speed congenics18 were used to backcross the targeted Stat5a (N3 generation, approximately 94% balb/c) or Stat5b (N4 generation, approximately 99% balb/c) allele in singly deficient mice to balb/c (Taconic Farms).
Methylcellulose colony formation
Bone marrow transduction of whole bone marrow (Stat5a and Stat5b singly deficient mouse strains backcrossed to balb/c) was performed as previously described.8 Briefly, whole bone marrow was harvested from 150 mg/kg 5-fluorouracil–treated mice, and the red blood cells were removed by brief incubation in hypotonic lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.4). Unfractionated cells were prestimulated in media containing pen/strep, fetal bovine serum, SCF, IL3, FLT3, and Tpo for 48 hours. Cells were transduced with retroviral supernatants of equivalent titer by 2 rounds of centrifugation at 2500g for 90 minutes in the presence of 8 μg/mL polybrene (American Bioanalytical, Natick, MA). Twenty-four hours following the second transduction, cells were washed thrice with cold PBS and resuspended in methylcellulose with cytokines (2 × 104 cells/mL in M3434 methylcellulose; Stem Cell Technologies, Vancouver, BC) or without cytokines (105 cells/mL in M3234; Stem Cell Technologies) in the presence or absence of 1 mg/mL G418 (Gibco/Invitrogen, Grand Island, NY; from a 20-mg/mL stock solution in 0.1 M Hepes [Cambrex, Walkersville, MD]) and plated in triplicate. Colonies containing at least 30 cells were counted 10 days after plating. Transduction efficiency for each genotype and experimental condition was determined by plating cells in the presence of cytokines and dividing the average number of G418-resistant colonies per plate by the average number of colonies per plate in a duplicate set of plates not containing G418.
Murine 14.5-dpc fetuses were harvested from Stat5ab+/null × Stat5ab+/null timed pregnancies and were washed in sterile PBS (Gibco/Invitrogen). Fetal livers were placed into RPMI containing penn/strep and 10% fetal bovine serum and brought to a single-cell suspension by passing through a 27-gauge needle. Red blood cells were lysed as in the paragraph above, and the remaining cells from each fetal liver were resuspended into fetal liver transplant media (RPMI containing pen/strep, 10% fetal bovine serum, 10 ng/mL IL6, 100 ng/mL SCF, 6 ng/mL IL3, 50 ng/mL FLT3, and 10 ng/mL Tpo) and placed at 37°C in 5% CO2. Fetal liver cell DNA was isolated using Puregene DNA purification kit (Gentra Systems, Minneapolis, MN) and genotyped as described.16 Cells of each genotype were pooled and replated in fetal liver transplant media and incubated at 37°C in 5% CO2. Beginning 36 hours after harvest, cells were transduced every 6 hours with retroviral supernatants of equivalent titer by centrifugation at 2500g for 10 minutes in the presence of 8 μg/mL polybrene (American Bioanalytical) for a total of 4 transductions. Twenty-four hours following the final transduction, cells were washed and plated in methylcellulose as described in the preceding paragraph. Colonies containing at least 30 cells were counted 10 days after plating. Transduction efficiency was determined as described above for TPNeo and MSCV-Neo experiments, while detection of GFP-positive cells by flow cytometry immediately prior to plating cells in methylcellulose was used to evaluate MSCV-Myc transduction efficiency.
Retroviral transduction and transplantation
Transduction and transplantation were performed as previously described.8 Syngeneic recipient mice were lethally irradiated and were injected with 106 bone marrow cells in 750 μL Hanks buffered saline solution via lateral tail-vein injection. Doses of irradiation were as follows: Lyn−/−Hck−/−Fgr−/−: 450 cGy C57/Black6 (Taconic Farms); Stat1−/−: 1000 cGy C57BL/6 × 129S6/SvEv F1 (B6129; Taconic Farms); outbred Stat5a−/−: 1000 cGy B6129 (Taconic Farms); balb/c backcrossed Stat5a−/−: 900 cGy wild-type littermate control; outbred Stat5b−/−: 1000cGy B6129 (Taconic Farms); balb/c backcrossed Stat5b−/−: 900 cGy wild-type littermate control.
Mouse analysis
Peripheral blood was obtained from the saphenous vein of moribund mice and blood counts were analyzed (Hemavet; CDC Technologies, Oxford, CT). Moribund mice were killed and spleen weight was recorded. Heart, lungs, tibia, spleen, kidney, and liver were fixed using 10% neutral buffered formalin (Sigma-Aldrich, St Louis, MO). Slides of peripheral blood and formalin-fixed tissue were hematoxylin and eosin (H&E) stained and imaged using an Olympus BX40 F4 microscope with an oil-immersion 50×/0.90 or 100×/1.30 objective lens (Olympus Optical, Tokyo, Japan) using an Olympus DP70 digital camera using DPController software (Olympus Optical). Statistical analyses were generated with StatView (SAS Institute, Cary, NC). Kaplan-Meier plots were generated using Excel (Microsoft, Redmond, WA).
Flow cytometry
Bone marrow cells and splenocytes were brought to a single-cell suspension in RPMI-1640 (Cambrex) and the red blood cells were removed by brief incubation in hypotonic lysis buffer. Cells were washed once with flow buffer (PBS, 0.5% BSA, 0.1 mM EDTA), and 106 cells were resuspended in 100 μL flow buffer and incubated with 1 μL each of the antibodies recognizing murine Gr-1 and Mac-1 conjugated to PE and PECy7, respectively (eBiosciences, San Diego, CA) for 1 hour on ice. Cells were then washed twice with flow buffer and resuspended in 250 μL flow buffer. Data were collected using either MoFlo (Dako, Carpinteria, CA) or Cytomics FC 500 (Beckman Coulter, Fullerton, CA). Figures were prepared using FloJo software (Tree Star, San Carlos, CA).
Progenitor analysis methods
As described under “Methylcellulose colony formation.” 14.5-dpc fetal liver cells were harvested, brought to a single-cell suspension in PBS (Gibco/Invitrogen), and stored at 4°C during DNA isolation (REDExtract; Sigma-Aldrich) and genotyping (as described16 ). Unfractionated fetal liver cells of each genotype were pooled into Stat5ab−/− (n = 8 fetal livers), Stat5ab+/− (n = 6), and Stat5ab+/+ (n = 7) cohorts, filtered through a 50-μM filter (Partec, Münster, Germany), and stained for flow cytometric analysis of progenitor populations as described.19 Antibody staining for lineage markers was performed using FITC-conjugated CD3e (145-2C11), CD4 (RM4-5), CD8a (53-6.7), CD19 (1D3), CD45R (RA3-6B2), and GR1 (RB6-8C5) acquired from BD PharMingen (San Diego, CA) and TER119 (TER-119) and CD127 (A7R34) from eBiosciences. Cells were also stained using biotin-CD34 (RAM34), PE-streptavidin, APC-Sca-1 (D7), PE-Cy7-CD16/32 (93), and APC-Alexa750-CD117 (2B8) antibodies obtained from eBiosciences. Data were collected from 5 × 106 cells per genotype using MoFlo flow cytometer (Dako), and figures were prepared using FloJo software (Tree Star).
Results
Stat5 is required in primary murine hematopoietic cells for TEL-PDGFRB–mediated growth
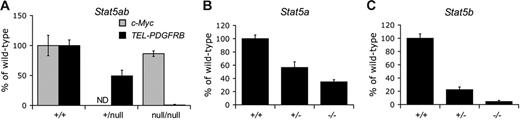
Mice harboring targeted deletions of both Stat5a and Stat5b genes (Stat5abnull/null) rarely survive past weaning.16,20 Therefore, we evaluated TEL-PDGFRB transformation in methylcellulose colony formation assays using fetal liver hematopoietic stem cells (HSCs) and progenitor cells. We transduced unfractionated Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null16 fetal liver cells with retroviral supernatants encoding either TEL-PDGFRB with a neomycin resistance cassette (TPNeo) or Neo alone (MSCV-Neo) and plated them in methylcellulose in the absence of cytokines. Transduction efficiency was equivalent among Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null cells. TEL-PDGFRB induced G418-resistant colony formation in Stat5ab+/+ and Stat5ab+/null cells. In contrast, Stat5abnull/null cells were completely resistant to TEL-PDGFRB–mediated transformation (Figure 1A). MSCV-Neo did not cause cytokine independent colony formation in Stat5ab+/+, Stat5ab+/null, or Stat5abnull/null cells (data not shown). As a positive control for the transduction of hematopoietic stem and progenitor cells from these mice, we transduced Stat5ab+/+ and Stat5abnull/null cells with c-Myc. c-Myc readily transformed both Stat5ab+/+ and Stat5abnull/null cells to generate cytokine-independent colonies (Figure 1A), confirming efficient retroviral transduction of Stat5-deficient cells.
Stat5 is required in primary murine hematopoietic cells for TEL-PDGFRB–mediated growth. (A) Cytokine-independent colony formation of 14.5-dpc fetal liver cells expressing either TEL-PDGFRB or c-Myc. Data shown are the mean percentage of colonies formed by Stat5abnull/null and Stat5ab+/null cells relative to Stat5ab+/+ cells for a given experimental condition. Error bars represent standard deviation. TPNeo induced formation of 195 ± 18 Stat5ab+/+, 194 ± 19 Stat5ab+/null, and 3 ± 1 Stat5abnull/null G418-resistant colonies per 105 transduced fetal liver cells. Positive control c-Myc induced formation of 159 ± 8 colonies per 105Stat5abnull/null fetal liver cells plated and 184 ± 31 colonies in Stat5ab+/+ cells. Cells transduced with negative control MSCV-Neo did not form colonies in the absence of cytokines (data not shown). Transduction efficiency of Stat5ab+/+ and Stat5abnull/null fetal liver cells with MSCV-Myc was 14% and 12% for 2 independent experiments. For TPNeo and MSCV-Neo, transduction efficiency of Stat5ab+/+ cells averaged 25% ± 15% and Stat5abnull/null cells averaged 45% ± 16% for 7 independent transductions of each genotype. (B) TEL-PDGFRB induced cytokine independent colony formation in Stat5a+/+ (469 ± 26 colonies per 105 transduced cells), Stat5a+/− (266 ± 40), and Stat5a−/− (163 ± 16) whole bone marrow cells. Data shown are the mean percentage of colonies formed by Stat5a−/− and Stat5a+/− cells relative to Stat5a+/+ cells for a given experimental condition. Error bars represent standard deviation. Transduction efficiency of Stat5a−/− cells averaged 98% ± 17% and 93% ± 18% for Stat5a+/+ bone marrow cells in 3 independent transductions. (C) TEL-PDGFRB induced cytokine-independent colony formation in Stat5b+/+ (602 ± 39 colonies per 105 transduced cells), Stat5b+/− (135 ± 25), and Stat5b−/− (27 ± 11) whole bone marrow cells. Data shown are the mean percentage of colonies formed by Stat5b−/− and Stat5b+/− cells relative to Stat5b+/+ cells for a given experimental condition. Error bars represent standard deviation. Transduction efficiency of Stat5b−/− cells was 99% and 81% for Stat5b+/+ bone marrow cells. (TPNeo indicates TEL-PDGFRB-PGK Neo; MSCV-Myc, MSCV-c-Myc ires GFP; and ND, not done.)
Stat5 is required in primary murine hematopoietic cells for TEL-PDGFRB–mediated growth. (A) Cytokine-independent colony formation of 14.5-dpc fetal liver cells expressing either TEL-PDGFRB or c-Myc. Data shown are the mean percentage of colonies formed by Stat5abnull/null and Stat5ab+/null cells relative to Stat5ab+/+ cells for a given experimental condition. Error bars represent standard deviation. TPNeo induced formation of 195 ± 18 Stat5ab+/+, 194 ± 19 Stat5ab+/null, and 3 ± 1 Stat5abnull/null G418-resistant colonies per 105 transduced fetal liver cells. Positive control c-Myc induced formation of 159 ± 8 colonies per 105Stat5abnull/null fetal liver cells plated and 184 ± 31 colonies in Stat5ab+/+ cells. Cells transduced with negative control MSCV-Neo did not form colonies in the absence of cytokines (data not shown). Transduction efficiency of Stat5ab+/+ and Stat5abnull/null fetal liver cells with MSCV-Myc was 14% and 12% for 2 independent experiments. For TPNeo and MSCV-Neo, transduction efficiency of Stat5ab+/+ cells averaged 25% ± 15% and Stat5abnull/null cells averaged 45% ± 16% for 7 independent transductions of each genotype. (B) TEL-PDGFRB induced cytokine independent colony formation in Stat5a+/+ (469 ± 26 colonies per 105 transduced cells), Stat5a+/− (266 ± 40), and Stat5a−/− (163 ± 16) whole bone marrow cells. Data shown are the mean percentage of colonies formed by Stat5a−/− and Stat5a+/− cells relative to Stat5a+/+ cells for a given experimental condition. Error bars represent standard deviation. Transduction efficiency of Stat5a−/− cells averaged 98% ± 17% and 93% ± 18% for Stat5a+/+ bone marrow cells in 3 independent transductions. (C) TEL-PDGFRB induced cytokine-independent colony formation in Stat5b+/+ (602 ± 39 colonies per 105 transduced cells), Stat5b+/− (135 ± 25), and Stat5b−/− (27 ± 11) whole bone marrow cells. Data shown are the mean percentage of colonies formed by Stat5b−/− and Stat5b+/− cells relative to Stat5b+/+ cells for a given experimental condition. Error bars represent standard deviation. Transduction efficiency of Stat5b−/− cells was 99% and 81% for Stat5b+/+ bone marrow cells. (TPNeo indicates TEL-PDGFRB-PGK Neo; MSCV-Myc, MSCV-c-Myc ires GFP; and ND, not done.)
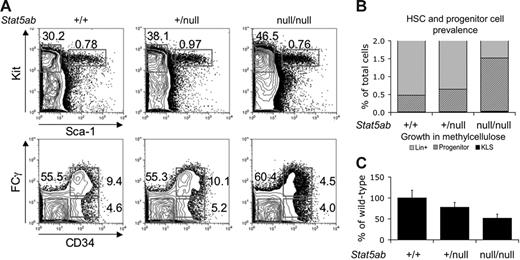
We were impressed by the dramatic reduction of TEL-PDGFRB–induced cytokine-independent colonies in the absence of Stat5 and were concerned that our data might be explained by reduced hematopoietic stem and progenitor cells in these mutant mice. To further assess the presence and quantity of hematopoietic stem and progenitor cells in Stat5abnull/null fetal livers, we performed multiparameter immunophenotypic analysis using high-speed flow cytometry.19 Less mature, lineage-negative cells were twice as prevalent among Stat5abnull/null fetal liver cells compared with Stat5ab+/+ cells (data not shown). Among lineage-negative cells, there were equivalent numbers of HSC (KLS) cells present in Stat5abnull/null and Stat5ab+/+ fetal livers (Figure 2A). Together, these data indicate that HSCs are twice as prevalent among Stat5abnull/null fetal liver cells, consistent with the findings of other investigators.21 The incidence of common myeloid progenitor (CMP) was the same among Stat5abnull/null, Stat5ab+/null, and Stat5ab+/+ Kit+Lin−Sca− fetal liver cells (Figure 2A). Megakaryocyte-erythrocyte progenitor (MEP) populations were moderately increased in Stat5abnull/null fetal liver cells (Figure 2A). Notably, granulocyte-monocyte progenitors (GMPs) were markedly decreased (Figure 2A). When evaluated as subsets of all fetal liver cells, immunophenotypically defined hematopoietic stem cell–enriched and progenitor populations were moderately increased in Stat5abnull/null fetal liver cells compared with wild-type littermate controls (summarized in Figure 2B).
Hematopoietic stem and progenitor-cell populations are preserved in fetal livers of Stat5abnull/null mice. (A) Flow cytometric analysis of Stat5abnull/null, Stat5ab+/null, and Stat5ab+/+ 14.5-dpc fetal livers for early hematopoietic cells. Top panel shows Kit and Sca-1 expression of lineage-negative fetal liver cells. Bottom panel shows CMP (FcγRloCD34+), MEP (FcγRloCD34−), and GMP (FcγRhiCD34+) populations, with percentage of Kit+Lin−Sca− falling into each progenitor population as indicated. (B) Proportion of cells that is identified as either KLS (black), progenitors (CMPs, MEPs, or GMPs, diagonal stripes), or lineage-committed (gray) hematopoietic cells within Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null 14.5-dpc fetal livers as determined by flow cytometric detection of cell-surface markers. (C) Cytokine-dependent colony formation of vector-transduced 14.5-dpc fetal liver cells when cells were plated in the presence of SCF, IL3, IL6, and EPO. Data shown are the mean percentage of colonies formed by Stat5abnull/null and Stat5ab+/null cells relative to Stat5ab+/+ cells. Error bars represent standard deviation. Vector control Stat5ab+/+ cells generated 688 ± 18 G418-resistant colonies per 105 transduced fetal liver cells, while Stat5ab+/null and Stat5abnull/null cells formed 535 ± 12 and 355 ± 10 colonies. TPNeo-transduced Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null cells formed 337 ± 33, 559 ± 43, and 321 ± 30 G418 colonies, respectively. Transduction efficiency of Stat5ab+/+ cells averaged 25% ± 15% and Stat5abnull/null cells averaged 45% ± 16% for 7 independent transductions of each genotype. (KLS indicates Kit+Sca+Lin−; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; and GMP, granulocyte-monocyte progenitor.)
Hematopoietic stem and progenitor-cell populations are preserved in fetal livers of Stat5abnull/null mice. (A) Flow cytometric analysis of Stat5abnull/null, Stat5ab+/null, and Stat5ab+/+ 14.5-dpc fetal livers for early hematopoietic cells. Top panel shows Kit and Sca-1 expression of lineage-negative fetal liver cells. Bottom panel shows CMP (FcγRloCD34+), MEP (FcγRloCD34−), and GMP (FcγRhiCD34+) populations, with percentage of Kit+Lin−Sca− falling into each progenitor population as indicated. (B) Proportion of cells that is identified as either KLS (black), progenitors (CMPs, MEPs, or GMPs, diagonal stripes), or lineage-committed (gray) hematopoietic cells within Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null 14.5-dpc fetal livers as determined by flow cytometric detection of cell-surface markers. (C) Cytokine-dependent colony formation of vector-transduced 14.5-dpc fetal liver cells when cells were plated in the presence of SCF, IL3, IL6, and EPO. Data shown are the mean percentage of colonies formed by Stat5abnull/null and Stat5ab+/null cells relative to Stat5ab+/+ cells. Error bars represent standard deviation. Vector control Stat5ab+/+ cells generated 688 ± 18 G418-resistant colonies per 105 transduced fetal liver cells, while Stat5ab+/null and Stat5abnull/null cells formed 535 ± 12 and 355 ± 10 colonies. TPNeo-transduced Stat5ab+/+, Stat5ab+/null, and Stat5abnull/null cells formed 337 ± 33, 559 ± 43, and 321 ± 30 G418 colonies, respectively. Transduction efficiency of Stat5ab+/+ cells averaged 25% ± 15% and Stat5abnull/null cells averaged 45% ± 16% for 7 independent transductions of each genotype. (KLS indicates Kit+Sca+Lin−; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; and GMP, granulocyte-monocyte progenitor.)
We plated transduced fetal liver cells in methylcellulose containing SCF, IL3, IL6, and EPO to determine if Stat5abnull/null progenitor cells form colonies in methylcellulose. MSCV-Neo–transduced Stat5abnull/null cells generated half the number of colonies found on Stat5ab+/+ plates (Figure 2C). Expression of TEL-PDGFRB rescued the colony formation defect in Stat5abnull/null fetal liver cells, suggesting that TEL-PDGFβR activated Stat5-independent signaling pathways to restore colony formation capacity to that of Stat5ab+/+ cells (data not shown). Taken together, these data suggest that the inability of TEL-PDGFRB to transform Stat5abnull/null fetal liver cells was not due to the absence of HSC or progenitor-cell populations.
Stat5a and Stat5b share 96% similarity on an amino acid level22 and serve largely redundant functions in hematopoiesis.15–16,17,23 Therefore, we hypothesized that TEL-PDGFRB–mediated transformation would not be affected by deficiency of either Stat5a or Stat5b alone (ie, either Stat5a−/−Stat5b+/+ or Stat5a+/+Stat5b−/−). Stat5a−/− and Stat5b−/− mice have no defects in basal hematopoiesis15,17 (and data not shown), however Stat5a and Stat5b possess distinct DNA-binding specificities24–25 and have unique roles in response to high doses of Flt3 ligand.26 We tested the role of individual Stat5 genes in TEL-PDGFRB transformation using methylcellulose colony assays and bone marrow transduction/transplantation assays. We transduced whole bone marrow cells harboring targeted disruption of either Stat5a or Stat5b with either TPNeo or MSCV-Neo retroviral supernatants and plated them in the absence of cytokines. TEL-PDGFRB transformed wild-type cells to cytokine-independent growth (Figure 1B-C). Remarkably, there was a downward trend of TEL-PDGFRB–transformed colonies when either Stat5a+/− singly targeted or Stat5b+/− singly targeted bone marrow cells were used. Finally, deficiency in either Stat5a or Stat5b alone caused a significant reduction in the incidence of TEL-PDGFRB–transformed colonies (Figure 1B-C), indicating that both Stat5a and Stat5b contribute to transformation. Transduction efficiency was equivalent in all cohorts. Taken together, these data demonstrate that Stat5 is critical to TEL-PDGFRB–induced transformation.
Stat5a gene dosage mediates the latency of TEL-PDGFRB–induced myeloproliferation
Given the surprising attenuation of TEL-PDGFRB–mediated transformation in the absence of Stat5a, we sought to determine the impact of Stat5a deficiency upon the myeloproliferative disease induced by TEL-PDGFRB in vivo. Unlike Stat5abnull/null mice, Stat5a-deficient mice have abnormal lactogenesis, but no defects in steady-state myelopoiesis,15 and can be used as donors for bone marrow transplant assays. We assessed the relative contribution of the Stat5a gene to TEL-PDGFRB–mediated transformation in bone marrow transduction/transplantation experiments using Stat5a singly deficient (Stat5a−/−), heterozygous (Stat5a+/−), and wild-type (Stat5a+/+) donor mice with retroviral supernatants encoding TEL-PDGFRB with eGFP (TPiGFP). Survival of TPiGFP→Stat5a−/− mice was significantly prolonged in comparison with TPiGFP→Stat5a+/+ mice (144 versus 31 days, P < .001; Figure 3A). Surprisingly, disease latency in TPiGFP→Stat5a+/− mice was similar to that of TPiGFP→Stat5a−/− mice (145 versus 144 days, P = .867) and was significantly prolonged compared with survival of TPiGFP→Stat5a+/+ mice (145 versus 31 days, P < .001; Figure 3A). Therefore, the latency of TEL-PDGFRB–induced disease was significantly affected by the loss of even a single wild-type Stat5a allele.
Stat5a gene dosage mediates the latency of TEL-PDGFRB–induced myeloproliferation. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring homozygous or heterozygous inactivation of Stat5a. TPiGFP→Stat5a+/+ mice (circles) developed severe MPD (spleen weight median, 790 ± 90 mg; range, 620-910 mg; n = 6). TPiGFP→Stat5a+/− mice (▵) developed moderate or severe MPD (spleen weight median, 770 ± 300 mg; range, 350-1120 mg; n = 7). TPiGFP→Stat5a−/− mice (squares) developed moderate MPD with splenomegaly (spleen weight, 710 ± 550 mg; range, 350-2070 mg; n = 8) and leukocytosis. Black shapes indicate death due to severe (fatal) MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (B) Median peripheral blood cell count at time of death due to disease. Error bars represent standard deviation. TEL-PDGFRB induced severe leukocytosis in TPiGFP→Stat5a+/+ mice (359 ± 225 × 109/L [359 000 ± 225 000/μL]; range, 60-737 × 109/L [60 000-737 000/μL]; n = 6) and moderate leukocytosis in TPiGFP→Stat5a+/− mice (28 ± 15 × 109/L [28 000 ± 15 000/μL]; range, 12-131 × 109/L [12 000-131 000/μL]; n = 7) as well as TPiGFP→Stat5a−/− mice (29 ± 27 × 109/L [29 000 ± 27 000/μL];range, 10-94 × 109/L [10 000-94 000/μL]; n = 8). (C) Three mice from each cohort were analyzed 28 days after transplantation. Median of peripheral blood counts is shown. Error bars represent standard deviation. Peripheral blood counts of GFP→Stat5a−/− control mice are also shown. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)
Stat5a gene dosage mediates the latency of TEL-PDGFRB–induced myeloproliferation. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring homozygous or heterozygous inactivation of Stat5a. TPiGFP→Stat5a+/+ mice (circles) developed severe MPD (spleen weight median, 790 ± 90 mg; range, 620-910 mg; n = 6). TPiGFP→Stat5a+/− mice (▵) developed moderate or severe MPD (spleen weight median, 770 ± 300 mg; range, 350-1120 mg; n = 7). TPiGFP→Stat5a−/− mice (squares) developed moderate MPD with splenomegaly (spleen weight, 710 ± 550 mg; range, 350-2070 mg; n = 8) and leukocytosis. Black shapes indicate death due to severe (fatal) MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (B) Median peripheral blood cell count at time of death due to disease. Error bars represent standard deviation. TEL-PDGFRB induced severe leukocytosis in TPiGFP→Stat5a+/+ mice (359 ± 225 × 109/L [359 000 ± 225 000/μL]; range, 60-737 × 109/L [60 000-737 000/μL]; n = 6) and moderate leukocytosis in TPiGFP→Stat5a+/− mice (28 ± 15 × 109/L [28 000 ± 15 000/μL]; range, 12-131 × 109/L [12 000-131 000/μL]; n = 7) as well as TPiGFP→Stat5a−/− mice (29 ± 27 × 109/L [29 000 ± 27 000/μL];range, 10-94 × 109/L [10 000-94 000/μL]; n = 8). (C) Three mice from each cohort were analyzed 28 days after transplantation. Median of peripheral blood counts is shown. Error bars represent standard deviation. Peripheral blood counts of GFP→Stat5a−/− control mice are also shown. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)
Despite significant prolongation of survival, all mice eventually succumbed to disease. We characterized TEL-PDGFRB disease in each Stat5a genotype cohort at time of death. Histopathologic analysis of TPiGFP→Stat5a+/+ mice revealed severe extramedullary hematopoiesis with accumulation of large numbers of mature myeloid cells in the liver sinusoids and perivascular regions of the liver and profoundly disrupted splenic architecture (data not shown). Blood vessels and capillaries were filled with granulocytes with multifocal congestion and hemorrhage in the lungs of TPiGFP→Stat5a+/+ mice. Consistent with the prolonged survival of both TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− mice, only mild extramedullary hematopoiesis was detected in both TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− mice (data not shown), although all cohorts had splenomegaly. There were slight to moderate accumulations of granulocytes in the livers and diffuse expansion of granulocytes in the spleens of both TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− cohorts at time of death due to disease (data not shown). Well-differentiated granulocytes infiltrated the lungs of TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− recipient mice, with mild to moderate multifocal congestion in the lungs. TPiGFP→Stat5a+/+ mice displayed severe leukocytosis, while leukocytosis in TPiGFP→Stat5a+/− and TPiGFP→Stat5a−/− mice was significantly reduced (median WBC, 29 × 109/L [29 000/μL] and 28 × 109/L [28 000/μL], respectively; Figure 3B). Taken together, these data show that TEL-PDGFRB–induced MPD was attenuated in both disease latency and severity in TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/− mice, suggesting that TEL-PDGFRB–induced MPD was significantly reduced by absence or haploinsufficiency of Stat5a.
While transduction efficiency was equivalent among Stat5a−/−, Stat5a+/−, and Stat5a+/+ cells in a methylcellulose assay for TEL-PDGFRB–mediated transformation, we sought to confirm that we had efficiently transduced and transplanted Stat5a−/− and Stat5a+/+ cells in our in vivo model. To address the possibility that pretreatment with 5-FU reduces the Stat5a−/− transduction/transplantation target population relative to that in Stat5a+/+ cells, we transduced Stat5a−/− donor cells with retroviral supernatants encoding a constitutively activated Stat5a, Stat51*6.27. As expected, Stat51*6→Stat5a−/− recipient mice developed a rapidly fatal disease (median survival, 19 ± 1 days; data not shown), similar to disease seen in wild-type mice.28 We further verified equivalent integration of retroviral construct among transplanted Stat5a−/− and Stat5a+/+ donor cells in TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/+ recipient mice as detected by linear amplification-mediated (LAM)–PCR (Figure S2, available on the Blood website; see the Supplemental Figures link at the top of the online article). These data are consistent with a previous report demonstrating that 5-FU affects Stat5a−/− and Stat5a+/+ myeloid donor-cell populations equally and that clonal integrations of Stat5a−/− and Stat5a+/+ transduced and transplanted cells are equivalent.29
We were struck by the dramatic reduction in disease severity afforded by loss of a single Stat5a allele in TPiGFP→Stat5a+/− mice. We analyzed TPiGFP→Stat5a+/+, TPiGFP→Stat5a+/−, and TPiGFP→Stat5a−/− mice 28 days after transplantation to assess the effects of Stat5a deficiency and haploinsufficiency on TEL-PDGFRB–induced myeloproliferation early after transduction/transplantation. All TPiGFP→Stat5a+/+ mice developed severe MPD (median WBC, 547 × 109/L [547 000/μL]; spleen weight, 860 mg; Figure 3C), as expected. TEL-PDGFRB–mediated myeloproliferation was blunted in TPiGFP→Stat5a+/− mice (median WBC, 56 × 109/L [56 000/μL]; spleen weight, 790 mg) (Figure 3C). TPiGFP→Stat5a−/− mice showed no evidence of disease (median WBC, 5 × 109/L [5000/μL]; spleen weight, 53 mg; Figure 3C). These data support the conclusion that TEL-PDGFRB–induced myeloproliferation responds significantly to the loss of even a single Stat5a allele.
TEL-PDGFRB–mediated MPD is incompletely penetrant in the absence of Stat5b
TPiGFP→Stat5a+/− and TPiGFP→Stat5a−/− mice were protected against TEL-PDGFRB–mediated MPD despite intact Stat5b. To directly address a role for Stat5b in TEL-PDGFRB–mediated MPD, we used donor mice with homozygous inactivation of Stat5b. Both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− mice developed fatal MPD (Figure 4A) marked by leukocytosis and extramedullary hematopoiesis. Mature myeloid cells invaded the spleens of both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− mice as detected by both flow cytometric (Figure 4B) and histopathologic (Figure 4C) analysis. Further, histopathologic analysis revealed that sheets of these mature myeloid cells (predominately mature neutrophils) disrupted splenic architecture and destroyed the normal pattern of white pulp nodules in a matrix of red pulp evident in eGFP control mice (Figure 4C). Mature myeloid cells infiltrated sinusoids and perivascular regions of the liver in both TPiGFP→Stat5b−/− and TPiGFP→Stat5b+/+ mice, unlike in their eGFP control mice counterparts (Figure 4C). The bone marrow of both cohorts was hypercellular, with atypical myeloid cells observed by histopathologic analysis (data not shown).
Severity and penetrance of TEL-PDGFRB–induced myeloproliferation are reduced in the absence of Stat5b. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring targeted deletion of Stat5b (TPiGFP→Stat5b−/−, squares) or wild-type control bone marrow cells (TPiGFP→Stat5b+/+, ○). TPiGFP→Stat5b+/+ mice developed severe MPD (spleen weight median, 890 ± 274 mg; range, 670-1460 mg [n = 6]; WBC median, 139 ± 87 × 109/L [139 000 ± 87 000/μL]; range, 13-237 × 109/L [13 000-237 000/μL] [n = 6]; median survival ± standard error, 33 ± 2.9 days). Severe MPD was detected in 5 of 7 TPiGFP→Stat5b−/− mice at time of death due to disease (spleen weight median, 1220 ± 401 mg; range, 420-1440 mg [n = 7]; WBC median, 103 ±151 × 109/L [103 000 ± 151 000/μL]; range, 6-446 × 109/L [6000-446 000/μL] [n = 7]; median survival ± standard error, 57 ± 60.2 days). Experiment was performed twice, once in the original outbred strain with wild-type littermate control and again with the targeted Stat5b allele backcrossed to balb/c resulting in similar disease latency and severity. Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. GFP was detected in bone marrow, spleen, and peripheral blood of TPiGFP→Stat5b−/− (37% ± 4% [n = 7]; 50% ± 15% [n = 7]; and 58% ± 32% [n = 7], respectively) and TPiGFP→Stat5b+/+ (44% [n = 1]; 50% ± 15% [n = 3]; and 58% ± 32% [n = 3], respectively) recipient mice (data not shown). (B) Flow cytometric detection of mature myeloid (Gr-1+Mac-1+) cells in the spleens of both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− mice 33 days after transplantation. (C) Microscopic images of representative mice at time of death (GFP→Stat5b+/+, 151 days after transplantation; TPiGFP→Stat5b+/+, 29 days after transplantation; and TPiGFP→Stat5b−/−, 29 days after transplantation). Panels show Wright-Giemsa–stained smears of peripheral blood (Ci-iii; 100×), and H&E–stained histologic sections of spleen (Civ-vi; 20×) and liver (Cvii-ix; 40×). These illustrate a normal morphology of peripheral blood, spleen, and liver in the GFP→Stat5b+/+ group, whereas both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− show marked leukocytosis of maturing neutrophils in the peripheral blood, pronounced red pulp expansion by myeloid hyperplasia composed of predominantly maturing forms in the spleen, and sinusoidal and perivascular infiltrates of maturing granulocytic cells in the liver. Arrows indicate clusters of maturing myeloid cells in the liver. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; WBC, peripheral white blood cell count; PB, peripheral blood; Sp, spleen; and Li, liver.)
Severity and penetrance of TEL-PDGFRB–induced myeloproliferation are reduced in the absence of Stat5b. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring targeted deletion of Stat5b (TPiGFP→Stat5b−/−, squares) or wild-type control bone marrow cells (TPiGFP→Stat5b+/+, ○). TPiGFP→Stat5b+/+ mice developed severe MPD (spleen weight median, 890 ± 274 mg; range, 670-1460 mg [n = 6]; WBC median, 139 ± 87 × 109/L [139 000 ± 87 000/μL]; range, 13-237 × 109/L [13 000-237 000/μL] [n = 6]; median survival ± standard error, 33 ± 2.9 days). Severe MPD was detected in 5 of 7 TPiGFP→Stat5b−/− mice at time of death due to disease (spleen weight median, 1220 ± 401 mg; range, 420-1440 mg [n = 7]; WBC median, 103 ±151 × 109/L [103 000 ± 151 000/μL]; range, 6-446 × 109/L [6000-446 000/μL] [n = 7]; median survival ± standard error, 57 ± 60.2 days). Experiment was performed twice, once in the original outbred strain with wild-type littermate control and again with the targeted Stat5b allele backcrossed to balb/c resulting in similar disease latency and severity. Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. GFP was detected in bone marrow, spleen, and peripheral blood of TPiGFP→Stat5b−/− (37% ± 4% [n = 7]; 50% ± 15% [n = 7]; and 58% ± 32% [n = 7], respectively) and TPiGFP→Stat5b+/+ (44% [n = 1]; 50% ± 15% [n = 3]; and 58% ± 32% [n = 3], respectively) recipient mice (data not shown). (B) Flow cytometric detection of mature myeloid (Gr-1+Mac-1+) cells in the spleens of both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− mice 33 days after transplantation. (C) Microscopic images of representative mice at time of death (GFP→Stat5b+/+, 151 days after transplantation; TPiGFP→Stat5b+/+, 29 days after transplantation; and TPiGFP→Stat5b−/−, 29 days after transplantation). Panels show Wright-Giemsa–stained smears of peripheral blood (Ci-iii; 100×), and H&E–stained histologic sections of spleen (Civ-vi; 20×) and liver (Cvii-ix; 40×). These illustrate a normal morphology of peripheral blood, spleen, and liver in the GFP→Stat5b+/+ group, whereas both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− show marked leukocytosis of maturing neutrophils in the peripheral blood, pronounced red pulp expansion by myeloid hyperplasia composed of predominantly maturing forms in the spleen, and sinusoidal and perivascular infiltrates of maturing granulocytic cells in the liver. Arrows indicate clusters of maturing myeloid cells in the liver. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; WBC, peripheral white blood cell count; PB, peripheral blood; Sp, spleen; and Li, liver.)
Our data demonstrated that Stat5b was not absolutely required for TEL-PDGFRB–mediated MPD. However, we noted that 5 of 13 TPiGFP→Stat5b−/− mice appeared to escape rapidly fatal MPD induced by TEL-PDGFRB and were longer lived (Figure 4A). We confirmed equivalent GFP expression in TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− recipient bone marrow cells and splenocytes (data not shown), showing that transduced and transplanted cells persist in surviving TPiGFP→Stat5b−/− mice. We evaluated myeloproliferation in the longer-lived subset of TPiGFP→Stat5b−/− mice. We assessed these mice by gross and microscopic pathology, peripheral blood cell count, and flow cytometric analysis of spleen and bone marrow. However, we did not detect significant myeloproliferation in long-lived (> 100 days after transplantation) TPiGFP→Stat5b−/− mice by histopathology and detected only normal to mildly elevated peripheral blood cell counts in 2 of 2 mice analyzed (mean WBC, 11 × 109/L [11 000/μL]). However, we noted that leukocytosis at time of death due to disease was somewhat reduced in TPiGFP→Stat5b−/− mice (median WBC, 80 × 109/L [80 000/μL]) compared with TPiGFP→Stat5b+/+ littermate controls (median WBC, 213 × 109/L [213 000/μL]). While dispensable for TEL-PDGFRB–induced myeloproliferation, Stat5b was required for complete penetrance of rapidly fatal disease.
Stat5b can restore MPD when coexpressed with TEL-PDGFRB in Stat5a-deficient bone marrow cells
We found significant differences in disease latencies between TPiGFP→Stat5a−/− and TPiGFP→Stat5a+/+ mice. To address the possibility that Stat5a and Stat5b are distinct in their ability to transmit myeloproliferative signals in response to TEL-PDGFRB, we repeated murine bone marrow transduction/transplantation assays using retroviral constructs to coexpress TEL-PDGFRB with either Stat5a (TPiStat5a) or Stat5b (TPiStat5b) (Figure S1). We transduced unfractionated bone marrow cells harvested from Stat5a−/− or Stat5a+/− mice with Stat5 add-back constructs and evaluated mice for survival and evidence of disease. TEL-PDGFRB–mediated myeloproliferation was restored by either Stat5a or Stat5b expression in TPiStat5a→Stat5a−/−, TPiStat5a→Stat5a+/−, and TPiStat5b→Stat5a−/− mice, with all cohorts developing fatal MPD characterized by leukocytosis and extramedullary hematopoiesis (Table 1; Figure S3). In these add-back experiments, Stat5a and Stat5b appeared to be equivalent in their ability to restore MPD with similar disease latencies (TPiStat5a→Stat5a−/− 96 days versus TPiStat5b→Stat5a−/− 97 days, P = .793). Also, TPiStat5a→Stat5a+/− mice developed MPD (median WBC, 66 × 109/L [66 000/μL]; spleen weight, 520 mg) with the shortest disease latency (median survival, 28 days versus 31 days in TPiGFP→Stat5a+/+ mice, P = .732), demonstrating again the correlation of disease latency and Stat5a gene dosage. Histopathologic analysis of both TPiStat5a→Stat5a−/− and TPiStat5b→Stat5a−/− mice revealed red pulp expansion with myeloid hyperplasia in the spleen and small clusters of well-differentiated granulocytic cells accumulated in the sinusoids and perivascular regions of the liver (data not shown). These data suggest that both Stat5a and Stat5b can contribute to TEL-PDGFRB–mediated myeloproliferation when expressed at high levels in bone marrow mononuclear cells.
Stat5a and Stat5b both restore myeloproliferation when coexpressed with TEL-PDGFRB in Stat5a-deficient bone marrow cells
| Construct . | Phenotype . | Median survival, d . | P . | n . |
|---|---|---|---|---|
| TPiGFP into Stat5a+/+ | Fatal MPD: 6 | 31 ± 2.6 | — | 15 |
| TPiGFP into Stat5a+/− | Fatal MPD: 2; MPD: 5 | 145 ± 16.0 | < .001 | 19 |
| TPiGFP into Stat5a−/− | Fatal MPD: 1; MPD: 8 | 144 ± 4.1 | < .001 | 17 |
| TPiStat5a add-back into Stat5a+/− | Fatal MPD: 1 | 28 ± 9.0 | .732 | 6 |
| TPiStat5a add-back into Stat5a−/− | Fatal MPD: 6; MPD: 2 | 96 ± 35.5 | < .001 | 12 |
| TPiStat5b add-back into Stat5a−/− | MPD: 2 | 97 ± 7.5 | < .001 | 8 |
| Construct . | Phenotype . | Median survival, d . | P . | n . |
|---|---|---|---|---|
| TPiGFP into Stat5a+/+ | Fatal MPD: 6 | 31 ± 2.6 | — | 15 |
| TPiGFP into Stat5a+/− | Fatal MPD: 2; MPD: 5 | 145 ± 16.0 | < .001 | 19 |
| TPiGFP into Stat5a−/− | Fatal MPD: 1; MPD: 8 | 144 ± 4.1 | < .001 | 17 |
| TPiStat5a add-back into Stat5a+/− | Fatal MPD: 1 | 28 ± 9.0 | .732 | 6 |
| TPiStat5a add-back into Stat5a−/− | Fatal MPD: 6; MPD: 2 | 96 ± 35.5 | < .001 | 12 |
| TPiStat5b add-back into Stat5a−/− | MPD: 2 | 97 ± 7.5 | < .001 | 8 |
Median survival of mice in each cohort is shown with standard error. Disease phenotype at time of death was characterized as either fatal MPD (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) or MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) with number of analyzed mice indicated. Statistical significance in survival between TPiGFP → Stat5+/+ and each cohort is shown by P value (determined by Mantel-Cox log rank). Statistical significance in survival between cohorts is determined by log rank (Mantel-Cox) such that the difference in survival of TPiGFP→Stat5a−/− and TPiStat5b→Stat5a−/− has a P value of .01 and the difference in TPiStat5a→Stat5a−/− and TPiStat5b→Stat5a−/− cohort survival has a P value of .79. TPiStat5a → Stat5a+/− mice developed severe MPD (spleen weight, 520 mg [n = 1]; WBC, 66 × 109/L [66 000/μL] [n = 1]). Moderate MPD was detected in TPiStat5a → Stat5a−/− mice (spleen weight median, 800 ± 170 mg; range, 550-970 mg [n = 8]; WBC median, 82 ± 106 × 109/L [82 000 ± 106 000/μL]; range, 40-108 × 109/L [40 000-108 000/μL] [n = 7]) and TPiStat5b → Stat5a−/− mice (spleen weight median, 470 ± 184 mg; range, 340-600 mg [n = 2]; WBC median, 23 ± 10 × 109/L [23 000 ± 10 000/μL]; range, 16-30 × 109/L [16 000-30 000/μL] [n = 2]). TPiGFP indicates TEL-PDGFRB ires eGFP; TPiStat5a, TEL-PDGFRB ires mStat5a; TPiStat5b, TEL-PDGFRB ires Stat5b; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count; and —, not applicable.
Stat1 and Src family kinases are dispensable for TEL-PDGFRB–mediated myeloproliferative disease
We evaluated the contribution of the remaining candidate signaling molecules, the Src family kinases, and Stat1 to TEL-PDGFRB–mediated MPD using mice either triply deficient in myelomonocytic Src family kinases Lyn, Hck, and Fgr (Lyn−/−Hck−/−Fgr−/−) or deficient for Stat1 (Stat1−/−). We transduced unfractionated bone marrow mononuclear cells from Lyn−/−Hck−/−Fgr−/− or strain-matched wild-type mice with TPiGFP retroviral supernatants and transplanted them into lethally irradiated syngeneic recipient mice. We analyzed mice from each cohort for survival and myeloproliferative disease burden at time of death by peripheral blood cell count (WBC), as well as gross and microscopic pathology. All recipient mice (100%) that received TEL-PDGFRB–transduced Lyn−/−Hck−/−Fgr−/− bone marrow cells and either GFP (TPiGFP→Lyn−/−Hck−/−Fgr−/−) or c-Src (TPiSrc→Lyn−/−Hck−/−Fgr−/−) (Figure 5A) developed rapidly fatal MPD characterized by elevated peripheral blood counts, extramedullary hematopoiesis, and splenomegaly (Figure 5B and data not shown). Surprisingly, coexpression of c-Src with TEL-PDGFRB to restore Src kinase activity as an add-back control was associated with longer disease latency (median survival, 71 days versus TPiGFP→Lyn−/−Hck−/−Fgr−/− 39 days, P = .007). Add-back c-Src may have prolonged survival of TPiSrc→Lyn−/−Hck−/−Fgr−/− mice by competing with other relevant signaling molecules for binding to TEL/PDGFβR juxtamembrane tyrosines, or to other relevant signaling molecules. The development of rapidly fatal disease in TPiGFP→Lyn−/−Hck−/−Fgr−/− and TPiSrc→Lyn−/−Hck−/−Fgr−/− mice suggests that these Src kinases are dispensable for TEL-PDGFRB–mediated MPD.
TEL-PDGFRB–mediated myeloproliferative disease does not require Src family members Lyn, Hck, and Fgr.(A) Retroviral constructs encode TEL-PDGFRB followed by an internal ribosomal entry site to allow expression of a second cDNA, either GFP or c-Src. (B) Kaplan-Meier plot of recipient mouse survival when TEL-PDGFRB is coexpressed with either GFP (□) or c-Src (○) in Hck-, Lyn-, and Fgr-deficient cells. The 4 TPiGFP→Lyn−/−Hck−/−Fgr−/− mice had varying degrees of disease severity (spleen weight, 445 ± 273 mg; range, 270-590 mg; median WBC, 38 ± 66 × 109/L [38 000 ± 66 000/μL]; range, 14-156 × 109/L [14 000-156 000/μL]). MPD was detected in 3 of 3 TPiSrc→ Lyn−/−Hck−/−Fgr−/− mice analyzed for disease severity at time of death (spleen weight median, 590 ± 190 mg; range, 480-850 mg; median WBC, 86 ±839 × 109/L [86 000 ± 839 000/μL]; range, 14-1502 × 109/L [14 000-1 502 000 000/μL]). Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (ires indicates internal ribosomal entry site; GFP, green fluorescent protein; LTR, long terminal repeat; PNT, pointed domain; TK, tyrosine kinase; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)
TEL-PDGFRB–mediated myeloproliferative disease does not require Src family members Lyn, Hck, and Fgr.(A) Retroviral constructs encode TEL-PDGFRB followed by an internal ribosomal entry site to allow expression of a second cDNA, either GFP or c-Src. (B) Kaplan-Meier plot of recipient mouse survival when TEL-PDGFRB is coexpressed with either GFP (□) or c-Src (○) in Hck-, Lyn-, and Fgr-deficient cells. The 4 TPiGFP→Lyn−/−Hck−/−Fgr−/− mice had varying degrees of disease severity (spleen weight, 445 ± 273 mg; range, 270-590 mg; median WBC, 38 ± 66 × 109/L [38 000 ± 66 000/μL]; range, 14-156 × 109/L [14 000-156 000/μL]). MPD was detected in 3 of 3 TPiSrc→ Lyn−/−Hck−/−Fgr−/− mice analyzed for disease severity at time of death (spleen weight median, 590 ± 190 mg; range, 480-850 mg; median WBC, 86 ±839 × 109/L [86 000 ± 839 000/μL]; range, 14-1502 × 109/L [14 000-1 502 000 000/μL]). Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (ires indicates internal ribosomal entry site; GFP, green fluorescent protein; LTR, long terminal repeat; PNT, pointed domain; TK, tyrosine kinase; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)
To determine the role of Stat1 in the development of TEL-PDGFRB–induced MPD, we expressed TEL-PDGFRB in whole bone marrow harvested from Stat1-deficient (TPiGFP→Stat1−/−) or wild-type control (TPiGFP→Stat1+/+) mice. Again, all mice (100%) in both groups developed rapidly fatal MPD characterized by leukocytosis, extramedullary hematopoiesis, and splenomegaly (data not shown). TPiGFP→Stat1−/− mice developed MPD with a shorter latency than their TPiGFP→Stat1+/+ counterparts (45 versus 53 days, P = .013), consistent with the role of Stat1 as a tumor suppressor.30 The significantly shortened survival of TPiGFP→Stat1−/− mice indicates that Stat1 is not required for TEL-PDGFRB disease development. Taken together, these data demonstrate that, although TEL/PDGFβR induces both Stat5-dependent and -independent signaling, Stat5 is critical for TEL-PDGFRB–mediated transformation of primary hematopoietic cells.
Discussion
Using a genetic approach, we found that the signaling pathways activated by TEL/PDGFβR in cell culture do not contribute equally to transformation. The combined loss of both Stat5a and Stat5b (Stat5abnull/null) eliminated TEL-PDGFRB–mediated transformation as measured by growth of cytokine-independent colony formation in methylcellulose (Figure 1A). Confirming the presence of hematopoietic stem and progenitor populations among Stat5abnull/null fetal liver cells, we found that they formed colonies in the presence of cytokines and that immunophenotypically-defined stem and progenitor cells were moderately increased among Stat5abnull/null over Stat5ab+/+ fetal liver cells. Future comprehensive analysis of the repopulating capacity of Stat5abnull/null hematopoietic stem and progenitor cells will clarify the contribution of Stat5 to normal hematopoiesis.
Although Stat5a and Stat5b serve largely redundant functions in baseline hematopoiesis,15,17 they made independent contributions to TEL-PDGFRB–mediated transformation in methylcellulose such that the loss of even a single wild-type Stat5a or Stat5b allele diminished the transformative properties of TEL-PDGFRB, and homozygous inactivation of either gene further reduced transformation (Figure 1B-C). The survival of TPiGFP→Stat5a+/− mice (145 ± 16 days) was closer to that of TPiGFP→Stat5a−/− mice (144 ± 4.1 days) than to TPiGFP→Stat5a+/+ mice (31 ± 2.6 days). Although the normal breast development in Stat5a+/− mice15 argues against this, we were initially concerned that undocumented expression of a truncated Stat5a protein with dominant-negative activity might explain the dramatic protection afforded TPiGFP→Stat5a+/− mice. We examined TPiGFP→Stat5a+/−, TPiGFP→Stat5a−/−, and TPiGFP→Stat5a+/+ mice at early time points and confirmed that disease burden was tightly correlated with Stat5a gene dosage such that TEL-PDGFRB–mediated disease was sensitive to the loss of even a single wild-type Stat5a allele. We also addressed the concern that an underlying defect in Stat5a−/− cells might have impaired transduction and/or transplantation in our model, indirectly causing reduced sensitivity of Stat5a+/− and Stat5a−/− cells to TEL-PDGFRB–mediated transformation. We found that transduction efficiency was equivalent between Stat5a−/− and Stat5a+/+ cells and that Stat51*6→Stat5a−/− recipient mice develop a rapidly fatal disease (median survival, 19 ± 1 days), indicating that Stat5a-deficient cells are transduction and transplantation competent. These data are consistent with previous results.29
We were surprised to find that the 2 Stat5 genes appeared to make independent contributions to disease development in our system. In contrast to the significant decrease in myeloproliferation afforded by Stat5a loss, TPiGFP→Stat5b−/− mice rapidly developed fatal MPD. Unlike the uniform induction of disease in TPiGFP→Stat5b+/+ mice, however, 5 of 13 TPiGFP→Stat5b−/− mice failed to develop rapidly fatal MPD (Figure 4). We repeated bone marrow transduction/transplantation experiments using Stat5a and Stat5b knock-out alleles backcrossed into the balb/c background and found similar results. Therefore, the differences in disease latency and severity we found between Stat5a and Stat5b singly deficient mice cannot be explained by differences in background strain. We performed a series of add-back experiments using Stat5a and Stat5b in a transduction/transplantation assay to confirm and compare the roles these factors play in MPD development. Stat5a add-back restored disease development in TPiStat5a→Stat5a−/− mice, and Stat5b also restored disease development when expressed in TPiStat5b→Stat5a−/− mice (Table 1). Taken together, these data suggest that our observation that the Stat5a and Stat5b genes play unequal roles in TEL-PDGFRB–induced MPD may be explained by exquisite sensitivity to subtle differences in Stat5a and Stat5b protein levels in different hematopoietic progenitor cells. Flow cytometric analysis of Stat5 phosphorylation will allow us to quantitatively evaluate Stat5 activation in different subsets of hematopoietic cells including stem and progenitor populations, although that work is beyond the scope of this study.
Although no signs of disease were found in TPiGFP→Stat5a−/− mice at early time points, these animals eventually succumbed to a “mild MPD” (splenomegaly with increased splenic myeloproliferation without significant peripheral leukocytosis) following a long latency (median survival, 144 days; Figure 3). The mechanism for the development of long-latency disease seen in TPiGFP→Stat5a−/− mice might be compensatory up-regulation of Stat5b expression or to the accumulation of additional genetic events that cooperate with TEL/PDGFβR independently of Stat5. Using Stat5abnull/null cells in a transduction and transplantation model of disease would allow us to discriminate between the consequences of Stat5-dependent and Stat5-independent signaling in TEL-PDGFRB–mediated disease. However, vector control–transduced Stat5abnull/null fetal liver cells were undetectable by 9 weeks after transplantation in recipient mice (data not shown), prior to the development of TEL-PDGFRB disease when donor fetal liver cells are used. Future transduction and transplantation studies will require alternative strategies such as conditional deletion of the Stat5 locus in recipient mice.
While Src family kinases are activated in native PDGFβR signaling9 and are important for S-phase induction in fibroblasts,31 our data suggest that they are dispensable for induction of MPD by TEL/PDGFβR. Our findings are consistent with a previous report that Lyn, Hck, and Fgr are not required for myeloid disease induced by BCR/ABL in mice.32 It is possible that Src activation is not functionally relevant in the context of TEL/PDGFβR due to different subcellular localization of the fusion protein and native PDGFβR. However, coexpression of c-Src with TEL-PDGFRB (TPiSrc→Lyn−/−Hck−/−Fgr−/−) was associated with longer disease latency than expression of TEL-PDGFRB alone (TPiGFP→Lyn−/−Hck−/−Fgr−/−). Add-back c-Src may have competed with Stat5 at PDGFβR tyrosines 579/581, or c-Src kinase activity may have directly suppressed myeloproliferation in TPiSrc→Lyn−/−Hck−/−Fgr−/− mice.33 We considered the possibility that MPD detected in Lyn−/−Hck−/−Fgr−/− mice was not TEL-PDGFRB dependent, as Lyn and Fgr are known to have inhibitory functions,32 but Lyn−/−Hck−/−Fgr−/− mice that did not undergo transplantation did not develop fatal myeloproliferation in the 3-month time frame of our experiments. While our data demonstrate that Lyn, Hck, and Fgr are not required for TEL-PDGFRB–mediated disease, we cannot exclude the possibility that a redundant Src family kinase was expressed in these cells and contributed in some way to disease development.
TEL/PDGFβR induces the expression of Stat1 and multiple Stat1 target genes in Ba/F3 cells (Wilbanks et al7 ; and M.H.T., Golub, and Gilliland, unpublished data, May 2002). However, we found that Stat1 is dispensable for TEL-PDGFRB–mediated disease. These data are consistent with the role of Stat1 as an antiproliferative, proapoptotic protein30 and earlier findings that TEL/JAK2 does not require Stat1 to induce MPD or lymphoblastic lymphoma.28
Our data show that Stat5 plays a central role in TEL-PDGFRB–driven myeloproliferation. The JAK-STAT pathway is commonly activated in leukemia,34,35 and activating JAK2 mutations have recently been discovered in association with human myeloproliferative diseases polycythemia vera (PV), essential thrombocytopenia (ET), and myeloid metaplasia and myelofibrosis (MMF) (reviewed by Tefferi and Gilliland36 ). Stat5 is required for the development of TEL-JAK2–induced myeloproliferative and lymphoproliferative disease in mice28 as well as BCR-ABL–mediated transformation in methylcellulose colony assays.20 Our data contribute to a model that suggests that Stat5 may be the key mediator of myeloproliferation in response to a diverse set of oncogenic tyrosine kinases.
Authorship
Contribution: J.A.C. performed research (major part), designed research, and wrote the paper; Z.X. and J.O. performed research; F.K. analyzed data; A.C. performed research; L.H. provided valuable reagents; M.H.T. designed research, performed research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael H. Tomasson, Division of Oncology, Department of Medicine, 660 S Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail: tomasson@im.wustl.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by NIH T32 HL07088-31A1 (J.A.C.) and NIH PO1 CA101937 (M.H.T.). This work was supported in part by the intramural program of NIDDK/NIH (L.H.).
We are grateful to C. Lowell, R. Schreiber, and H. Davey for generously providing mice harboring targeted deletion of Src and Stat family members; G. Gilliland for magnanimous support and backcross genotyping; S. Courtneidge and W. Leonard for sharing valuable reagents; T. Graubert, J. Hsieh, T. Ley, D. Link, J. Milbrandt, J. Weber, and K. Weilbaecher for valuable discussions; and W. Eades and J. Hughes for flow cytometric progenitor analysis.

![Figure 3. Stat5a gene dosage mediates the latency of TEL-PDGFRB–induced myeloproliferation. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring homozygous or heterozygous inactivation of Stat5a. TPiGFP→Stat5a+/+ mice (circles) developed severe MPD (spleen weight median, 790 ± 90 mg; range, 620-910 mg; n = 6). TPiGFP→Stat5a+/− mice (▵) developed moderate or severe MPD (spleen weight median, 770 ± 300 mg; range, 350-1120 mg; n = 7). TPiGFP→Stat5a−/− mice (squares) developed moderate MPD with splenomegaly (spleen weight, 710 ± 550 mg; range, 350-2070 mg; n = 8) and leukocytosis. Black shapes indicate death due to severe (fatal) MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (B) Median peripheral blood cell count at time of death due to disease. Error bars represent standard deviation. TEL-PDGFRB induced severe leukocytosis in TPiGFP→Stat5a+/+ mice (359 ± 225 × 109/L [359 000 ± 225 000/μL]; range, 60-737 × 109/L [60 000-737 000/μL]; n = 6) and moderate leukocytosis in TPiGFP→Stat5a+/− mice (28 ± 15 × 109/L [28 000 ± 15 000/μL]; range, 12-131 × 109/L [12 000-131 000/μL]; n = 7) as well as TPiGFP→Stat5a−/− mice (29 ± 27 × 109/L [29 000 ± 27 000/μL];range, 10-94 × 109/L [10 000-94 000/μL]; n = 8). (C) Three mice from each cohort were analyzed 28 days after transplantation. Median of peripheral blood counts is shown. Error bars represent standard deviation. Peripheral blood counts of GFP→Stat5a−/− control mice are also shown. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-07-036335/4/m_zh80090700460003.jpeg?Expires=1769096090&Signature=ZtIMM80TWWKuCEtG9avuvx636QOifecItENcX0H-S14hOEKt4OwF86j516Klep-QVMy90Cved1bU1bFkveKxUm6SGpoN1sT2Pl8DQpGD-v~~Mx8N6G7Sz09uMNnj~sHzo5RuaVttglgIxpyYhQl-5WuUrRhXnNxGgQNM3rOXYMe8zO6xUXItO8l~moXfVI6~OMXrqb2lzXoWnud3hzlwBA5btSBlRk8srYIxMSKdYVcg9LXgKIgyJLaUgRosE1i-gHNYF~9uwN3v1wmZ2wzlkLYcy1CNPMPE3TpoNRVLP5VQ~CTyE3Ei8~3rMSNr5xCeoQbCuIKLLzEeRmMya-GZkw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Severity and penetrance of TEL-PDGFRB–induced myeloproliferation are reduced in the absence of Stat5b. (A) Kaplan-Meier plot shows survival of recipient mice when TEL-PDGFRB is expressed in bone marrow cells harboring targeted deletion of Stat5b (TPiGFP→Stat5b−/−, squares) or wild-type control bone marrow cells (TPiGFP→Stat5b+/+, ○). TPiGFP→Stat5b+/+ mice developed severe MPD (spleen weight median, 890 ± 274 mg; range, 670-1460 mg [n = 6]; WBC median, 139 ± 87 × 109/L [139 000 ± 87 000/μL]; range, 13-237 × 109/L [13 000-237 000/μL] [n = 6]; median survival ± standard error, 33 ± 2.9 days). Severe MPD was detected in 5 of 7 TPiGFP→Stat5b−/− mice at time of death due to disease (spleen weight median, 1220 ± 401 mg; range, 420-1440 mg [n = 7]; WBC median, 103 ±151 × 109/L [103 000 ± 151 000/μL]; range, 6-446 × 109/L [6000-446 000/μL] [n = 7]; median survival ± standard error, 57 ± 60.2 days). Experiment was performed twice, once in the original outbred strain with wild-type littermate control and again with the targeted Stat5b allele backcrossed to balb/c resulting in similar disease latency and severity. Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. GFP was detected in bone marrow, spleen, and peripheral blood of TPiGFP→Stat5b−/− (37% ± 4% [n = 7]; 50% ± 15% [n = 7]; and 58% ± 32% [n = 7], respectively) and TPiGFP→Stat5b+/+ (44% [n = 1]; 50% ± 15% [n = 3]; and 58% ± 32% [n = 3], respectively) recipient mice (data not shown). (B) Flow cytometric detection of mature myeloid (Gr-1+Mac-1+) cells in the spleens of both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− mice 33 days after transplantation. (C) Microscopic images of representative mice at time of death (GFP→Stat5b+/+, 151 days after transplantation; TPiGFP→Stat5b+/+, 29 days after transplantation; and TPiGFP→Stat5b−/−, 29 days after transplantation). Panels show Wright-Giemsa–stained smears of peripheral blood (Ci-iii; 100×), and H&E–stained histologic sections of spleen (Civ-vi; 20×) and liver (Cvii-ix; 40×). These illustrate a normal morphology of peripheral blood, spleen, and liver in the GFP→Stat5b+/+ group, whereas both TPiGFP→Stat5b+/+ and TPiGFP→Stat5b−/− show marked leukocytosis of maturing neutrophils in the peripheral blood, pronounced red pulp expansion by myeloid hyperplasia composed of predominantly maturing forms in the spleen, and sinusoidal and perivascular infiltrates of maturing granulocytic cells in the liver. Arrows indicate clusters of maturing myeloid cells in the liver. (TPiGFP indicates TEL-PDGFRB ires eGFP; MPD, myeloproliferative disease; WBC, peripheral white blood cell count; PB, peripheral blood; Sp, spleen; and Li, liver.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-07-036335/4/m_zh80090700460004.jpeg?Expires=1769096090&Signature=JkGWW3WfhY0JpJc-8cc0qjddwtwNUG~NNUSpvkGdhOrnOCyIog-I~9TfNmgSXlMb3ZDz51sXt4~hZQqO6AJE~GS8CrmsGhq35WwQxR3prshD3TZqLHv2NWC3y4sQ6H1PQdg5tESt91ilM4PbbBjL~EJK~sz5e8M9nYYGkdg1mhVXuFTFytYlOn6bIno3VHfYRHUqWkyhBB13U08xZY4e7laz~cGR2xwu9bWcVZC3R-pGnEy5AbVE4noG1Gz8gAGH1MaWReOqMgW9As9uhwRW8FKtwQ7m6f--4QIGWr9RLhMjM-kjJhV5HJnTpbHxtwdHfqf2Hw6eDEWPzHkqhjnlzg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. TEL-PDGFRB–mediated myeloproliferative disease does not require Src family members Lyn, Hck, and Fgr.(A) Retroviral constructs encode TEL-PDGFRB followed by an internal ribosomal entry site to allow expression of a second cDNA, either GFP or c-Src. (B) Kaplan-Meier plot of recipient mouse survival when TEL-PDGFRB is coexpressed with either GFP (□) or c-Src (○) in Hck-, Lyn-, and Fgr-deficient cells. The 4 TPiGFP→Lyn−/−Hck−/−Fgr−/− mice had varying degrees of disease severity (spleen weight, 445 ± 273 mg; range, 270-590 mg; median WBC, 38 ± 66 × 109/L [38 000 ± 66 000/μL]; range, 14-156 × 109/L [14 000-156 000/μL]). MPD was detected in 3 of 3 TPiSrc→ Lyn−/−Hck−/−Fgr−/− mice analyzed for disease severity at time of death (spleen weight median, 590 ± 190 mg; range, 480-850 mg; median WBC, 86 ±839 × 109/L [86 000 ± 839 000/μL]; range, 14-1502 × 109/L [14 000-1 502 000 000/μL]). Black shapes indicate death due to severe MPD marked by leukocytosis and splenomegaly (WBC, > 50 × 109/L [50 000/μL] and spleen weight, > 450 mg) at time of death due to disease. Shaded shapes represent moderate MPD (WBC, > 15 × 109/L [15 000/μL] and/or spleen weight, > 450 mg) at time of death due to disease. Open shapes indicate animal found dead. (ires indicates internal ribosomal entry site; GFP, green fluorescent protein; LTR, long terminal repeat; PNT, pointed domain; TK, tyrosine kinase; MPD, myeloproliferative disease; and WBC, peripheral white blood cell count.)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-07-036335/4/m_zh80090700460005.jpeg?Expires=1769096090&Signature=oCIETktrajoS3Apu840qkzrUGVFxzrumRHL4ozeX4NYYBZToYd-l~3htGfQafUgZD2lsP36Vfj2ErWtJIf2FAgwoNGOLJiT1R1Y~zX3NKBiEhQ-sj40y0vYuxlO9S3GWaNO3XicM9wl70n-yyW6isu7-W5sMY4BuFKtbCfevpc-4wwFXOh9~Rhr1kMHuACvT4NO8pCaoVx~7FSiVSTzmuc7FpE5P~aERS22Zc5UpVa3Sp3PyA5L7nocpXjt8v8A1FMmLGPx46no0ouupIp-Gn338DGtmcq6lyQRfm9~S4GZlOFKI0XdcftaNPXIwxRj8g8Gpkr~rCQxtb56Z~zA2CA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal