Abstract
A recent phase 1 trial has demonstrated that the generation of tumor-reactive T lymphocytes by transfer of specific T-cell receptor (TCR) genes into autologous lymphocytes is feasible. However, compared with results obtained by infusion of tumor-infiltrating lymphocytes, the response rate observed in this first TCR gene therapy trial is low. One strategy that is likely to enhance the success rate of TCR gene therapy is the use of tumor-reactive TCRs with a higher capacity for tumor cell recognition. We therefore sought to develop standardized procedures for the selection of well-expressed, high-affinity, and safe human TCRs. Here we show that TCR surface expression can be improved by modification of TCR alpha and beta sequences and that such improvement has a marked effect on the in vivo function of TCR gene-modified T cells. From a panel of human, melanoma-reactive TCRs we subsequently selected the TCR with the highest affinity. Furthermore, a generally applicable assay was used to assess the lack of alloreactivity of this TCR against a large series of common human leukocyte antigen alleles. The procedures described in this study should be of general value for the selection of well- and stably expressed, high-affinity, and safe human TCRs for subsequent clinical testing.
Introduction
In the majority of human cancers, the shared tumor-associated antigens that are available for immunotherapy are nonmutated self-antigens. While T-cell reactivity against certain self-antigens can be observed and is in fact rather common for melanoma-associated antigens,1 the boosting of such reactivity by vaccination has largely been unsuccessful to date.2 Work over the past years by Dudley et al has demonstrated that objective tumor regressions can be obtained in approximately 50% of patients with metastatic melanoma by infusion of ex vivo–expanded tumor-infiltrating lymphocytes (TILs) after pretreatment with lymphodepleting chemotherapy.3,4 While the demonstration of the clinical effectiveness of TIL infusion can be considered one of the major breakthroughs of cellular immunotherapy, the isolation of antigen-specific cells from resected tumor material and the in vitro expansion of these cells is feasible only for a fraction of melanoma patients. In addition, the scarcity of tumor-reactive T cells in tumor material of other human cancers precludes clinical application for most other human malignancies
As an alternative to the use of naturally occurring tumor-reactive T cells, tumor-reactive cells may be generated by the transfer of tumor-specific T-cell receptor (TCR) genes. Such TCR gene transfer circumvents the requirements for isolation and in vitro expansion of T cells, and would allow the use of high-affinity tumor-reactive TCRs in larger patient groups.5,6 Retrovirus-mediated delivery of TCR genes has been used to convey virus and tumor reactivity to human T cells,7-11 and TCR-modified cytotoxic T cells and helper T cells have displayed in vivo functionality in several mouse models.12-15
Recently, the feasibility of TCR gene transfer was assessed in a clinical phase 1 study.16 In this trial, peripheral blood lymphocytes of patients with metastatic melanoma were transduced with a Mart-1–specific T-cell receptor, and reinfused following lymphodepleting chemotherapy. While the infusion of TCR-transduced cells was effective in that these cells persisted for more than 2 months in most individuals, objective tumor regression was observed in only 2 of 15 patients. These data indicate that TCR gene transfer is feasible in a clinical setting, but that a substantial improvement of this approach is required to become a clinically meaningful treatment strategy.
One possible factor in the low response rate observed in this trial is a suboptimal capacity of the TCR-modified T cells for tumor cell recognition. First, tumor-specific T cells can differ widely in their capacity for tumor cell recognition, even when recognizing the same antigen.17,18 Second, the expression of retrovirally introduced human TCRs in human T cells is suboptimal,19 often requiring cotransduction of vectors encoding TCR alpha or beta genes together with a selectable marker, rather than with the bicistronic vectors that have been used in the clinic. Here, we set out to develop and test generally applicable strategies for the isolation of TCRs for clinical use. The resulting 3-step approach has been used to select a melanoma-reactive TCR with improved expression, a higher capacity for antigen recognition, and lack of detectable alloreactivity.
Materials and methods
Mice
Six- to 10-week-old female C57BL/6 mice (H-2b) and pmel-1 TCR transgenic mice20 were obtained from the Experimental Animal Department of the Netherlands Cancer Institute. All animal experiments were performed in accordance with institutional and national guidelines and were approved by the Experimental Animal Committee of the Netherlands Cancer Institute (DEC).
Cell lines and peripheral blood mononuclear cells
FLYRD18 is a human fibrosarcoma retroviral packaging cell line (ECACC no. 95091902). The Jurkat/MA cell line is a Jurkat cell line lacking endogenous TCR expression.21 T2 is an HLA-A2+ cell line that is deficient for TAP (transporter associated with antigen presentation). The panel of single major histocompatibility complex (MHC) class I allele–expressing K562 cell lines (the SAL panel) has been described previously.22 All cell lines were cultured in Iscove modified Dulbecco medium (IMDM) (GIBCO, Invitrogen, Carlsbad, CA) with 5% fetal calf serum (PAA, Pasching, Austria), penicillin (100 U/mL), and streptomycin (100 μg/mL).
Melanoma cell lines Mel 526 (HLA-A2+, Gp100+, Mart+) and Mel 938 (HLA-A2−, Gp100+, Mart+) were described previously.23 Mel GDO and Mel AKR (HLA-A2+, Gp100+, Mart+) were established in the NKI from resected tumor lesions. Melanoma cell lines were maintained in RPMI (GIBCO, Invitrogen) in the presence of 5% FCS, penicillin (100 U/mL), and streptomycin (100 μg/mL).
Peripheral blood mononuclear cells PBMCs from anonymous healthy donors derived via the local blood bank were isolated by leukapheresis and subsequent Ficoll-Isopaque density centrifugation. Following transduction, PBMCs were cultured in Yssels medium24 supplemented with 20 U/mL IL-2 (Proleukin; Chiron, Emeryville, CA). Every 14 days, transduced PBMCs were stimulated with irradiated JY cells and allogeneic PBMCs, plus 100 ng/mL phytohemagglutinin and 20 U/mL IL-2.
TCR gene optimalization and cloning
Sequences of the Gp100-specific pmel-1 TCR were kindly provided by N. Restifo (NIH, Bethesda, MD). Sequences of the Mart-1–specific 1D3 and 2C2 TCRs have been described previously.25 The Mart-1–specific DMF4 TCR26 and the Gp100-specific R6C12 TCR27 were isolated at the NIH. Modified TCR genes were designed and produced by GeneArt (Regensburg, Germany). DNA sequences are provided in Figure S2, available on the Blood website; see the Supplemental Figures link at the top of the online article. Wild-type wt and gene-optimized TCR alpha and beta chains were cloned into the retroviral vector pMX28 containing an internal ribosomal entry site, or in the indicated vector.
Production of retroviral supernatants and retroviral transduction
FLYRD18 packaging cells were plated in 6-well plates at 1.5 × 105 cells/well. After one day, cells were transfected with 2.5 μg retroviral vector DNA using FuGENE TM6 (Roche Diagnostics, Indianapolis, IN). After 48 hours, retroviral supernatant was pooled, centrifuged, and frozen at − 80°C. PBMCs were activated with 20 U/mL IL-2 and 2 mg/mL phytohemagglutinin, at 1 × 106 cells/mL. Forty-eight hours after stimulation, PBMCs were resuspended in retroviral supernatant, transferred to RetroNectin-coated plates at 0.5 × 106 cells/mL, and centrifuged for 90 minutes at 430g. Jurkat/MA cells were transduced without the centrifugation step. Mouse splenocytes were transduced as described previously.12
Flow cytometric analysis
Surface expression of pmel-1 TCR-transduced murine splenocytes was measured using Db-tetramers, or by double staining with fluorescein isothiocyanate (FITC)-labeled anti-Vβ13 monoclonal antibody (mAb) and PE-labeled anti-Vβ2, 3, 4, 5, 8, 9, 10, and 11 mAb (anti-Vβ-pool), in combination with anti-CD8α mAb (all mAbs from BD Pharmingen, San Jose, CA). Surface expression of TCR-transduced PBMCs was measured by staining with MHC-tetramers, using MHC-tetramers generated through ultraviolet-peptide exchange,29,30 or by staining with anti-Vβ12 (for DMF4), anti-Vβ14 (for 1D3 and 2C2), or anti-Vβ8 (for R6C12) antibody (Immunotech, Westbrook, ME), in combination with anti-CD8 or anti-CD4 antibody (Becton Dickinson, San Jose, CA). Cells were analyzed and sorted using a FACSCalibur and FACSAria (Becton Dickinson).
Adoptive transfer and viral infection
Mice received an intravenous adoptive transfer of transduced splenocytes, nontransduced splenocytes, or in vitro–activated splenocytes from pmel-1 transgenic mice. To induce lymphodepletion, mice received total body irradiation (TBI) of 5 Gy, one day before adoptive transfer. Mice were vaccinated at the day of adoptive transfer by intraperitoneal injection of 1 × 107 plaque-forming units of recombinant vaccinia virus encoding hGp100(25-33), kindly provided by N. Restifo.31 For the measurement of T-cell responses, peripheral blood samples were taken at the indicated days after treatment.
IFN-γ assay
T2 cells were pulsed with peptides for 1 to 2 hours at 37°C. Next, 0.5 × 106 TCR-transduced PBMCs were incubated with 0.5 × 106 peptide-pulsed T2 cells or 0.5 × 106 SALs, in the presence of 20 U/mL IL-2 and 1 μL/mL Golgiplug (BD Biosciences, Basel, Switzerland). After 4- to 5-hour incubation at 37°C, cells were washed and stained with FITC-labeled anti-CD8 antibody and phycoerythrin (PE)-labeled anti-CD4 antibody, and analyzed for IFN-γ production by intracellular cytokine staining.
Chromium release assay
Target cells were labeled for 1 hour at 37°C with 100 μCi (3.7 MBq) 51Cr (Amersham, Gent, Belgium). Labeled target cells were incubated with effector cells at indicated ratios for 4 hours at 37°C in 200 μL medium, in the presence of a 50-fold excess of unlabeled K562 cells.
Results
Increased expression and in vivo function of gene-optimized TCRs
A specific issue in TCR gene transfer has been the relatively low level of transgene expression that is obtained after retroviral modification of human T lymphocytes. This is at least partially due to competition of introduced and endogenous TCR gene products for the limited pool of CD3 components, and inefficient heterodimer formation of the introduced TCR chains.32,33
It has previously been demonstrated that in vitro TCR expression can be improved by the use of synthetic genes with an optimized codon usage.34 However, the in vivo consequences of TCR gene optimization—which will determine its utility in a clinical setting—have not been assessed. To address this issue, the nucleic acid sequence of the murine Gp100–specific pmel-1 TCR was modified to conform to the codon bias observed in highly expressed mammalian genes,35 thereby avoiding cis-acting sequence motifs.36
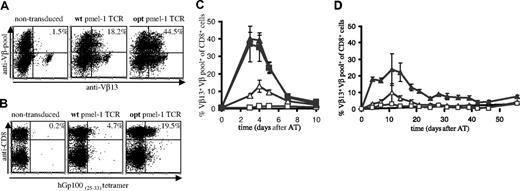
To distinguish pmel-1 expression from endogenous TCR expression, mouse splenocytes, transduced with retroviruses containing the wt or optimized (opt) pmel-1 TCR genes, were stained with a combination of anti-Vβ13 antibodies and a pool of anti-Vβ antibodies, thereby allowing detection of T cells with dual TCR expression.15 Analysis of TCR-transduced cells via this strategy (Figure 1A) or with MHC-tetramers (Figure 1B) revealed a substantial increase in the percentage of cells expressing the pmel-1 TCR upon gene optimization (Vβ staining 44.5% versus 18.2%; MHC-tetramer staining 19.5% versus 4.7%). The level of expression was affected to a lesser extent (mean fluorescence intensity [MFI]: 63 versus 52). The markedly lower percentage of MHC-tetramer+ cells compared with the percentage of Vβ+ cells can be explained by the fact that T cells with relatively low TCR expression can escape detection by MHC-tetramer staining. Furthermore, T cells in which the introduced TCR beta chain is expressed as a mixed heterodimer with endogenous alpha chains are detected by Vβ staining, but not MHC-tetramer staining.32,33 In addition, when single anti-Vβ staining is used (as for human cells; Figure 2A), T cells with endogenous TCRs expressing this beta chain will also be detected.
To test whether gene optimization affected the in vivo function of TCR-modified T cells, B6 mice received an adoptive transfer of wt or opt TCR-transduced cells. As a control, mice received nontransduced cells or splenocytes of pmel-1 transgenic mice. Remarkably, although mice received an equal number of TCR-transduced cells, T-cell responses upon infection with a recombinant vaccinia strain encoding the hGp100(25-33) epitope were markedly higher in recipients of opt pmel-1 TCRs than in recipients of wt pmel-1 TCRs, and comparable with responses in recipients of pmel-1 TCR-transgenic cells (Figure 1C, peak T-cell responses of 13% and 39% for wt and opt, respectively).
In vivo antigen expansion of wild-type and gene-optimized TCR-transduced murine splenocytes. Activated B6 splenocytes were transduced with vectors encoding wild-type or gene-optimized pmel-1 TCR. (A) TCR expression was determined by flow cytometry using anti-CD8α, anti-Vβ13, and a pool of anti-Vβ2, anti-Vβ3, anti-Vβ4, anti-Vβ5, anti-Vβ6, anti-Vβ8, anti-Vβ10, and anti-Vβ11. All fluorescence-activated cell sorting (FACS) plots show events that are gated for CD8 expression. The numbers indicate the percentage of CD8+ T cells with detectable pmel-1 TCR expression, calculated as follows: %Vβ13+Vβpool+ cells/%Vβpool+ cells × 100%. (B) Detection of TCR expression using anti-CD8 antibodies and Db-hGp10025-33 tetramer. The numbers indicate the percentage of tetramer+ CD8+ cells. (C,D) B6 recipients received splenocytes containing either 1 × 106 pmel-1 TCR transgenic CD8+ cells (■, only in panel C), 1.5 × 106 wild-type TCR-transduced CD8+ cells (▵), or 1.5 × 106 optimized TCR-transduced CD8+ cells (▴). Control mice received 20 × 106 activated, nontransduced cells (□). Mice received an intraperitoneal injection of 1 × 107 pfu rVV-hGp100(25-33) virus at the day of adoptive transfer (C), or mice received a sublethal dose of total body irradiation one day before adoptive transfer (D). At the indicated time points, peripheral blood was collected and analyzed by flow cytometry using APC anti-CD8α, FITC anti-Vβ13, and PE anti-Vβpool. The percentage of Vβ13+Vβpool+ CD8+ cells of total Vβpool+ CD8+ cells is plotted. For mice that received pmel-1 TCR transgenic cells, the percentage of Vβ13+ CD8+ cells is plotted. Error bars represent standard deviations (n = 4).
In vivo antigen expansion of wild-type and gene-optimized TCR-transduced murine splenocytes. Activated B6 splenocytes were transduced with vectors encoding wild-type or gene-optimized pmel-1 TCR. (A) TCR expression was determined by flow cytometry using anti-CD8α, anti-Vβ13, and a pool of anti-Vβ2, anti-Vβ3, anti-Vβ4, anti-Vβ5, anti-Vβ6, anti-Vβ8, anti-Vβ10, and anti-Vβ11. All fluorescence-activated cell sorting (FACS) plots show events that are gated for CD8 expression. The numbers indicate the percentage of CD8+ T cells with detectable pmel-1 TCR expression, calculated as follows: %Vβ13+Vβpool+ cells/%Vβpool+ cells × 100%. (B) Detection of TCR expression using anti-CD8 antibodies and Db-hGp10025-33 tetramer. The numbers indicate the percentage of tetramer+ CD8+ cells. (C,D) B6 recipients received splenocytes containing either 1 × 106 pmel-1 TCR transgenic CD8+ cells (■, only in panel C), 1.5 × 106 wild-type TCR-transduced CD8+ cells (▵), or 1.5 × 106 optimized TCR-transduced CD8+ cells (▴). Control mice received 20 × 106 activated, nontransduced cells (□). Mice received an intraperitoneal injection of 1 × 107 pfu rVV-hGp100(25-33) virus at the day of adoptive transfer (C), or mice received a sublethal dose of total body irradiation one day before adoptive transfer (D). At the indicated time points, peripheral blood was collected and analyzed by flow cytometry using APC anti-CD8α, FITC anti-Vβ13, and PE anti-Vβpool. The percentage of Vβ13+Vβpool+ CD8+ cells of total Vβpool+ CD8+ cells is plotted. For mice that received pmel-1 TCR transgenic cells, the percentage of Vβ13+ CD8+ cells is plotted. Error bars represent standard deviations (n = 4).
Adoptive transfer of transduced cells to irradiated recipients, a setting resembling TCR gene transfer in lymphodepleted patients, confirmed the marked difference between wt and opt pmel-1–transduced cells and revealed long-term persistence of cells transduced with gene-optimized TCRs (Figure 1D). Furthermore, an increase in in vivo antigen-specific T-cell responses upon TCR gene optimization is likewise observed for a second TCR (OT-I, de Witte et al, unpublished observations, December 2006).
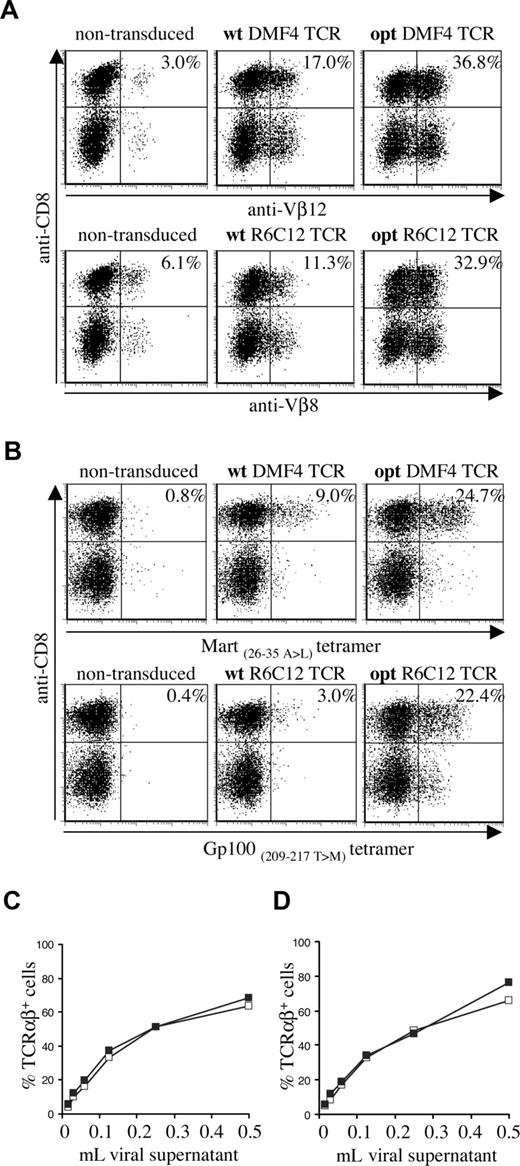
To assess the effect of TCR gene optimization of human melanoma–specific TCRs, we generated wild-type and gene-optimized versions for 2 human TCRs (Mart-1–specific DMF4 and Gp100-specific R6C12). As for the murine TCR, gene optimization led to a substantial increase in the percentage of cells with detectable transgene expression in transduced PBMCs (Figure 2A: Vβ staining, Figure 2B: MHC-tetramer staining). Gene optimization of the DMF4 TCR resulted in a 2-fold increase in the percentage of Vβ12-expressing cells (36.8% compared with 17.0%) and an almost 3-fold increase in tetramer-binding cells (24.7% compared with 9.0%). In R6C12 TCR–transduced cells, gene modification resulted in a 3-fold increase in Vβ8-expressing cells (32.9% versus 11.3%) and a more than 7-fold increase in tetramer-binding cells (22.4% versus 3.0%). Gene optimization predominantly affected the percentage of T cells with detectable transgene expression, whereas the level of TCR expression was enhanced to a lesser extent (MFI MHC-tetramer+ cells opt versus wt TCR: DMF4, 127 versus 100; R6C12, 98 versus 63). This effect of gene optimization was observed using a series of independent transductions (n = 4) and independent DNA batches (n = 2).
Expression of wild-type and gene-optimized human melanoma–specific TCRs. Activated PBMCs were transduced with wild-type or codon-optimized Gp100-specific R6C12 and Mart-1–specific DMF4 TCRs. TCR expression was determined 4 days after transduction by flow cytometric analysis using (A) anti-CD8 and anti-Vβ12 (top panels) or anti-Vβ8 (bottom panels), or (B) anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) tetramer (top panels) or A2.1-Gp100(209-217, 210 T>M) tetramer (bottom panels). The numbers indicate the percentage of Vβ+ or tetramer+ CD8+ cells. (C,D) Retrovirus encoding wt (□) or opt (■) DMF4 (C) or R6C12 TCR (D) was titrated on Jurkat/MA cells by adding the indicated amount of viral supernatant to a total volume of 1 mL. Four days after transduction, cells were analyzed by flow cytometry for TCR expression by anti-TCRαβ antibody staining.
Expression of wild-type and gene-optimized human melanoma–specific TCRs. Activated PBMCs were transduced with wild-type or codon-optimized Gp100-specific R6C12 and Mart-1–specific DMF4 TCRs. TCR expression was determined 4 days after transduction by flow cytometric analysis using (A) anti-CD8 and anti-Vβ12 (top panels) or anti-Vβ8 (bottom panels), or (B) anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) tetramer (top panels) or A2.1-Gp100(209-217, 210 T>M) tetramer (bottom panels). The numbers indicate the percentage of Vβ+ or tetramer+ CD8+ cells. (C,D) Retrovirus encoding wt (□) or opt (■) DMF4 (C) or R6C12 TCR (D) was titrated on Jurkat/MA cells by adding the indicated amount of viral supernatant to a total volume of 1 mL. Four days after transduction, cells were analyzed by flow cytometry for TCR expression by anti-TCRαβ antibody staining.
As the effect of gene optimization is thought to be in part due to enhanced RNA stability, it was possible that the improved expression of gene-modified TCRs was partially caused by more efficient production of retroviral particles. To be able to distinguish between an effect of gene optimization on TCR protein production and on viral titers, transductions were performed in a cell line that is devoid of endogenous TCR cell surface expression. In these Jurkat/MA cells, exogenous TCR alpha and beta chains are expressed at the cell surface in the absence of competition with endogenous TCR chains.
Whereas transduction with wild-type and gene-optimized TCRs resulted in a marked difference in expression in PBMCs, transduction of Jurkat/MA cells with serial dilutions of retrovirus revealed comparable expression for gene-optimized and wild-type TCRs, both for the DMF4 (Figure 2C) and the R6C12 TCR (Figure 2D). This indicates that gene optimization has an effect primarily on TCR protein production, whereas retroviral titers are not measurably influenced.
Expression patterns and activity of a panel of melanoma-specific TCRs
Having established the in vitro and in vivo benefit of TCR gene optimization, we sought to select the TCR with the highest expression and functional activity from a panel of 4 gene-optimized TCRs. This panel consisted of 3 Mart-1–specific TCRs (1D3, 2C2, and DMF4) and a Gp100-specific TCR (R6C12). The 1D3 and 2C2 TCRs were derived from highly tumor-reactive cytotoxic T lymphocyte (CTL) clones isolated from a melanoma patient vaccinated with Mart-126-35 peptide.25 The DMF4 TCR was isolated from a dominant T-cell clone in a patient who experienced an objective tumor regression upon adoptive transfer of autologous TILs.3,26 This receptor was used in the recent TCR gene transfer trial. The R6C12 TCR was derived from a CTL clone of a melanoma patient vaccinated with Gp100209-217 peptide.27
First, we assessed the relative efficacy with which the 4 different TCRs were expressed at the cell surface. To rule out differences in retroviral titers of the TCR panel, retroviral supernatants were titrated in the Jurkat/MA cell line.
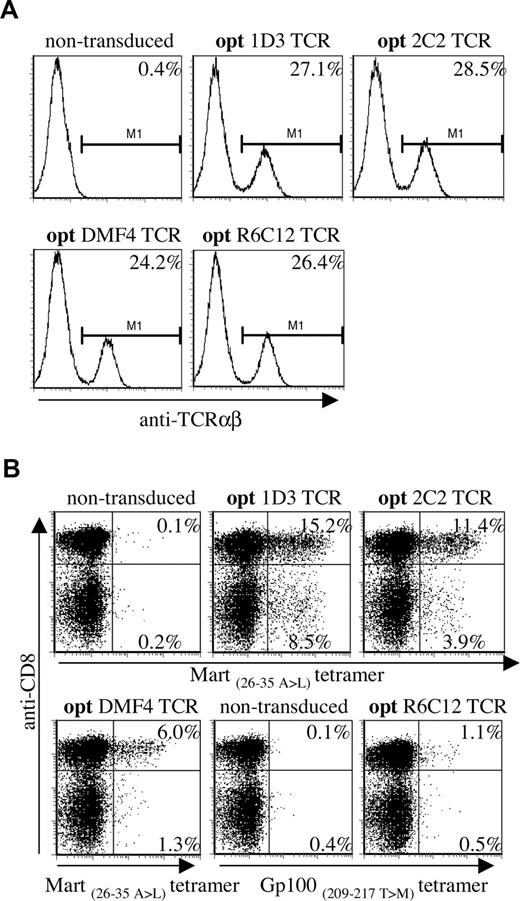
Virus dilutions resulting in comparable expression of the 4 different TCRs in Jurkat/Ma (Figure 3A, titrations shown in Figure S1A,B) were used to transduce PBMCs, and TCR expression was determined by MHC-tetramer staining (Figure 3B; note, transduction efficiencies using diluted virus are lower than when using untitrated virus). A marked hierarchy in expression of the introduced TCRs in CD8+ cells was reproducibly detected in 4 independent experiments, with 1D3 giving the highest percentage of MHC-tetramer+ cells (15.2%, MFI: 233), followed by 2C2 (11.4%, MFI: 225) and DMF4 (6.0%, MFI: 152). Transduction with the Gp100-specific R6C12 TCR resulted in only a small fraction of MHC-tetramer+ cells (1.1%, MFI: 75), even though expression of this TCR in Jurkat/Ma was efficient.
Differential expression of a panel of optimized melanoma-specific TCRs in peripheral blood T cells. (A) Jurkat/MA cells were transduced with titrated aliquots (Figure S1) of viral supernatants of vectors encoding the gene-optimized Mart-1–specific TCRs 1D3, 2C2, and DMF4 or the Gp100-specific TCR R6C12. Transduced cells were stained with anti-TCRαβ 4 days after transduction. Histograms show levels of TCR expression for the different TCRs. (B) Retroviral aliquots as used in panel A were used to transduce human peripheral blood T cells. TCR expression was determined by staining with anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) or A2.1-Gp100(209-217, 210 T>M) tetramers. The numbers in the upper-right and lower-right corners indicate the percentage of tetramer+ CD8+ and tetramer+ CD8− cells, respectively.
Differential expression of a panel of optimized melanoma-specific TCRs in peripheral blood T cells. (A) Jurkat/MA cells were transduced with titrated aliquots (Figure S1) of viral supernatants of vectors encoding the gene-optimized Mart-1–specific TCRs 1D3, 2C2, and DMF4 or the Gp100-specific TCR R6C12. Transduced cells were stained with anti-TCRαβ 4 days after transduction. Histograms show levels of TCR expression for the different TCRs. (B) Retroviral aliquots as used in panel A were used to transduce human peripheral blood T cells. TCR expression was determined by staining with anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) or A2.1-Gp100(209-217, 210 T>M) tetramers. The numbers in the upper-right and lower-right corners indicate the percentage of tetramer+ CD8+ and tetramer+ CD8− cells, respectively.
Interestingly, transduction with 1D3 and 2C2 resulted in a substantial population of MHC-tetramer+ CD4+ cells (as confirmed by staining with anti-CD4 antibody, data not shown). Expression of the introduced TCR in CD4+ cells was most efficient for the 1D3 TCR (8.5%, MFI: 205), followed by the 2C2 TCR (3.9%, MFI: 120). MHC-tetramer staining of CD4+ cells following transduction with DMF4 and R6C12 was close to background levels (1.3% and 0.5%, respectively).
Together, these results show that viral titers that yield equal TCR expression in the absence of competition with endogenous TCR chains yield substantially different expression levels in human PBMCs. Of the 4 melanoma-specific TCRs tested here, 1D3 shows the highest expression in both CD8+ and CD4+ cells.
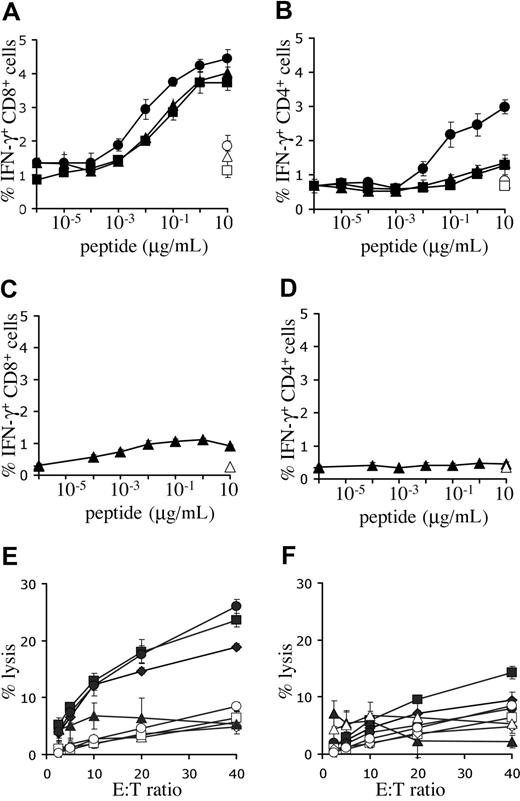
To determine the capacity for antigen recognition of T cells expressing the melanoma-specific TCRs, intracellular IFN-γ production of PBMCs, transduced with titrated virus, was determined. Incubation with Mart-1(26-35) peptide–loaded targets resulted in cytokine production in CD8+ cells transduced with either one of the Mart-1–specific receptors (Figure 4A). However, the 1D3 receptor was approximately 10-fold more sensitive than 2C2 or DMF4 (EC50 1D3: 3-10 nM, EC50 2C2 and DMF4: 30-100 nM). Furthermore, modification of PBMCs with the 1D3 receptor also provided CD4+ cells with the capacity for IFN-γ production upon antigen recognition (EC50: 30-100 nM), whereas this was not observed for the 2C2 or DMF4 receptor (Figure 4B). Transduction of PBMCs with the R6C12 receptor resulted in only low levels of IFN-γ production in CD8+ cells upon incubation with Gp100(209-217) peptide–loaded targets (Figure 4C, EC50: 1 nM), and no detectable production in CD4+ cells (Figure 4D). Of the Mart-1–specific TCRs, the 1D3 TCR was selected for further study, as it was well expressed, had a higher sensitivity than the 2 other Mart-1–specific receptors, and induced IFN-γ production in both CD8+ and CD4+ cells.
Functional analysis of melanoma-specific TCRs. (A,B) Human PBMCs were transduced with titrated aliquots of virus as detailed in Figure 3. Five days after transduction, cell cultures transduced with the Mart-1–specific TCRs DMF4 (▴), 1D3 (●), or 2C2 (■) were incubated with T2 cells loaded with the indicated concentrations of Mart-1(26-35) peptide. As a control, transduced cells were incubated with T2 cells loaded with the highest concentration of Gp100(209-217) peptide (open symbols). (C,D) Five days after transduction, cell cultures transduced with Gp100 R6C12 TCR (♦) were incubated with T2 cells loaded with the indicated concentrations of Gp100(209-217) peptide. As a control, transduced cells were also incubated with T2 cells loaded with the highest concentration of Mart-1(26-35) peptide (open symbols). After 5 hours of incubation, cells were stained with FITC anti-CD8 and PE anti-CD4, and intracellular cytokine production was determined using APC anti-IFN-γ. The percentage of IFN-γ–positive CD8 cells (A,C) and CD4 cells (B,D) is shown. Error bars in panels A-D represent standard deviations (n = 3). (E,F) Lysis of melanoma cell lines in a 51Cr-release assay. Twelve days after transduction, 1D3 TCR–transduced (E) or R6C12 TCR–transduced (F) cells were cocultured with different HLA-A2.1+, Mart+, and Gp100+ cell lines: AKR (●), GDO (■), and 526 (♦). The HLA-A2.1−, Mart+, and Gp100+ cell line 938 (▴) was used as a control. Coculture of nontransduced cells with melanoma cell lines is indicated by open symbols: AKR (○), GDO (□), 526 (◇), and 938 (▵). Cells were incubated at the indicated effector-target ratios for 4 hours, after which the percentage of lysis was determined. Error bars represent standard deviations (n = 3).
Functional analysis of melanoma-specific TCRs. (A,B) Human PBMCs were transduced with titrated aliquots of virus as detailed in Figure 3. Five days after transduction, cell cultures transduced with the Mart-1–specific TCRs DMF4 (▴), 1D3 (●), or 2C2 (■) were incubated with T2 cells loaded with the indicated concentrations of Mart-1(26-35) peptide. As a control, transduced cells were incubated with T2 cells loaded with the highest concentration of Gp100(209-217) peptide (open symbols). (C,D) Five days after transduction, cell cultures transduced with Gp100 R6C12 TCR (♦) were incubated with T2 cells loaded with the indicated concentrations of Gp100(209-217) peptide. As a control, transduced cells were also incubated with T2 cells loaded with the highest concentration of Mart-1(26-35) peptide (open symbols). After 5 hours of incubation, cells were stained with FITC anti-CD8 and PE anti-CD4, and intracellular cytokine production was determined using APC anti-IFN-γ. The percentage of IFN-γ–positive CD8 cells (A,C) and CD4 cells (B,D) is shown. Error bars in panels A-D represent standard deviations (n = 3). (E,F) Lysis of melanoma cell lines in a 51Cr-release assay. Twelve days after transduction, 1D3 TCR–transduced (E) or R6C12 TCR–transduced (F) cells were cocultured with different HLA-A2.1+, Mart+, and Gp100+ cell lines: AKR (●), GDO (■), and 526 (♦). The HLA-A2.1−, Mart+, and Gp100+ cell line 938 (▴) was used as a control. Coculture of nontransduced cells with melanoma cell lines is indicated by open symbols: AKR (○), GDO (□), 526 (◇), and 938 (▵). Cells were incubated at the indicated effector-target ratios for 4 hours, after which the percentage of lysis was determined. Error bars represent standard deviations (n = 3).
Peptide titrations are not useful to compare the relative effectiveness of T-cell receptors recognizing distinct epitopes (ie, the 1D3 and R6C12 TCRs recognizing the Mart-1 and Gp100 epitope, respectively), as differences in epitope density on target cells are not taken into account. To address this issue, the cytotoxic activity of peripheral blood T cells modified with either the Mart-1–specific 1D3 or the Gp100-specific R6C12 receptor was tested against several HLA-A2+ human melanoma cell lines that express both target antigens. Transduction of human PBMCs with the 1D3 TCR endowed these cells with the capacity to lyse HLA-A2–positive melanoma cell lines, whereas HLA-A2–negative target cells were not killed (Figure 4E). In contrast, lysis by cells transduced with the R6C12 TCR was substantially lower, and only slightly above background (Figure 4F).
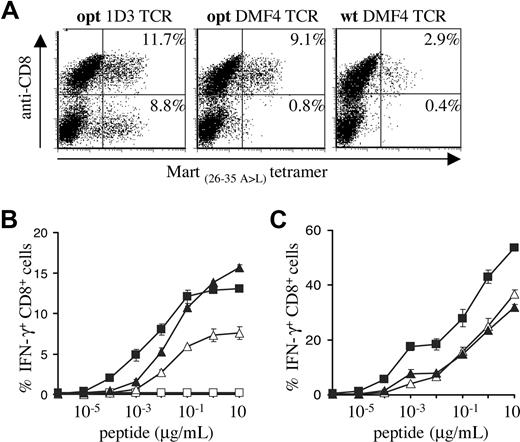
The use of gene optimization and subsequent in vitro comparison of different TCRs yielded a synthetic 1D3 alpha-beta gene pair that is well expressed and highly active. To formally test the value of these optimizations, PBMCs transduced with titrated virus encoding the opt 1D3 receptor were compared with PBMCs transduced with the wt DMF4 TCR that was recently used in a clinical trial.17 In addition, the opt DMF4 TCR was included to determine whether gene optimization affects primarily the yield of cells showing detectable transgene expression, or also T-cell sensitivity.
MHC-tetramer staining confirmed expression patterns observed in previous experiments (Figure 5A), with gene-optimized 1D3 showing the highest percentage of MHC-tetramer+ CD8+ cells (11.7%), and gene-optimized DMF4 showing a markedly higher percentage than its wild-type counterpart (9.1% vs 2.9%). Furthermore, only 1D3 showed a substantial population of MHC-tetramer+ CD4+ cells (8.8%).
Comparison of the gene-optimized 1D3 TCR and wild-type DMF4 TCR. Activated PBMCs were transduced with titrated viral supernatants of pMX vectors encoding the DMF4 wild-type or gene-optimized receptor or the 1D3 gene-optimized receptor. (A) Four days after transduction, expression in PBMCs was determined by staining with anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) tetramer. The numbers in the top-right and bottom-right corners indicate the percentage of tetramer+ CD8+ and tetramer+ CD8− cells, respectively. (B) Transduced cells were incubated with T2 cells loaded with the indicated Mart-1(26-35) peptide concentrations. After 5 hours of incubation, cells were stained with anti-CD8, and intracellular cytokine production was determined by anti-IFN-γ staining. The percentage of IFN-γ+ CD8+ cells is shown for wild-type DMF4 (▵), gene-optimized DMF4 (▴), gene-optimized 1D3 (■), or nontransduced lymphocytes (□). Error bars represent standard deviations (n = 2). (C) Tetramer+ CD8+ cells were sorted and 1 week later incubated with T2 cells loaded with the indicated Mart-1(26-35) peptide concentrations. After 5 hours of incubation, cells were stained with anti-CD8, and intracellular cytokine production was determined by anti-IFN-γ staining. The percentage of IFN-γ+ CD8+ cells is shown for sorted wild-type DMF4 (▵), gene-optimized DMF4 (▴), and gene-optimized 1D3 (■) transduced PBMCs.
Comparison of the gene-optimized 1D3 TCR and wild-type DMF4 TCR. Activated PBMCs were transduced with titrated viral supernatants of pMX vectors encoding the DMF4 wild-type or gene-optimized receptor or the 1D3 gene-optimized receptor. (A) Four days after transduction, expression in PBMCs was determined by staining with anti-CD8 and A2.1-Mart-1(26-35, 27 A>L) tetramer. The numbers in the top-right and bottom-right corners indicate the percentage of tetramer+ CD8+ and tetramer+ CD8− cells, respectively. (B) Transduced cells were incubated with T2 cells loaded with the indicated Mart-1(26-35) peptide concentrations. After 5 hours of incubation, cells were stained with anti-CD8, and intracellular cytokine production was determined by anti-IFN-γ staining. The percentage of IFN-γ+ CD8+ cells is shown for wild-type DMF4 (▵), gene-optimized DMF4 (▴), gene-optimized 1D3 (■), or nontransduced lymphocytes (□). Error bars represent standard deviations (n = 2). (C) Tetramer+ CD8+ cells were sorted and 1 week later incubated with T2 cells loaded with the indicated Mart-1(26-35) peptide concentrations. After 5 hours of incubation, cells were stained with anti-CD8, and intracellular cytokine production was determined by anti-IFN-γ staining. The percentage of IFN-γ+ CD8+ cells is shown for sorted wild-type DMF4 (▵), gene-optimized DMF4 (▴), and gene-optimized 1D3 (■) transduced PBMCs.
In line with the data shown in Figure 4A, optimized 1D3 TCR–transduced cells were approximately 10-fold more sensitive than cells transduced with the optimized DMF4 TCR. Comparison of wild-type and optimized DMF4 revealed equal sensitivity, indicating that peptide sensitivity is not measurably affected by gene optimization (Figure 5B).
The percentage of T cells transduced with the unmodified DMF4 TCR that was detected in functional assays was high compared with the percentage of transgene expression as detected by MHC-tetramer staining. This could either reflect the fact that MHC-tetramer staining underestimates the frequency of TCR-modified T cells or that T cells expressing the unmodified DMF4 are more likely to be functionally active compared with T cells expressing other Mart-1–specific TCRs. To test this, 1D3 opt, DMF4 opt, and DMF4 wt TCR-transduced cells were sorted on the basis of MHC-tetramer and CD8 staining (resulting in 84%, 80%, and 82% tetramer+ CD8+ cells, respectively; data not shown), and IFN-γ production was determined upon peptide stimulation (Figure 5C). Sorted PBMCs transduced with either wild-type or gene-optimized DMF4 receptor showed similar percentages of IFN-γ–producing cells. Furthermore, PBMCs transduced with gene-optimized 1D3 maintained their higher sensitivity after sorting.
Recognition of allogeneic MHC molecules
As discussed previously,5 TCR gene transfer in the clinical setting entails a partial MHC mismatch between the TCR recipient and the original “TCR donor.” Consequently, during thymic selection, the introduced TCR has not been selected against reactivity toward all of the MHC alleles expressed by the recipient, and recognition of allogeneic MHC molecules complexed with self-antigens could result in autoimmune pathology. Although alloreactivity has not been observed for the TCR used in a first phase 1 trial,16 the high frequency of T-cell alloreactivity in other settings37 warrants the development of standardized methods to screen for such reactivity.
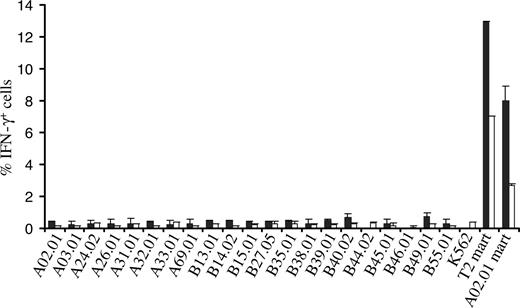
To mimic a setting in which there is an MHC mismatch between TCR recipient and donor, PBMCs transduced with the 1D3 receptor were tested against a large panel of K562 cell lines each expressing single HLA-A and -B alleles (the SAL panel)22 by determining intracellular IFN-γ production upon coculture. Of the 21 SALs tested, none induced detectable IFN-γ production in 1D3-expressing CD8+ or CD4+ cells, whereas incubation with Mart-1(26-35) peptide–loaded T2 cells or HLA-A2+ SALs did result in a substantial population of CD8+ and CD4+ cells producing IFN-γ (Figure 6). These results show that for the receptor and MHC alleles tested, alloreactivity does not seem to play an important role.
Assessment of alloreactivity of TCR-transduced PBMCs against single MHC allele–expressing cells. 1D3 TCR–transduced cells were incubated with 21 single MHC allele-expressing cell lines for a period of 5 hours. T2 cells and A02.01 K562 cells loaded with the Mart-1(26-35) peptide were used as a positive control. After incubation, cells were stained with anti-CD8 and PE CD4, and intracellular cytokine production was determined using anti-IFN-γ. The percentage of IFN-γ+ CD8+ cells (■) and CD4+ cells (□) is depicted. Bars represent range (n = 2).
Assessment of alloreactivity of TCR-transduced PBMCs against single MHC allele–expressing cells. 1D3 TCR–transduced cells were incubated with 21 single MHC allele-expressing cell lines for a period of 5 hours. T2 cells and A02.01 K562 cells loaded with the Mart-1(26-35) peptide were used as a positive control. After incubation, cells were stained with anti-CD8 and PE CD4, and intracellular cytokine production was determined using anti-IFN-γ. The percentage of IFN-γ+ CD8+ cells (■) and CD4+ cells (□) is depicted. Bars represent range (n = 2).
Long-term in vitro culture and vector comparison
As long-term persistence of adoptively transferred cells is thought to be important for clinical antitumor efficacy,38 we set out to determine whether PBMCs transduced with the gene-optimized 1D3 receptor would maintain TCR expression and cytotoxic capacity after prolonged in vitro culture.
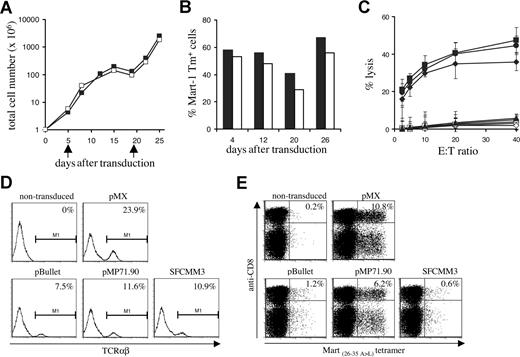
Every 3 or 4 days, total cell numbers (Figure 7A) and percentage of MHC-tetramer+ cells (Figure 7B) were assessed. A high percentage of MHC-tetramer+ cells was maintained over 4 weeks of culture, even after a 3-log expansion. Interestingly, in line with previous data on the effect of T-cell activation state on activity of the retroviral LTR,39 the fraction of MHC-tetramer+ cells was highest in the period following T-cell restimulation. After prolonged in vitro culture, 1D3 TCR–transduced cells remained capable of lysing HLA-A2–positive melanoma cell lines (Figure 7C).
Long-term in vitro culture and vector comparison. (A) PBMCs transduced with the 1D3 TCR (■) and nontransduced (□) PBMCs were restimulated every 14 days (indicated by arrows) by addition of irradiated feeder cells and phytohemagglutinin. At the time points indicated, total cell numbers were determined. (B) TCR expression in 1D3-transduced cells was determined by staining with anti-CD8, anti-CD4, and A2.1-Mart-1(26-35, 27 A>L) tetramer. The numbers indicate the percentage of tetramer+ CD8+ (■) and tetramer+ CD4+ (□) cells. (C) Four weeks after transduction, cytolytic activity of 1D3 TCR–transduced cells against different HLA-A2.1+, Mart+ cell lines—AKR (●), GDO (■), and 526 (♦)—was determined. The HLA-A2.1−, Mart+ cell line 938 (▴) was used as a control. Cytolytic activity of nontransduced cells is indicated by open symbols: AKR (○), GDO (□), 526 (◇), and 938 (▵). Error bars represent standard deviations (n = 3) (D,E) Transient viral supernatant of the indicated vectors all encoding the 1D3 TCR was used to transduce Jurkat/MA cells (D) or PBMCs (E). Four days after transduction, TCR expression in Jurkat/MA cells was determined by staining with anti-TCRαβ. Expression in PBMCs was determined by staining with anti-CD8 and A2.1-Mart-1(26-35,27 A>L) tetramer. The numbers indicate the percentage of tetramer+ CD8+ cells.
Long-term in vitro culture and vector comparison. (A) PBMCs transduced with the 1D3 TCR (■) and nontransduced (□) PBMCs were restimulated every 14 days (indicated by arrows) by addition of irradiated feeder cells and phytohemagglutinin. At the time points indicated, total cell numbers were determined. (B) TCR expression in 1D3-transduced cells was determined by staining with anti-CD8, anti-CD4, and A2.1-Mart-1(26-35, 27 A>L) tetramer. The numbers indicate the percentage of tetramer+ CD8+ (■) and tetramer+ CD4+ (□) cells. (C) Four weeks after transduction, cytolytic activity of 1D3 TCR–transduced cells against different HLA-A2.1+, Mart+ cell lines—AKR (●), GDO (■), and 526 (♦)—was determined. The HLA-A2.1−, Mart+ cell line 938 (▴) was used as a control. Cytolytic activity of nontransduced cells is indicated by open symbols: AKR (○), GDO (□), 526 (◇), and 938 (▵). Error bars represent standard deviations (n = 3) (D,E) Transient viral supernatant of the indicated vectors all encoding the 1D3 TCR was used to transduce Jurkat/MA cells (D) or PBMCs (E). Four days after transduction, TCR expression in Jurkat/MA cells was determined by staining with anti-TCRαβ. Expression in PBMCs was determined by staining with anti-CD8 and A2.1-Mart-1(26-35,27 A>L) tetramer. The numbers indicate the percentage of tetramer+ CD8+ cells.
Finally, having selected a well- and stably expressed, highly affine, melanoma-specific TCR with no detectable alloreactivity against a large series of HLA alleles, we aimed to assess which retroviral vector is most suitable for expression of TCR transgenes in human lymphocytes. To this purpose, 3 different retroviral vectors that have previously been used in clinical trials (pBullet,40,41 pMP71.90,42 and SFCMM343,44 ) were compared with the pMX vector used in in vivo mouse studies for their ability to yield TCR transgene expression in human PBMCs, and the same set of retroviral vectors was also evaluated in the Jurkat/MA system.
The Mart 1D3 receptor was cloned into each vector in either an α-IRES-β or an α-SV40EP-β configuration. All vectors were capable of inducing expression of the 1D3 receptor in the Jurkat/MA system (Figure 7D). In contrast, only transduction with pMX- and pMP71.90-based vectors yielded efficient expression of the 1D3 TCR in PBMCs, resulting in 10.8% (MFI: 261) and 6.2% (MFI: 323) tetramer-binding cells, respectively (Figure 7E). Transduction with both these vectors resulted in stable expression of the 1D3 TCR during a period of at least 3 weeks (data not shown).
These data support 2 points, first evaluation of TCR expression in cell lines such as Jurkat/MA cells has little predictive value for the capacity of these vectors to yield detectable TCR expression in peripheral blood T cells. Second, for the expression of TCR alpha and beta genes from a single vector, pMP71.90 is the most effective of those vectors previously used in a clinical setting, and comparable with the pMX vector that has successfully been used in a series of in vivo mouse studies.
Discussion
Here, we describe a 3-step approach for the selection of well-expressed, high affine, and safe TCRs. An important step in this approach is the improvement of TCR expression via gene modification, since expression of introduced TCR alpha and beta genes is generally low due to formation of heterodimers of endogenous and introduced chains, resulting in lower expression of the correctly paired heterodimer. Furthermore, correctly assembled chains compete with other heterodimers for binding to the CD3 complex32,33 that forms the limiting component in the TCR assembly process.
We modified TCR genes to improve stability and translation of the messenger RNA while leaving the amino acid sequence unaltered, thus enabling efficient protein expression. We were able to show that cell surface expression of 2 of 3 murine TCRs and 2 of 2 human TCRs benefits substantially from gene optimalization (this paper and A.J., unpublished observations, September 2006).
Furthermore, gene modification of the murine pmel-1 TCR distinctly improves in vivo antigen recognition, as determined by expansion of the transduced T-cell population upon vaccination. Interestingly, this marked effect on the in vivo expansion of TCR-modified cells was observed even though the increase in the level of expression upon gene optimization was only modest. As a comparable increase in expression is observed upon gene optimization of 2 human TCRs, these data suggest that the in vivo behavior of human TCR-transduced PBMCs may also benefit substantially from gene optimization. Likewise, the current data suggest that the modest increase in expression of exogenous TCRs upon remodeling of the TCRαβ interface45 may also result in a substantially improved engraftment of TCR-modified cells in vivo.
Notably, the effect of gene optimization was more pronounced for the human Gp100–specific R6C12 receptor than for the Mart-1–specific DMF4 receptor, both with respect to the percentage of cells showing detectable transgene expression and the level of expression. To assess whether it may be possible to predict the value of gene optimization for different TCRs, we determined the percentage of codons that was modified in the variable regions of these receptors. In the gene-optimized R6C12 receptor, 52% of codons were modified in the alpha chain and 47% in the beta chain. In contrast, modifications of DMF4 alpha and beta chains were 32% and 21%, respectively. This may suggest that for TCRs for which a lower level of gene optimization is required, this process results in a smaller increase in TCR expression. However, a substantially larger dataset will be required to rigorously test this notion.
In addition to demonstrating the in vivo effect of TCR gene optimization, the data described in this study show that selection of both viral platforms and individual TCRs should occur by analysis of expression in peripheral blood T cells rather than cell systems that lack endogenous TCR expression, in which the requirements for expression of exogenous TCRs are substantially lower. As an example, while expression of R6C12 and 1D3 receptors is comparable in Jurkat/MA cells, only the latter is efficiently expressed in human PBMCs. Likewise, while all 4 retroviral vector systems tested yielded substantial TCR expression in Jurkat/MA cells, pMX and pMP71 were superior in yielding TCR expression in PBMCs.
Clinical application of TCR gene transfer should be preceded by an evaluation of the possible side effects, caused by either on-target reactivity (mediated via recognition of target antigens on normal tissues) or off-target reactivity (mediated by mechanisms described in the next paragraph). The risk of on-target autoimmunity will primarily depend on the expression pattern of the antigen involved. Melanoma-differentiation antigens, such as Mart-1 and Gp100, are expressed on melanoma cells as well as normal melanocytes, and targeting of these antigens is known to induce autoimmune melanocyte destruction resulting in vitiligo and uveitis.3,20 TCR gene transfer targeting these antigens has not resulted in severe side effects.16 However, it remains possible that increased autoimmunity will occur when more potent TCRs or conditioning regimens are used.
Off-target autoimmunity by TCR-transduced cells theoretically can be induced via 3 different mechanisms.5 First, introduction of exogenous TCR chains can lead to the formation of heterodimers with endogenous alpha and beta chains, which might be reactive toward self-peptides. Second, if ignorant self-reactive T cells are transduced, triggering of these cells via the introduced TCR can result in an expanded population of autoreactive cells. Off-target autoreactivity via these 2 mechanisms would occur irrespective of MHC disparities between TCR donor and recipient. Both mouse models of TCR gene transfer12,15 and the recent phase 1 clinical trial16 do not provide evidence that these mechanisms form a substantial reason for concern.
As a third possibility for off-target autoimmunity, MHC mismatches between TCR donor and recipients may result in recognition of allogeneic MHC molecules complexed to self-antigens by the TCR-modified cells. With the aim to develop a generally applicable strategy to screen for such alloreactivity, we developed a simple assay to test TCR-modified T cells against a set of cell lines expressing defined single HLA-A and -B alleles with a high prevalence in the human population. Lack of reactivity against any of the MHC alleles expressed by this panel provides evidence that for this T-cell receptor the risk of type III off-target autoimmunity may be little. However, it may be worthwhile to further expand the set of class I alleles that is tested for a more complete evaluation of MHC alloreactivity, and possibly such an evaluation should also include reactivity against MHC class II alleles. Furthermore, although allorecognition by TCRs is often less peptide dependent,46 it remains possible that a TCR that is unreactive toward an MHC allele on the cell line used for in vitro testing does recognize this MHC allele when complexed with a tissue-specific antigen. Based on this latter consideration, it may remain useful to set up a database for allowed MHC mismatches for every TCR used in coming clinical trials.
We have used this evaluation and optimization strategy to select a melanoma-specific receptor with improved expression, a higher affinity compared with other receptors, and a lack of detectable alloreactivity. CD4+ T cells transduced with this MHC class I–restricted 1D3 receptor are capable of MHC-tetramer binding and production of IFN-γ. Prior data have shown that the provision of CD4+ T-cell help contributes to both primary CD8+ responses and CD8+ T cell memory formation.47 Furthermore, recent studies in mouse models have shown that transfer of a CD8+-dependent MHC class I–restricted TCR into CD4+ T cells can be used to generate MHC class I–restricted CD4+ T-cell help.13,14 However, as optimal function of the modified CD4+ T cells requires the presence of the CD8αβ coreceptor,13 the selection of TCRs that can function independent of CD8 is considered attractive.27,48,49 Perhaps more importantly, compared with a TCR evaluated in a previous clinical trial, CD8+ T cells expressing the 1D3 TCR recognize antigen at a 10-fold lower concentration. Furthermore, the high level of expression of the 1D3 TCR may predict a more effective in vivo persistence as based on analogy with the murine melanoma–specific pMel TCR.
The 3-step approach we describe in this study may be of use for the selection of well-expressed, high affine, and safe T-cell receptors for future clinical trials, thereby enhancing the clinical development of TCR gene therapy. Such selection may involve the evaluation either of naturally occurring TCRs or—by analogy with antibody development—of TCRs obtained by technologies that circumvent the limitations of the naturally occurring immune repertoire.11,49,50
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Netherlands Organisation for Health Research and Development (ZonMW grant 4310005).
We thank R. Debets for the pBullet vector, M. Griffioen for the pMP71.90 vector, S. Ebeling for the SFCMM3 vector, M. Heemskerk for useful suggestions, the NKI FACS-facility and B-lab for technical support, and J. Borst and M. de Witte for reading the paper.
Authorship
Contribution: A.J. designed and performed research, analyzed data, and wrote the paper; R.G.-E. designed and performed research and analyzed data; M.D. performed research and analyzed data; W.K., Y.M.Z., I.I.N.D., N.R., P.R., and R.A.M. provided important reagents; T.N.M.S. and J.B.A.G.H. designed research and wrote the paper. A.J. and R.G.-.E. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John B. A. G. Haanen, Division of Immunology, the Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX, Amsterdam, the Netherlands; e-mail:j.haanen@nki.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal