Abstract
The 8;21 chromosomal translocation occurs in 15% to 40% of patients with the FAB M2 subtype of acute myeloid leukemia (AML). This chromosomal abnormality fuses part of the AML1/RUNX1 gene to the ETO/MTG8 gene and generates the AML1-ETO protein. We previously identified a C-terminal truncated AML1-ETO protein (AEtr) in a mouse leukemia model. AEtr is almost identical to the AML1-ETO exon 9a isoform expressed in leukemia patients. Here, we describe a novel function of AEtr in the development of aneuploidy through spindle checkpoint attenuation. AEtr cells had a reduced mitotic index following nocodazole treatment, suggesting a failure in a subset of cells to arrest in mitosis with a functional spindle checkpoint. Additionally, primary leukemia cells and cell lines expressing AEtr were aneuploid. Moreover, AEtr cells had reduced levels of several spindle checkpoint proteins including BubR1 and securin following treatment with the spindle poison nocodazole. These results suggest that inactivation of the spindle checkpoint may contribute to the development of aneuploidy described in t(8;21) leukemia patients.
Introduction
The 8;21 chromosomal translocation occurs in approximately 15% to 40% of patients with the FAB M2 subtype of acute myeloid leukemia (AML) and joins part of the genes encoding AML1/RUNX1 and ETO/MTG8.1 As a result of this genetic alteration, AML1-ETO fusion transcripts are generated. Introduction of full-length AML1-ETO encoding a 752–amino acid protein into the murine AML1 locus results in embryonic lethality and disrupts definitive hematopoiesis, similar to deletion of the murine AML1 gene or the DNA-binding partner of AML1, CBFβ, suggesting that AML1-ETO disrupts AML1 function.2–5
Murine models that mimic human t(8;21) leukemia have been difficult to establish because expression of full-length AML1-ETO by itself is not sufficient to induce leukemia development. A variety of approaches to express AML1-ETO in mice were taken, and the conclusion drawn from these studies is that additional mutations cooperate with AML1-ETO to promote leukemogenesis.6–11 We previously reported that a truncated AML1-ETO protein (AEtr) lacking 200 amino acids at the C-terminus of full-length AML1-ETO is sufficient to promote leukemia, which indicates that one type of additional mutation to promote AML1-ETO–associated leukemogenesis is to inhibit C-terminal functions of the protein.12 AEtr is almost identical to the recently identified AML1-ETO exon 9a splice form that is widely expressed in t(8;21) leukemia patients and is sufficient to induce leukemia in mice.13 Given that t(8;21) patients express an AML1-ETO isoform with a C-terminal deletion, it is important to understand how this protein contributes to leukemia development.
A feature of t(8;21) leukemia is the loss of one sex chromosome in approximately half of all patients that occurs at a much higher frequency compared with AML patients with other chromosomal translocations.14–21 Autosomal changes, including trisomy 8, trisomy 4, and deletion of chromosome 7, have also been detected in approximately 13% to 16% of t(8;21) patients and in 2 t(8;21) patient–derived cell lines at the time of establishment.18,21–24 Although it was suggested that centrosome abnormalities may contribute to aneuploidy in AML patients, t(8;21) patients had fewer centrosomal abnormalities compared with patients with other translocations.25 These data suggest that another mechanism regulating chromosome segregation leads to the initial loss and gain of chromosomes in t(8;21) leukemia patients.
Aneuploidy can result from chromosome missegregation during mitosis and is a feature of many cancers. A major mechanism to prevent aneuploidy is the spindle checkpoint, which inhibits the anaphase-promoting complex (APC). APC is an E3 ubiquitin ligase that targets cell-cycle–regulatory proteins for proteasomal degradation and promotes cell-cycle progression.26 One APC-activating subunit during mitosis is cdc20, which is inhibited upon association with either Mad2 or BubR1 until alignment of chromosomes in metaphase.26 Loss of either Mad2 or BubR1 in cell lines or in mice results in aneuploidy due to chromosome missegregation, underscoring the importance of this checkpoint.27–30 APCcdc20 activation results in the degradation of the anaphase inhibitor securin.31,32 Securin prevents anaphase by binding to the separase protease, which is responsible for loss of sister chromatid cohesion through cleavage of the Scc1/hRad21 cohesin subunit.33 Although the spindle checkpoint is normally activated in cells during every mitosis, it can be chemically induced by several microtubule-altering drugs including nocodazole, colchicine, and paclitaxel, which inhibit chromosome movements prior to metaphase.34
We report in this study that expression of the leukemogenic AEtr protein promotes mitotic progression in the presence of a spindle checkpoint–activating agent, nocodazole, indicating that this checkpoint is attenuated. AEtr-expressing cells contained reduced levels of the BubR1 checkpoint protein and the anaphase inhibitor securin following microtubule disruption with nocodazole. As predicted for cells that have lost spindle checkpoint activity, chromosomal analyses in cell lines as well as in primary leukemia cells reflected that AEtr expression results in aneuploidy. These results demonstrate that attenuation of the spindle checkpoint may be a novel mechanism initiating aneuploidy associated with t(8;21) leukemia.
Materials and methods
Cell culture
Kasumi-1, SKNO-1, and K562 cells were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Omega, Tarzana, CA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). 293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin.
Primary bone marrow cells were isolated from femurs of MF-1 mice as described.12 Cells were cultured in DMEM supplemented with 10% FBS, 2% SCF conditional media prepared from BHK/MKL cells stably expressing a cDNA encoding the secretory form of mouse SCF (provided by Dr Schickwannn Tsai), 2% IL-3 conditional media prepared from X63Ag8-653 myeloma cells, and 1% WEHI-3B conditional media.35
Nocodazole (M1404; Sigma, St Louis, MO) was dissolved in DMSO and was used at either 0.1 μg/mL or 1 μg/mL as indicated; colchicine (C9754; Sigma) was dissolved in sterile dH2O and used at 0.1 μg/mL; paclitaxel (193532; MP Biomedicals, Solon, OH) was dissolved in DMSO and used at 100-nM final concentration.
Retrovirus infection
Approximately 5 × 106 293T cells in 100-mm dishes was cotransfected with 15 μg MSCV-IRESpuro vector, MSCV-HA-AEtr-IRESpuro, or MSCV-HA-AEtr R174Q-IRESpuro along with 10 μg amphotrophic packaging vector pCL10A1 using calcium phosphate precipitation method. Media containing retrovirus were collected 48 hours after transfection and retroviral supernatant was added to 5 × 105 K562 cells/well of a 6-well plate with 16 μg/mL polybrene (Sigma). For infection, cells were centrifuged at 1400g for 2.5 hours at 32°C. Cells were cultured in 4 μg/mL puromycin 48 hours following retroviral transduction and resistant cells were pooled.
For construction of the MSCV-HA-AEtr-IRESpuro plasmid, an XbaI fragment of MSCV-HA-AEtr-IRES-EGFP was blunted with klenow and inserted into the HpaI site of MSCV-IRESpuro. MSCV-IRESpuro was constructed by ligating a HindIII fragment from MSCV-GFP-IRES-PURO containing the puromycin resistance gene (1.2 kb) to MSCV-IRES-EGFP digested with HindIII. Thus, the EGFP coding sequence is replaced with the puromycin resistance coding sequence. MSCV-HA-AEtr R174Q-IRESpuro was constructed by replacing a SpeI digestion fragment of MSCV-HA-AEtr-IRESpuro with a SpeI fragment from MSCV-HA-AE exon 9a R174Q-IRESEGFP. MSCV-HA-AE exon 9a R174Q-IRES EGFP was generated by insertion of a BglII and BamHI fragment from pCDNA6-HA-AML1-ETO R174Q. The R174Q mutation was cloned into pCDNA6-HA-AML1-ETO by polymerase chain reaction (PCR) mutagenesis with primers 5′-GATGGGCCCCAAGAACCTCGA-3′ and 5′-CTGGTGCCATTAGTTAACGTTG-3′, and the product was digested with ApaI/HpaI and cloned into pCMV5-AML1-ETO ApaI/HpaI. This construct was subsequently digested with HindIII/HpaI and cloned by a 3-way ligation of HindIII/HindIII and HpaI/HindIII fragments of pCDNA6-HA-AML1-ETO. The point mutation in MSCV-HA-AEtrR174Q-IRESpuro was confirmed by sequencing.
For primary bone marrow infections, 293T cells were transfected with 15 μg MSCV-IRES-EGFP or MSCV-AEtr-IRES-EGFP and 10 μg MCV-ecotropic packaging vector.36 Virus was collected 48 hours after transfection and approximately 1 × 106 bone marrow cells/well of a 6-well plate was infected by spinoculation with 8 μg/mL polybrene. Cells were cultured overnight and 0.1 μg/mL nocodazole was added the following day for 20 hours. Cells were stained with 1 μg/mL Hoechst 33342 (Cell Signaling, Beverly, MA) for 45 minutes and analyzed by flow cytometry with a BD LSR II flow cytometer (BD Biosciences, San Jose, CA). DNA histograms were acquired by UV excitation of Hoechst 33342 followed by doublet discrimination and gating of EGFP-positive cells.
Cell cycle analysis
Cells were fixed in 70% ethanol (in PBS) for 30 minutes and were washed with PBS. Cells were resuspended in 50 μg/mL propidium iodide (Sigma) and RNase A. DNA content was measured by flow cytometry (BD Biosciences, San Jose, CA) to detect propidium iodide fluorescence. DNA histograms were analyzed by ModFiT LT software (Verity, Topsham, ME) to accurately measure cell-cycle phases after doublet discrimination of the propidium iodide fluorescent signal.
Please see Document S1 for additional materials and methods (available on the Blood website; see the Supplemental Materials link at the top of the online article) for BrdU and phospho–histone H3 staining, protein immunoblotting, real-time PCR, Northern blot, mitotic chromosome analysis, and spectral karyotyping (SKY).
Results
A t(8;21) fusion protein, AEtr, reduces mitotic arrest in nocodazole
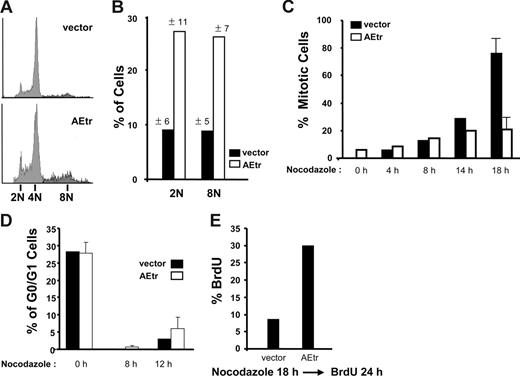
We previously reported that the AEtr protein promotes leukemia development in mice.12 In contrast to the growth-inhibitory effect reported for full-length AML1-ETO, expression of AEtr in several hematopoietic cell lines does not cause cell-cycle arrest in G0/G1 phase.12,37,38 Since cell proliferation is an important step during leukemia development, we wanted to better characterize the cell-cycle effects of AEtr expression in a hematopoietic cell line. Therefore, we used retroviral transduction to stably express AEtr or vector control in K562 hematopoietic cells. In our initial study, asynchronous K562 cells expressing AEtr proliferated similarly to vector control cells.12 Therefore in this study, we attempted to study the cell-cycle effects of AEtr in synchronous populations. Synchronization experiments were performed with nocodazole, a microtubule-disrupting agent that arrests cells in mitosis by activating the spindle checkpoint. Following nocodazole treatment, most vector control cells (Figure 1A) or parental K562 cells (data not shown) arrested in mitosis with 4N DNA content. Of interest, compared with control cells, there were more AEtr-expressing cells that did not arrest in mitosis with 4N DNA following nocodazole treatment and contained 2N or 8N DNA contents (Figure 1A). This result was highly reproducible in multiple independent AEtr-expressing pools and was statistically significant (Figure 1B, P = .027 [2N], P = .025 [8N]). Additionally, expression of the AML1-ETO exon 9a isoform, which is almost identical to AEtr, in K562 cells also resulted in fewer mitotic cells following nocodazole treatment (data not shown).
AEtr cells have reduced mitotic index in nocodazole. (A) K562 cells stably expressing AEtr or vector control cells were treated with nocodazole for 18 hours. Propidium iodide (PI)–stained cells were detected by flow cytometry. Representative PI histogram of vector control or AEtr cells following nocodazole treatment for 18 hours. (B) Graph indicates the percentages of vector control and AEtr cells that contained either 2N or 8N DNA after nocodazole treatment as determined by PI staining. The averages for 3 to 5 independent experiments with independent pools are indicated. The standard deviation is indicated above each bar. (C) Mitotic index of control or AEtr cells following nocodazole addition. Phospho–histone H3 staining was performed 0, 4, 8, 14, or 18 hours after nocodazole addition. The error bars in the 18-hour time point indicate the average of 3 independent experiments. (D) The graph indicates the percentage of cells in G0/G1 phase of the cell cycle determined by ModFiT analysis of histograms of PI-stained cells following nocodazole treatment for 0, 8, or 12 hours. (E) K562 vector or AEtr-expressing cells were treated with nocodazole for 18 hours followed by addition of 10 μM BrdU for 24 hours. Cells were fixed and stained with anti–BrdU-FITC antibody and analyzed by flow cytometry to determine the percentage of S phase cells. The average BrdU incorporation percentage is shown for 3 independent K562 vector control or AEtr-expressing pools. Error bars show standard deviation (SD) of 2 different AEtr pools.
AEtr cells have reduced mitotic index in nocodazole. (A) K562 cells stably expressing AEtr or vector control cells were treated with nocodazole for 18 hours. Propidium iodide (PI)–stained cells were detected by flow cytometry. Representative PI histogram of vector control or AEtr cells following nocodazole treatment for 18 hours. (B) Graph indicates the percentages of vector control and AEtr cells that contained either 2N or 8N DNA after nocodazole treatment as determined by PI staining. The averages for 3 to 5 independent experiments with independent pools are indicated. The standard deviation is indicated above each bar. (C) Mitotic index of control or AEtr cells following nocodazole addition. Phospho–histone H3 staining was performed 0, 4, 8, 14, or 18 hours after nocodazole addition. The error bars in the 18-hour time point indicate the average of 3 independent experiments. (D) The graph indicates the percentage of cells in G0/G1 phase of the cell cycle determined by ModFiT analysis of histograms of PI-stained cells following nocodazole treatment for 0, 8, or 12 hours. (E) K562 vector or AEtr-expressing cells were treated with nocodazole for 18 hours followed by addition of 10 μM BrdU for 24 hours. Cells were fixed and stained with anti–BrdU-FITC antibody and analyzed by flow cytometry to determine the percentage of S phase cells. The average BrdU incorporation percentage is shown for 3 independent K562 vector control or AEtr-expressing pools. Error bars show standard deviation (SD) of 2 different AEtr pools.
Prolonged mitotic arrest in the presence of nocodazole is an indication of a functional mitotic spindle checkpoint; conversely, a lower mitotic index is suggestive of an attenuated checkpoint. Therefore, a time course experiment following nocodazole treatment was performed to determine the mitotic index of cells by staining for phospho–histone H3, a mitosis marker. By 18 hours after nocodazole treatment, approximately 80% of control cells stained positive for phospho–histone H3 (Figure 1C). In contrast, only 20% to 30% of AEtr-expressing cells were phospho–histone H3 positive and contained 4N DNA content determined by PI staining.
Since a reduction of mitotic AEtr cells could be due to slower cell-cycle progression of cells following nocodazole treatment, rather than cells inappropriately completing mitosis, the effect of nocodazole on cell-cycle progression of control or AEtr cells was determined. Control and AEtr-expressing cells were plated at the same density 24 hours prior to nocodazole addition. At the time of nocodazole treatment, both cell types had similar percentages of cells in G0/G1 phase (Figure 1D). Cells were monitored at 8 and 12 hours after nocodazole addition by PI staining. AEtr-expressing cells proliferated similarly to control cells, and both cell lines exited G0/G1 by 8 hours after nocodazole addition (Figure 1D). These data indicate that the lower mitotic index observed following nocodazole treatment of AEtr-expressing cells was not due to cell-cycle arrest in G0/G1 phase.
Microtubule disruption with nocodazole or colchicine can trigger apoptosis of cells with a weakened mitotic checkpoint.39 Since apoptosis could also account for the reduced mitotic index, cell death was measured in both control and AEtr cells before and after nocodazole treatment. Annexin V staining revealed that there was no significant increase in apoptosis following nocodazole treatment in either vector or AEtr cells (2%-7% apoptosis, P = .43; data not shown), suggesting that cells were viable.
Continuous proliferation of cells in the presence of spindle poisons is another indication of an attenuated spindle checkpoint. Therefore, cells were monitored for proliferation by bromodeoxyuridine (BrdU) incorporation to measure the percentage of cells that continued into the next S phase following nocodazole treatment. Control or AEtr cells were treated with or without nocodazole for 18 hours followed by labeling with BrdU for 24 hours. Cells cultured without nocodazole had similar percentages of BrdU-positive cells (data not shown). Following nocodazole treatment, there was a 3-fold increase in percentage of BrdU-positive AEtr cells compared with control cells (8.6% in control cells and 30% in AEtr cells, Figure 1E). Together, these results indicate that nocodazole did not fully arrest AEtr cells in mitosis and that the cells continued to proliferate into the subsequent cell cycle.
Checkpoint attenuation in other spindle poisons and in primary cells
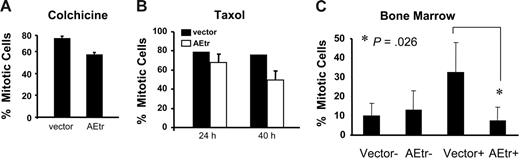
To determine if the lower mitotic index of AEtr cells was restricted to nocodazole treatment, cell-cycle profiles were analyzed following treatment with colchicine, another microtubule-destabilizing compound, or with paclitaxel, a microtubule stabilizer. Similar to nocodazole treatment, fewer AEtr cells arrested in mitosis when cultured in colchicine for 18 hours (Figure 2A). Following treatment of cells with paclitaxel, AEtr cells showed a reduction in mitotic index 40 hours after paclitaxel treatment but not at 24 hours after treatment compared with control (Figure 2B). These data indicate that AEtr cells have an attenuated spindle checkpoint reflected by the reduced mitotic arrest in response to several antimitotic drugs.
AEtr reduces mitotic arrest in several spindle poisons and in primary cells. (A) K562 cells stably expressing AEtr or vector control cells were treated with 0.1 μg/mL colchicine for 18 hours. PI-stained cells were detected by flow cytometry, and the percentage of mitotic cells is indicated in the bar graph for 2 independent experiments. (B) K562 cells stably expressing AEtr or vector control were treated with 100 nM paclitaxel for 24 and 40 hours. PI-stained cells were detected by flow cytometry, and the percentage of mitotic cells is indicated in the bar graph for 2 independent experiments. (C) Primary bone marrow cells transduced with control EGFP virus or virus encoding AEtr and EGFP were treated with (+) and without (−) 0.1 μg/mL nocodazole for 20 hours. Mitotic index was measured after Hoechst 33342 staining of cells to gate the cells in G2/M phase of the cell cycle. The bar graph shows the flow cytometry data averaged for 3 independent experiments. The asterisk indicates a significant difference between AEtr and control cells after nocodazole treatment. Error bars show SD of the mean from 2 or 3 independent experiments as indicated in each panel.
AEtr reduces mitotic arrest in several spindle poisons and in primary cells. (A) K562 cells stably expressing AEtr or vector control cells were treated with 0.1 μg/mL colchicine for 18 hours. PI-stained cells were detected by flow cytometry, and the percentage of mitotic cells is indicated in the bar graph for 2 independent experiments. (B) K562 cells stably expressing AEtr or vector control were treated with 100 nM paclitaxel for 24 and 40 hours. PI-stained cells were detected by flow cytometry, and the percentage of mitotic cells is indicated in the bar graph for 2 independent experiments. (C) Primary bone marrow cells transduced with control EGFP virus or virus encoding AEtr and EGFP were treated with (+) and without (−) 0.1 μg/mL nocodazole for 20 hours. Mitotic index was measured after Hoechst 33342 staining of cells to gate the cells in G2/M phase of the cell cycle. The bar graph shows the flow cytometry data averaged for 3 independent experiments. The asterisk indicates a significant difference between AEtr and control cells after nocodazole treatment. Error bars show SD of the mean from 2 or 3 independent experiments as indicated in each panel.
To examine whether mitotic progression in nocodazole also occurs in primary cells, AEtr was expressed in primary bone marrow cells. Primary bone marrow cells were infected with control retrovirus encoding EGFP alone or virus encoding AEtr and EGFP. Two days after infection, cells were treated with nocodazole for 20 hours. Cell-cycle analysis revealed that AEtr expression in these cells also resulted in a lower mitotic index following nocodazole treatment (Figure 2C). These data were statistically significant (P = .026) and indicate that AEtr disrupts the spindle checkpoint in primary hematopoietic cells.
To determine if the DNA-binding domain of AEtr is required for the continued proliferation in nocodazole, a mutation in the DNA-binding domain of AEtr was generated. The R174Q mutation in the DNA-binding domain of the AML1/RUNX1 gene was originally identified in a patient with familial platelet disorder.40 Subsequently, it was shown that this mutation prevents DNA binding but not nuclear localization and association with CBFβ.41 Therefore, the AEtr R174Q protein was stably expressed in K562 cells (Figure S1A). Nocodazole or paclitaxel treatment of these cells or control cells was performed for either 24 or 40 hours. Expression of AEtr R174Q was able to reduce the mitotic index in response to nocodazole and paclitaxel 40 hours after treatment (Figure S1B). To determine if the reduced mitotic index was due to cells exiting mitosis and continuing through the cell cycle to gain 2N or 8N DNA, multiple AEtr R174Q pools and control pools were analyzed. These data revealed that the AEtr R174Q mutant still resulted in 2N DNA content after nocodazole treatment but not in 8N DNA content (Figure S1C), suggesting that DNA binding of AEtr is required for the acquisition of 8N DNA content in nocodazole.
Abnormal mitosis and aneuploidy in AEtr cells
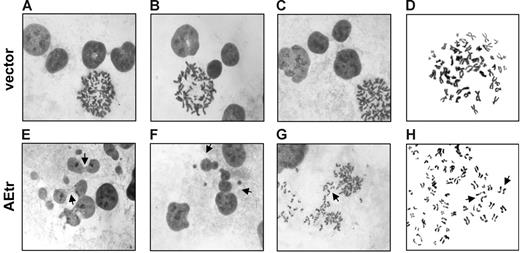
Alterations in the spindle checkpoint can lead to abnormal mitosis and aneuploidy.39 Therefore, we monitored mitosis in control and AEtr cells by microscopy after staining the interphase DNA and mitotic chromosomes with Giemsa. Control cells had normal interphase nuclear morphologies and normal sister chromatid cohesion at prometaphase (Figure 3A-D). Of interest, interphase nuclei of AEtr-expressing cells contained aberrant chromosome bridges/micronuclei and 15.4% were polyploid compared with only 3% in vector control (Figure 3E-F; Fisher exact test, P = .001). Additionally, AEtr mitotic cells with lagging chromosomes (Figure 3G) as well as cells with prematurely dissociated sister chromatids prior to anaphase were observed in 7.4% of prometaphase AEtr cells, but no vector control cells had separated chromatids prior to anaphase (Figure 3H). These data demonstrate that AEtr cells undergo abnormal chromosome segregation and are consistent with an attenuated mitotic checkpoint.
Abnormal morphology of interphase and mitotic AEtr cells. (A-C) Giemsa staining of interphase vector control nuclei. (D) Chromosome spread of a prometaphase vector control cell. (E) Giemsa staining of interphase AEtr nuclei. Arrows indicate chromosome bridges. (F) Polyploid AEtr interphase cells (indicated by arrows). (G) Lagging chromosomes (indicated by arrow) during anaphase of an AEtr cell. (H) Premature sister chromatid separation in an AEtr prometaphase cell. Images were captured with a Leica DMLB microscope with a 100×/_ oil objective and a Spot 2 digital camera (Diagnostic Instruments, Sterling Heights, MI) using Spot v.4.64 software. Images were processed with Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA).
Abnormal morphology of interphase and mitotic AEtr cells. (A-C) Giemsa staining of interphase vector control nuclei. (D) Chromosome spread of a prometaphase vector control cell. (E) Giemsa staining of interphase AEtr nuclei. Arrows indicate chromosome bridges. (F) Polyploid AEtr interphase cells (indicated by arrows). (G) Lagging chromosomes (indicated by arrow) during anaphase of an AEtr cell. (H) Premature sister chromatid separation in an AEtr prometaphase cell. Images were captured with a Leica DMLB microscope with a 100×/_ oil objective and a Spot 2 digital camera (Diagnostic Instruments, Sterling Heights, MI) using Spot v.4.64 software. Images were processed with Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA).
Abnormal mitosis and failure to undergo mitotic arrest in spindle poisons suggested that AEtr cells may be aneuploid due to an attenuated spindle checkpoint. Therefore, chromosome counts of control or AEtr cells were performed. Parental K562 cells have a modal chromosome number of 67.42 Chromosome counting of 54 cells from control or 2 independent AEtr-expressing pools indicated that the average chromosome number was 63 for vector and 58 and 60 for each AEtr line. Remarkably, the range of chromosome numbers for AEtr cells was much more diverse and ranged from 36 to 116 chromosomes per cell. In contrast, all control cells contained between 49 to 85 chromosomes (Figure 4). Furthermore, 17% to 20% of AEtr cells contained fewer than 50 chromosomes or more than 80 chromosomes per cell, whereas no control cells contained these chromosome numbers. These data support a model in which AEtr expression compromises the spindle checkpoint to initiate chromosome missegregation.
AEtr cells are aneuploid. Mitotic chromosome counts from vector control or AEtr cells. Cells from vector control cells or 2 AEtr lines (AEtr#1, AEtr#2) were used to generate metaphase chromosome spreads. Chromosomes from 54 metaphase cells per line were counted. The histogram indicates the number of vector control or 2 different AEtr pools containing various ranges of chromosomes per cell.
AEtr cells are aneuploid. Mitotic chromosome counts from vector control or AEtr cells. Cells from vector control cells or 2 AEtr lines (AEtr#1, AEtr#2) were used to generate metaphase chromosome spreads. Chromosomes from 54 metaphase cells per line were counted. The histogram indicates the number of vector control or 2 different AEtr pools containing various ranges of chromosomes per cell.
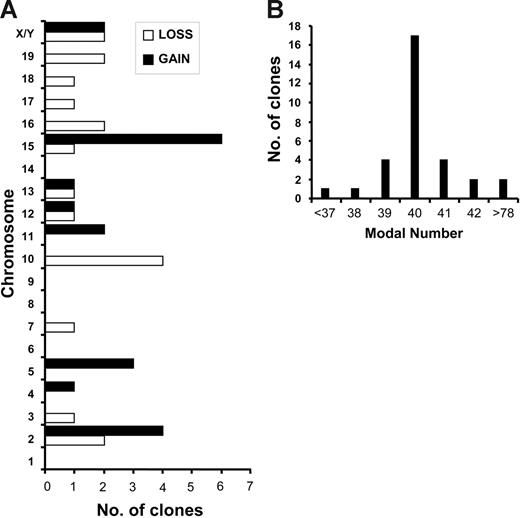
To determine if AEtr expression promoted aneuploidy in primary leukemia cells, we examined chromosomal alterations in either splenocytes or peripheral blood cells from 8 leukemias induced by giving mice transplants of AEtr-expressing cells. Spectral karyotyping analyses of at least 10 metaphase cells per leukemia sample showed clonal abnormalities in 6 of 8 cases, with complex karyotypes with multiple clones (Table 1). Clonal numeric abnormalities involving multiple chromosomes (Figure 5A), as well as structural abnormalities, were also observed (Table 1). Fourteen of 31 clones had modal numbers off the mode of 40 chromosomes (Figure 5B). Specifically, trisomy of chromosomes 2, 5, and 15 and loss of chromosome 10 were most often observed. These data indicate that aneuploidy occurs in primary leukemia cells expressing AEtr.
Chromosomal abnormalities in AEtr-induced leukemias
| Mouse . | Tissue . | Karyotype . |
|---|---|---|
| 6114* | Spleen | 42, XX, +5, der(7)t(7;12)(F4;B), +15 [2] |
| 39, XX, der(7)t(7;10)(F3;C3)/−10 [2] | ||
| 42, XX, +4, +15 [1] | ||
| 39,XX, der(10;16)(A1;A1) [1] | ||
| 39, XX, −10,+11,−19 [1] | ||
| 78, XXXX, −2,−2,der(7)t(2;7)(F3;F3)×2, i(16)(A1)×2 [1] | ||
| 38, XX, +5,der(7)t(7;10)(F3;C3),−10,+11,−15,−18,−19 [1] | ||
| 40, XY [1] | ||
| 6115* | Spleen | 41, XX, +2, −10, +15 [1] |
| 41, X, −X or −Y, der(7)t(7;15)(F3;C), idic(10)(A3), +13, +13 [1] | ||
| 40, XX, der(7)t(7;12)(F4;B)del(12)(BD3), der(8)t(8;12)(A1;F1) [1] | ||
| 40, X, −X, +2, −3, der(7)t(7;12)(F4;B)del(12)(BD3), del(9)(A4F3)+15 [1] | ||
| 40, XX, −2, der(7)t(7;12)(F4;B)del(12)(BD3), +15 [1] | ||
| 39, XY, +5, t(5;12)(B1;A1), −12, −16 [1] | ||
| 36, XX, −2,der(7)t(2;7)(H1;F3), −13, −16, −17 [1] | ||
| 40, XX [2] | ||
| 6266 | Spleen | 41, XY, +12 [4] |
| 40, XY [6] | ||
| 6107 | Spleen | 40, XY [10] |
| 3419 | Spleen | 80, XXXX [4] |
| 40, XX [6] | ||
| 2625 | Spleen | 41, XX, +15 [4] |
| 40, XX [6] | ||
| 1680* | Blood | 40, XY [8] |
| 40, XX [2] | ||
| 1347 | Spleen | 40, XY, t(1;15)(H1;C) [2] |
| 40, idem,t(6;11)(A1;B3) [2] | ||
| 40, XY, t(1;11)(H5;B3), t(2;4)(D;B1) [2] | ||
| 40, XY, t(2;7)(H3;F1) [1] | ||
| 40, XY, t(1;7;17;13)(F;F1;D;B) [1] | ||
| 39, XY, −7, t(9;11)(D;B3) [1] | ||
| 40, XY [1] |
| Mouse . | Tissue . | Karyotype . |
|---|---|---|
| 6114* | Spleen | 42, XX, +5, der(7)t(7;12)(F4;B), +15 [2] |
| 39, XX, der(7)t(7;10)(F3;C3)/−10 [2] | ||
| 42, XX, +4, +15 [1] | ||
| 39,XX, der(10;16)(A1;A1) [1] | ||
| 39, XX, −10,+11,−19 [1] | ||
| 78, XXXX, −2,−2,der(7)t(2;7)(F3;F3)×2, i(16)(A1)×2 [1] | ||
| 38, XX, +5,der(7)t(7;10)(F3;C3),−10,+11,−15,−18,−19 [1] | ||
| 40, XY [1] | ||
| 6115* | Spleen | 41, XX, +2, −10, +15 [1] |
| 41, X, −X or −Y, der(7)t(7;15)(F3;C), idic(10)(A3), +13, +13 [1] | ||
| 40, XX, der(7)t(7;12)(F4;B)del(12)(BD3), der(8)t(8;12)(A1;F1) [1] | ||
| 40, X, −X, +2, −3, der(7)t(7;12)(F4;B)del(12)(BD3), del(9)(A4F3)+15 [1] | ||
| 40, XX, −2, der(7)t(7;12)(F4;B)del(12)(BD3), +15 [1] | ||
| 39, XY, +5, t(5;12)(B1;A1), −12, −16 [1] | ||
| 36, XX, −2,der(7)t(2;7)(H1;F3), −13, −16, −17 [1] | ||
| 40, XX [2] | ||
| 6266 | Spleen | 41, XY, +12 [4] |
| 40, XY [6] | ||
| 6107 | Spleen | 40, XY [10] |
| 3419 | Spleen | 80, XXXX [4] |
| 40, XX [6] | ||
| 2625 | Spleen | 41, XX, +15 [4] |
| 40, XX [6] | ||
| 1680* | Blood | 40, XY [8] |
| 40, XX [2] | ||
| 1347 | Spleen | 40, XY, t(1;15)(H1;C) [2] |
| 40, idem,t(6;11)(A1;B3) [2] | ||
| 40, XY, t(1;11)(H5;B3), t(2;4)(D;B1) [2] | ||
| 40, XY, t(2;7)(H3;F1) [1] | ||
| 40, XY, t(1;7;17;13)(F;F1;D;B) [1] | ||
| 39, XY, −7, t(9;11)(D;B3) [1] | ||
| 40, XY [1] |
Spectral karyotype analysis of 8 primary murine leukemias induced by transplantation of hematopoietic cells expressing AEtr. A minimum of 10 metaphase cells per leukemia sample was analyzed. Mouse identification, tissue of origin, and karyotype are indicated for each sample.
Metaphase cells derived from both donor and recipient mice were observed.
Primary AEtr leukemia cells are aneuploid. (A) Gains and losses of chromosomes determined by SKY. A total of 88 metaphase cells from 8 independent leukemias was analyzed. The number of independent clones that displayed a particular loss or gain of each chromosome is indicated in the bar graph. (B) The number of clones displaying different modal numbers is indicated.
Primary AEtr leukemia cells are aneuploid. (A) Gains and losses of chromosomes determined by SKY. A total of 88 metaphase cells from 8 independent leukemias was analyzed. The number of independent clones that displayed a particular loss or gain of each chromosome is indicated in the bar graph. (B) The number of clones displaying different modal numbers is indicated.
AEtr alters levels of spindle checkpoint components
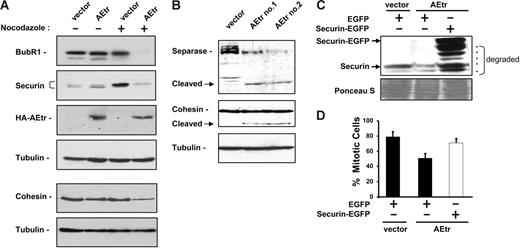
Mitotic spindle alteration by disruption or stabilization normally activates the spindle checkpoint and inhibits APC activity. Inappropriate APC activation could account for reduced mitotic arrest in nocodazole, therefore, we examined levels of several proteins involved in the spindle checkpoint, including 2 APC inhibitors, BubR1 and Mad2. Mad2 levels were similar in control and AEtr cells either prior to or after nocodazole addition (data not shown). In contrast, BubR1 levels were decreased in AEtr cells after nocodazole treatment (Figure 6A). BubR1 reduction can lead to inappropriate APC activation leading to the premature destruction of the APC substrate securin.31,32 Securin levels were similar in asynchronous control and AEtr cells (Figure 6A). Following nocodazole treatment, control cells had increased levels of securin, consistent with previous reports that APC activity is required to destabilize securin. In contrast, AEtr cells treated with nocodazole had lower securin levels, suggesting that APCcdc20 may be active in these cells (Figure 6A).
Spindle checkpoint proteins are reduced in AEtr cells. (A) Western blot analysis of BubR1 and securin in K562 vector or AEtr-expressing cells treated with 0.025% DMSO (−) or 1 μg/mL nocodazole (+) for 18 hours. Immunoblot for tubulin indicates relative protein loading. Anti-HA Western shows AEtr expression. The same lysates were also analyzed for cohesin subunit Rad21 and tubulin to show relative protein loading. (B) Western blot analysis of separase full length and the autocleaved form in control or 2 different AEtr pools treated with nocodazole. The same membrane was probed for cohesin, which indicates the cleaved cohesin form is detectable only after a long exposure compared with cohesin in panel A. (C) K562 vector or AEtr-expressing cells were transfected with plasmids encoding EGFP or securin-EGFP. At 48 hours after transfection, cells were treated with 1 μg/mL nocodazole for 18 hours. Cells were monitored for endogenous securin and transfected securin-EGFP expression by immunoblotting, which detected expression of full-length securin-EGFP and C-terminal degraded forms (indicated by asterisks) as well as endogenous securin. (D) Cells were stained with propidium iodide to measure DNA content by flow cytometry. The percentage of cells in mitosis following transfection of EGFP or securin-EGFP for 3 independent experiments is shown. Error bars show SD of the average from 3 independent experiments.
Spindle checkpoint proteins are reduced in AEtr cells. (A) Western blot analysis of BubR1 and securin in K562 vector or AEtr-expressing cells treated with 0.025% DMSO (−) or 1 μg/mL nocodazole (+) for 18 hours. Immunoblot for tubulin indicates relative protein loading. Anti-HA Western shows AEtr expression. The same lysates were also analyzed for cohesin subunit Rad21 and tubulin to show relative protein loading. (B) Western blot analysis of separase full length and the autocleaved form in control or 2 different AEtr pools treated with nocodazole. The same membrane was probed for cohesin, which indicates the cleaved cohesin form is detectable only after a long exposure compared with cohesin in panel A. (C) K562 vector or AEtr-expressing cells were transfected with plasmids encoding EGFP or securin-EGFP. At 48 hours after transfection, cells were treated with 1 μg/mL nocodazole for 18 hours. Cells were monitored for endogenous securin and transfected securin-EGFP expression by immunoblotting, which detected expression of full-length securin-EGFP and C-terminal degraded forms (indicated by asterisks) as well as endogenous securin. (D) Cells were stained with propidium iodide to measure DNA content by flow cytometry. The percentage of cells in mitosis following transfection of EGFP or securin-EGFP for 3 independent experiments is shown. Error bars show SD of the average from 3 independent experiments.
Securin is an inhibitor of the separase protease. Activated separase cleaves itself as well as centromeric cohesin following securin degradation by APCcdc20 activation.33,43–46 We therefore examined cohesin levels and found that full-length cohesin was decreased in AEtr cells following nocodazole treatment (Figure 6A).
Cohesin cleavage results from separase activation. One indication of separase activity is autocleavage of the enzyme. Indeed, cleaved separase was detectable in AEtr cells, but not in control cells (Figure 6B). Moreover, cleavage products of cohesin were also detected in AEtr cells after a long exposure of the immunoblot compared with Figure 6A, indicating that sister chromatid cohesion is disrupted in nocodazole-treated cells. These data suggest that APC activation in AEtr cells leads to separase-mediated cohesin cleavage.
AEtr contains C-terminal ETO domains that interact with transcriptional repressors, therefore we tested whether the reduction of BubR1, securin, and cohesin in AEtr cells could be due to transcriptional down-regulation.37,47,48 Real-time PCR analysis revealed no significant differences in either BubR1 or cohesin mRNA either before or after nocodazole treatment between control and AEtr cells (Figure S2A). Of interest, securin mRNA was down-regulated in AEtr cells following nocodazole treatment compared with vector control cells. This down-regulation was reproducible in several experiments and was significant (P = .008). Additionally, Northern blot also reflected the down-regulation of securin mRNA in nocodazole-treated AEtr cells (Figure S2B). These data suggest that securin transcription may be repressed in AEtr cells.
We attempted to restore the checkpoint to cells by expressing checkpoint components in cells. BubR1 transfection was reported to restore the spindle checkpoint in other studies, however transfection of K562-AEtr cells with a BubR1 expression plasmid did not increase the mitotic index after nocodazole treatment (data not shown).28,49 To determine if securin down-regulation contributed to spindle checkpoint attenuation, K562 control or AEtr cells were transfected with plasmids encoding EGFP or securin-EGFP. Two days following transfection, cells were treated with nocodazole for 18 hours. Immunoblotting revealed that securin-EGFP was expressed in AEtr cells, however degraded forms were also detected (Figure 6C). Cell-cycle profiles reflected that AEtr cells expressing EGFP had reduced mitotic index after nocodazole treatment (Figure 6D). In contrast, a majority of AEtr cells expressing securin-EGFP arrested in mitosis with 4N DNA content similar to vector control cells expressing EGFP. These data demonstrate that loss of securin contributes to the reduced mitotic arrest of AEtr cells in nocodazole.
Spindle checkpoint disruption in t(8;21) patient–derived cells
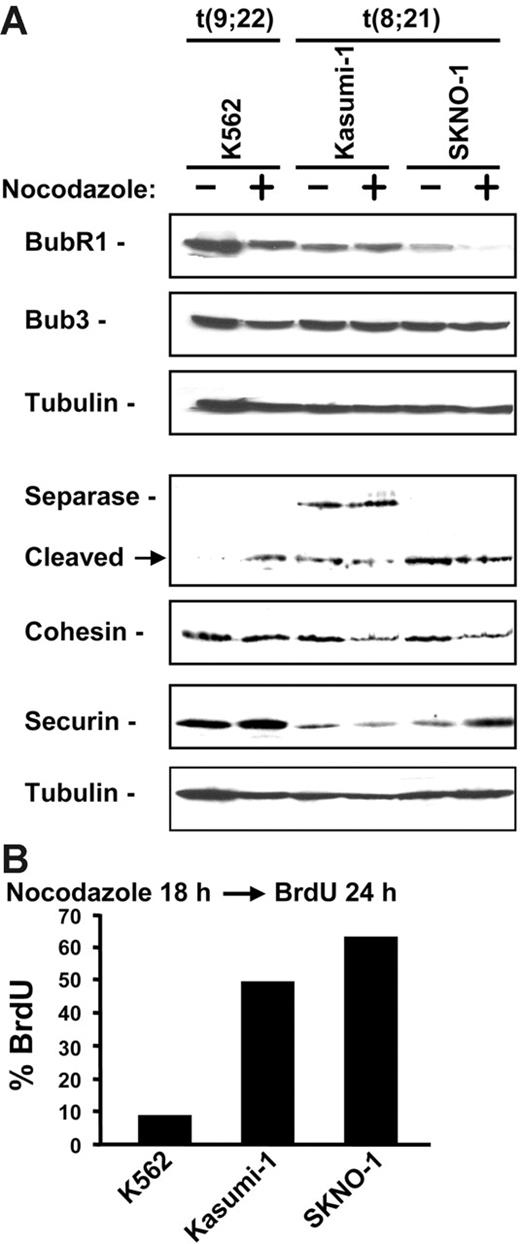
We next examined the spindle checkpoint in 2 t(8;21) patient–derived cell lines, Kasumi-1 and SKNO-1, which express predominantly full-length AML1-ETO compared with the AML1-ETO9a isoform, which is almost identical to AEtr.13 Kasumi-1 and SKNO-1 cells were compared with K562 cells, which are derived from a patient with t(9;22). Cell lines were treated with either nocodazole or colchicine and monitored for mitotic arrest. Of interest, more than half of Kasumi-1 or SKNO-1 cells underwent apoptosis in concentrations of either microtubule-disrupting agent that did not promote apoptosis of K562 cells (data not shown). Since cell death in response to spindle toxins reflects an inactive spindle checkpoint, the checkpoint in these t(8;21) cell lines was studied further.
Reduction of BubR1 by RNA interference has been reported to cause apoptosis of HeLa cells in response to microtubule disruption.28 Therefore, we examined the expression levels of BubR1 as well as other proteins involved in maintaining the spindle checkpoint. Both Kasumi-1 and SKNO-1 cells had reduced expression of BubR1 even in the absence of nocodazole compared with K562 cells (Figure 7A). However, only SKNO-1 cells contained reduced BubR1 following nocodazole treatment. No differences in Bub3, a BubR1-interacting protein, were observed. We next examined securin, separase, and cohesin levels in these cell lines. Securin levels were decreased in both Kasumi-1 and SKNO-1 cells compared with K562 cells in the absence of nocodazole treatment. Both Kasumi-1 and SKNO-1 expressed the cleaved autoprocessed form of separase (Figure 7A). Additionally, full-length cohesin was reduced in Kasumi-1 and SKNO-1 following nocodazole treatment compared with K562 cells. These data indicate that t(8;21) patient–derived cell lines have altered expression of proteins that contribute to the spindle checkpoint.
t(8;21) cell lines have disrupted spindle checkpoint. (A) Immunoblot of lysates from K562, Kasumi-1, and SKNO-1 cells treated without (−) or with (+) 1 μg/mL nocodazole for 18 hours. Membranes were probed with antibodies for BubR1, Bub3, separase, cohesin subunit Rad21/scc1, and securin. Tubulin immunoblot shows relative protein loading. (B) Kasumi-1, SKNO-1, and K562 cells were treated with nocodazole for 18 hours and labeled with 10 μM BrdU for 24 hours. Cells were fixed and stained with anti–BrdU-FITC and analyzed by flow cytometry. The percentage of cells stained positive for BrdU is shown.
t(8;21) cell lines have disrupted spindle checkpoint. (A) Immunoblot of lysates from K562, Kasumi-1, and SKNO-1 cells treated without (−) or with (+) 1 μg/mL nocodazole for 18 hours. Membranes were probed with antibodies for BubR1, Bub3, separase, cohesin subunit Rad21/scc1, and securin. Tubulin immunoblot shows relative protein loading. (B) Kasumi-1, SKNO-1, and K562 cells were treated with nocodazole for 18 hours and labeled with 10 μM BrdU for 24 hours. Cells were fixed and stained with anti–BrdU-FITC and analyzed by flow cytometry. The percentage of cells stained positive for BrdU is shown.
To determine the fate of the remaining viable Kasumi-1 and SKNO-1 cells after nocodazole treatment, cells were monitored for continued proliferation by measuring BrdU incorporation. Flow cytometry indicated that more than 50% of the remaining viable Kasumi-1 and SKNO-1 cells incorporated BrdU 24 hours following nocodazole treatment compared with 8.6% incorporation in K562 cells (Figure 7B). These data indicate that t(8;21) patient–derived cell lines have an attenuated spindle checkpoint.
Discussion
Aneuploidy is a common feature of human malignancies and results from chromosome missegregation during mitosis. The mechanism for aneuploidy in t(8;21)-associated AML has not been investigated prior to this study. Data presented in this study demonstrate that AEtr, which is almost identical to the AML1-ETO exon 9a isoform expressed in t(8;21) patients, influences the spindle checkpoint to promote aneuploidy. Attenuation of the spindle checkpoint as opposed to complete loss of the checkpoint may confer an advantage to cells to promote aneuploidy without inducing mitotic catastrophe.
Microtubule disruption led to 2 different populations of AEtr cells based on DNA content. The 2N DNA content indicates that a subset of AEtr cells resisted microtubule disruption by nocodazole or colchicine and was able to successfully segregate chromosomes and proceed through cytokinesis. The resulting daughter cells have the expected 2N DNA content, which results from a 4N mitotic cell undergoing mitosis to produce 2 daughter cells. Resistance to spindle poisons has been reported in T-ALL leukemia cells and subclones of the AML HL-60 cell line, but has not been reported in t(8;21) AML patients.50,51
2N DNA caused by nocodazole resistance in AEtr cells could occur through several mechanisms. First, it is possible that a subset of AEtr cells extrudes the drugs by up-regulating the ABC transporters. It was previously shown that full-length AML1-ETO represses the promoter activity of the MDR-1 gene, which encodes an ABC transporter. However, this repression occurred in C33A cells, a nonhematopoietic line, and required a domain missing in both AEtr and AML1-ETO exon 9a.52 Other mechanisms for nocodazole resistance include survivin overexpression, MAP overexpression, or increased microtubles.53 These latter possibilities seem more likely since a measurement of fluorescent paclitaxel did not indicate increased drug loss in AEtr cells compared with control cells (A.B. and D.-E.Z., unpublished data, January 2006). Additionally, we observed increased microtubules in AEtr cells compared with control cells before and after nocodazole treatment (A.B. and D.-E.Z., unpublished data, January 2006).
The second AEtr population following nocodazole treatment contained 8N DNA content. 8N DNA results from a 4N nucleus exiting mitosis and proceeding through the next cell cycle. Indeed AEtr cells continued to proliferate after nocodazole treatment. 8N cells result from spindle checkpoint loss and are a genetically unstable population that gives rise to aneuploidy and tumors in vivo.54 The 8N AEtr population was detectable by DNA content increase, chromosome number increase, and nuclear morphology, and required an intact DNA-binding domain in AEtr. Although K562 cells are reported to differentiate into polyploid megakaryocytes, Wright-Giemsa staining (data not shown) and nuclear morphology did not indicate that polyploid AEtr cells were undergoing megakaryocytic differentiation. Therefore, a subset of AEtr cells bypassed the mitotic spindle checkpoint to gain 8N DNA and become aneuploid.
In contrast to K562 cells expressing only AEtr, t(8;21) patient–derived cell lines (Kasumi-1 and SKNO-1) expressing predominantly full-length AML1-ETO compared with the AML1-ETO exon 9a isoform underwent mitotic catastrophe. We previously reported that full-length AML1-ETO causes G0/G1 arrest followed by apoptosis in a hematopoietic cell line.38 The proapoptotic effect of full-length AML1-ETO may account for mitotic catastrophe of t(8;21) patient–derived cell lines in response to spindle disruption. Alternatively, loss of the spindle checkpoint is also known to promote mitotic catastrophe and may account for the loss of highly aneuploid clones in t(8;21) patients.55 Since full-length AML1-ETO causes G0/G1 arrest in several hematopoietic lines, including K562, we were unable to assess accurately the effects of full-length AML1-ETO expression on the mitotic spindle checkpoint. We attempted to analyze the levels of various spindle checkpoint proteins after transient expression of AML1-ETO in K562 cells. However, the cells rapidly arrested in G0/G1, and therefore the changes in protein levels of BubR1, securin, and cohesin were an artifact of growth arrest rather than cells continuing through mitosis to enter G0/G1.12
Since a high percentage of t(8;21) patient–derived cell lines continued to proliferate in the presence of nocodazole, it is likely that AML1-ETO also disrupts the spindle checkpoint. The pattern of spindle checkpoint protein levels in the t(8;21) patient cell lines did not completely reflect the results obtained in K562 AEtr cells, with the exception of cohesin reduction in both Kasumi-1 and SKNO-1 following nocodazole treatment. This could be due to the fact that these cell lines express low levels of AML1-ETO exon 9a compared with the full-length protein or that these cells have additional genetic alterations that affect the spindle checkpoint. Thus, in t(8;21) patients, it is likely that both full-length and shorter AML1-ETO isoforms contribute to the attenuation of cell-cycle checkpoints resulting in leukemia development.
Cells that continue to proliferate with an attenuated spindle checkpoint should missegregate chromosomes and become aneuploid. Indeed, AEtr-expressing cell lines and primary leukemia cells were aneuploid. In addition to gross changes in chromosome modal number, chromosome bridges/micronuclei were observed in AEtr cells, whereas no control cells contained micronuclei. AEtr expression in primary leukemias resulted in metaphase cells with numeric and structural abnormalities. While structural anomalies of chromosomes would not be caused by disruption of the spindle checkpoint, gains and losses of entire chromosomes could be. A comparison of AEtr-induced aneuploidy with other mouse models suggests the AEtr leukemias contain more aneuploidy than AML induced by expression of HoxA9, HOXB6, c-myc, or PU.1 hypomorphic mice.56–58 The aneuploidy induced by AEtr is comparable with the numeric and structural changes observed in AML induced by PML-RARα expression.59
t(8;21)-associated leukemia is not the most aneuploid hematologic malignancy. However, the loss of a sex chromosome, trisomies 8 and 4, and loss of chromosome 7 are frequently observed in patients. Spindle checkpoint attenuation may initiate aneuploidy only during early stages of t(8;21) disease. Subsequent to checkpoint attenuation, loss and gain of specific chromosomes must be selected for in t(8;21) patients, leading to the karyotype of patients at the time of diagnosis. Most likely, the loss of differentiation-promoting genes and hematopoietic growth suppressors as well as the acquisition of genes involved in self-renewal are selected in patients.
Microtubule disruption in AEtr cells or in patient-derived cell lines led to the alteration of several components of the spindle checkpoint including BubR1, securin, separase, and cohesin. However, we cannot rule out alterations of other important spindle checkpoint proteins including chromosomal passenger kinases or proteins involved in the mitotic exit network. Although BubR1 re-expression was not able to increase the mitotic index of AEtr cells (data not shown), a downstream target of BubR1 reduction, securin, could rescue the defect. Some studies report that loss of securin can lead to karyotype changes in cell lines and in vivo, at least in initial passages.60–62 Therefore, it is possible that APC activity, through securin down-regulation, is responsible for the spindle checkpoint attenuation in AEtr cells. Additionally, the transcriptional down-regulation of securin in AEtr cells may also contribute to the loss of functional securin protein.
Our data provide a novel mechanism for the initiation of aneuploidy in t(8;21) leukemia patients. Normal chromosome segregation is inhibited when the spindle checkpoint is activated prior to metaphase alignment of chromosomes. This involves inhibition of APC activity resulting in stabilization of securin and other substrates. Separase is inactive thereby protecting centromeric cohesin from cleavage and preventing anaphase. In contrast, AEtr cells attenuate the spindle checkpoint resulting in loss of sister chromatid association. Loss of centromeric cohesin prior to metaphase alignment results in chromosome segregation in the absence of bioriented sister chromosomes, promoting aneuploidy. Based on data presented in this study, we propose that aneuploidy observed in t(8;21) leukemias results from spindle checkpoint attenuation. Inhibition of pathways promoting loss of chromatid cohesion may be novel therapeutic targets in the treatment of leukemia patients with disrupted spindle checkpoints.
Authorship
Contribution: A.B. designed and performed the research and wrote the paper; M.Y. prepared leukemia samples used for spectral karyotyping; L.F.P. provided experimental advice and assisted with paper preparation; J.R.B. assisted with chromosome counts, provided experimental advice, and assisted with paper preparation; M.M.L. performed SKY analysis and assisted with paper preparation; D.-E.Z. supervised experimental design, data analysis, and paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, The Scripps Research Institute, 10550 N Torrey Pines Rd, MEM-L51, La Jolla, CA 92037; e-mail: dzhang@scripps.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by National Institutes of Health research grants CA104509 and CA096735 (D.-E.Z.), NCI grant U01CA84221 and LLS SCOR 7019-04 (M.M.L.), and an NRSA fellowship 5F32HL079900 (A.B.). The Stein Endowment Fund has partially supported the departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
We wish to thank Drs Dan Foltz, Beth Weaver, and members of the Zhang lab for critical reading of the paper; Drs Nanao Kamada and Sachiko Matozaki for Kasumi-1 and SKNO-1 human leukemia cell lines; Dr Schickwann Tsai for the BHK/MKL cells; Drs Wei Dai, Guo-Wei Fang, Prasad Jallepalli, Shlomo Melmed, Debananda Pati, David Pellman, Hui Zou, and Don Cleveland for plasmids and reagents; and Dr Kevin Sullivan, Dr Martin Gullberg, and Oxana Malakhova for technical advice. This is manuscript 18109-MEM from The Scripps Research Institute.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal